Abstract
An α-l-arabinofuranosidase (α-l-arabinofuranoside arabinofuranohydrolase [EC 3.2.1.55]; referred to below as ArfI) from Cytophaga xylanolytica XM3 was purified 85-fold by anion-exchange and hydrophobic interaction column chromatography. The native enzyme had a pI of 6.1 and an apparent molecular mass of 160 to 210 kDa, and it appeared to be a trimer or tetramer consisting of 56-kDa subunits. With p-nitrophenyl-α-l-arabinofuranoside as the substrate, the enzyme exhibited a Km of 0.504 mM and a Vmax of 319 μmol · min−1 · mg of protein−1, and it had optimum activity at pH 5.8 and 45°C. ArfI was relatively stable over a pH range of 4 to 10 and at temperatures up to 45°C, and it retained nearly full activity when stored at 4°C for periods as long as 24 months. The enzyme also released arabinose from 4-methylumbelliferyl-α-l-arabinofuranoside, as well as from rye, wheat, corn cob, and oat spelt arabinoxylans and sugar beet arabinan, but not from arabinogalactan. ArfI showed no hydrolytic activity toward a range of p-nitrophenyl- or 4-methylumbelliferyl-glycosides other than arabinoside, for which it was entirely specific for the α-l-furanoside configuration. ArfI interacted synergistically with three partially purified endoxylanase fractions from C. xylanolytica in hydrolyzing rye arabinoxylan. However, cell fractionation studies revealed that ArfI was largely, if not entirely, cytoplasmic, so its activity in vivo is probably most relevant to hydrolysis of arabinose-containing oligosaccharides small enough to pass through the cytoplasmic membrane. Antibodies prepared against purified ArfI also cross-reacted with arabinofuranosidases from other freshwater and marine strains of C. xylanolytica, as well as with some proteins that did not possess arabinofuranosidase activity. To our knowledge, this is the first α-l-arabinofuranosidase to be purified and characterized from any gliding bacterium.
Xylans are included within the family of plant cell wall heteropolysaccharides referred to as hemicelluloses. They consist of a β-1,4-linked xylopyranose backbone, to which are often attached side groups of arabinose, (O-methyl-)glucuronic acid, ferulic or p-coumaric acid, and/or acetate, depending on the plant source (2, 43). Next to cellulose, xylans are the most abundant polysaccharides on earth (4), and it has been estimated that about 1010 metric tons are recycled annually, with the degradative arm occurring largely through the action of microbes (47).
In an effort to increase our understanding of the microbiology and biochemistry of xylan degradation, we initiated a study of the xylan-hydrolyzing system of a new, anaerobic, gliding bacterium, Cytophaga xylanolytica XM3 (12). This bacterium and other freshwater and marine strains similar to it were isolated from freshwater sediments on the basis of their ability to adhere to and dominate the fermentation of insoluble xylan particles in sulfidogenic and methanogenic enrichment cultures. Unlike the secreted xylanases found with most other organisms, the xylanase system of C. xylanolytica is almost entirely cell associated, but it can be easily extracted from whole cells by using 0.2% Triton X-100. Such Triton X-100 extracts were shown to possess activities important for xylan hydrolysis, including endo-1,4-β-d-xylanase (EC 3.2.1.8), β-d-xylosidase (EC 3.2.1.37), α-l-arabinofuranosidase (EC 3.2.1.55; referred to below as ARAF), and α-d- and β-d-glucopyranosidases (EC 3.2.1.20 and EC 3.2.1.21, respectively) (13). Triton X-100 extracts were also remarkably stable, retaining full xylanolytic activities for more than 6 months when stored at 4°C; however, they exhibited no activity toward microcrystalline cellulose, ball-milled Whatman no. 1 cellulose filter paper, or carboxymethyl cellulose (13). The latter observations were consistent with the inability of cells to grow on cellulose or carboxymethyl cellulose.
Efforts to resolve the nature and number of components of the xylanase system yielded several column chromatography fractions, each enriched with multiple endoxylanase (ENDOX) activities, as well one fraction enriched with a single ARAF activity. ARAFs are important because they catalyze the hydrolytic removal of arabinofuranosyl residues from hemicellulosic polysaccharides (18). Such residues are typically attached by α-1,2- and/or α-1,3-linkages to the backbones of arabinoxylans, arabinans, and arabinogalactans (4, 9), and their removal by ARAFs usually facilitates the attack of the xylan backbone by ENDOXs (11, 23, 35, 49). Here we report the purification and properties of an ARAF (ArfI) produced by C. xylanolytica XM3 when it is growing on arabinoxylan. Also included in this paper are the results of studies performed to (i) determine the cellular location of ArfI, (ii) examine the synergy between ArfI and ENDOXs of C. xylanolytica, and (iii) evaluate the occurrence of ArfI-like proteins in other freshwater and marine strains of C. xylanolytica. In another paper (19) we will report on the cloning and sequencing of the ArfI-encoding gene (i.e., arfI), as well as of an additional gene (arfII) that encodes an ARAF not synthesized by C. xylanolytica during growth on arabinoxylan.
(A preliminary report of the findings has been presented previously [36].)
MATERIALS AND METHODS
Growth of cells and preparation of cell extracts.
C. xylanolytica XM3 (= DSM 6779) and other freshwater strains were grown anoxically at 30°C in rubber-stoppered 2-liter Pyrex bottles nearly filled with Na2S-reduced, CO2–bicarbonate-buffered freshwater mineral medium no. 2 containing 0.2% (wt/vol) oat spelt arabinoxylan (preextracted with 70% [vol/vol] ethanol) as the sole fermentable substrate (12). Marine strains were grown in a similar manner, except that marine medium was used (12).
For enzyme purification, cells were harvested from late-exponential- to early-stationary-phase batch cultures by centrifugation at 10,000 × g for 20 min at 4°C. Cell pellets were resuspended in 0.2% (wt/vol) Triton X-100 to 1/40 the original culture volume and stirred for 2 h at 4°C. The treated cells were then removed by centrifugation at 10,000 × g for 30 min at 4°C, and the resulting supernatant fluid was centrifuged at 100,000 × g for 2 h at 4°C. The supernatant fluid from the latter centrifugation was placed into a 3,500-molecular-weight-cutoff dialysis membrane and dialyzed against 50 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES) buffer (pH 8.0; total volume, ca. 4 liters) at 4°C. The dialyzed material, referred to below as the Triton extract, was stored at 4°C until use.
Sonicated cell extracts were prepared from cells harvested by centrifugation at 18,000 × g and resuspended in 10 mM HEPES buffer (pH 6.8). The preparations were exposed to eight 30-s bursts from a sonic probe (0.125-in.-diameter tapered tip; output setting 5; 50% duty; model 450 Sonifier [Branson, Danbury, Conn.]), followed by recentrifugation.
Purification of ArfI.
Purification of ArfI from Triton extracts was done at room temperature (RT) by a low-pressure column chromatography procedure performed with an Econo System apparatus (Bio-Rad Laboratories, Hercules, Calif.) generally operating at a flow rate of 1 ml · min−1. Eluted fractions were monitored for protein content by measuring the A280 and for ARAF activity by using a semiquantitative microtiter plate assay (see below). Sequential column chromatography was performed by using the following steps. In step 1 Triton extract (204 ml, 430 mg of protein) was applied to a DEAE-cellulose column (2.5 by 50 cm) that had previously been equilibrated with 50 mM HEPES buffer (pH 8.0). The column was washed with 1 liter of equilibration buffer at a flow rate of 2 ml · min−1, and elution of ArfI was achieved by using a linear gradient consisting of 0 to 1 M NaCl in the same buffer. Fractions containing ArfI eluted between 50 and 200 mM NaCl; these fractions were pooled and concentrated by ultrafiltration through a type YM 10 membrane having a 10,000-molecular-weight cutoff (Amicon Co., Danverse, Mass.), and in the process the buffer was changed to 20 mM HEPES (pH 8.0). In step 2 the ArfI pool from step 1 (75 ml, 180 mg of protein) was applied to a DEAE-Sephadex A-50 column (1.5 by 50 cm) that had previously been equilibrated with 20 mM HEPES (pH 8.0), and elution was performed with a linear gradient consisting of 0 to 1 M NaCl in the same buffer. Fractions containing ArfI eluted from the column between 0.1 and 0.4 M NaCl; these fractions were pooled, dialyzed, and concentrated as described above, except that the dialysis buffer was 20 mM potassium phosphate (pH 6.5). In step 3 the ArfI pool from step 2 (75 ml, 74 mg of protein) was applied to a hydroxylapatite column (1.5 by 50 cm) that had previously been equilibrated with 20 mM potassium phosphate buffer (pH 6.5), and chromatography was performed with a linear gradient consisting of 20 to 300 mM potassium phosphate. Fractions containing ArfI eluted from the column between 170 and 225 mM phosphate and were pooled. In step 4 a 5 M NaCl solution was added to the ArfI pool from step 3 (43 ml, 7.7 mg of protein) to achieve a final NaCl concentration of 1 M, and the mixture was applied to a Phenyl-Sepharose CL-4B column (1.0 by 50 cm) that had previously been equilibrated with a solution containing 1 M NaCl in 5 mM 3-N-morpholinopropanesulfonic acid (MOPS) buffer (pH 6.5). Elution of ArfI was performed with a simultaneously decreasing linear gradient consisting of 1 to 0 M NaCl and increasing pH 6.5 to 8.0 gradient in 5 mM MOPS buffer. ArfI activity eluted between 250 and 0 mM NaCl and pH 7.5 to 8.0, and the relevant fractions were pooled. In step 5 the ArfI pool from step 4 was concentrated by ultrafiltration (as described above), and the buffer was changed to 20 mM MOPS (pH 7.5). This pool of ArfI (74.5 ml, 1.7 mg of protein) was then applied to a Macro Prep 50Q column (1.0 by 50 cm) that had previously been equilibrated with 20 mM MOPS (pH 7.5). Chromatography was performed with an increasing linear gradient consisting of 0 to 1 M NaCl in the same buffer. Active fractions eluted between 50 and 100 mM NaCl and were pooled. In step 6 the ArfI pool from step 5 was dialyzed, as described above, against 20 mM MOPS (pH 8.0) containing 0.5 M NaCl, and the dialyzed pool (91 ml, 1.6 mg of protein) was rechromatographed on a Phenyl-Sepharose CL-4B column (1.5 by 50 cm), but this time with a decreasing linear gradient consisting of 0.5 to 0 M NaCl in the same buffer. ArfI-containing fractions eluted between 25 and 0 mM NaCl and were pooled. Ultrafiltration was then used to concentrate the ArfI preparation to a volume of 28.8 ml (1.2 mg of protein); this preparation was stored at 4°C.
Partial purification of ENDOXs.
Components from three major electrophoretically separable zones of ENDOX activity (i.e., the top, middle, and bottom zones after native polyacrylamide gel electrophoresis [PAGE] of Triton extracts [see Fig. 2A, lane 1]) were partially purified from a Triton extract by using a four-step procedure. The first three steps involved sequential column chromatography with the following matrices and eluants by using the conditions described above for ArfI: (i) DEAE-cellulose and NaCl (step i); (ii) hydroxylapatite and PO4−3] (step ii); and (iii) Macro Prep 50Q and NaCl (step iii). A significant amount of ENDOX activity in the major peak eluting in step i did not bind to the hydroxylapatite column in step ii and eluted in the void volume. This material (referred to as ENDOX II) was enriched with ENDOX components that migrated in the middle zone of native PAGE gels and was saved. Steps ii and iii were effective in removing some additional contaminating proteins from the remaining ENDOX activity (which consisted of components from the top and bottom zones of native PAGE gels), but did not resolve these proteins. Therefore, an additional step, step iv, was included, which involved preparative native PAGE of the remaining ENDOX activity on a model 491 Prep-Cell column (Bio-Rad) containing a 2-cm (4% polyacrylamide) stacking gel and a 10-cm (10% polyacrylamide) resolving gel (7). Electrophoresis was done according to the manufacturer’s recommendations, and fractions were screened for ENDOX activity by measuring the release of reducing sugar from xylan (see below) and on native PAGE slab gels subsequently overlaid with oat spelt arabinoxylan zymogram gels (see below). Step iv separated the remaining ENDOX components into a group of activities that migrated slowly through the gel and represented the top zone from native PAGE gels (referred to as ENDOX I) and a group of activities that migrated rapidly and represented the bottom zone (referred to as ENDOX III).
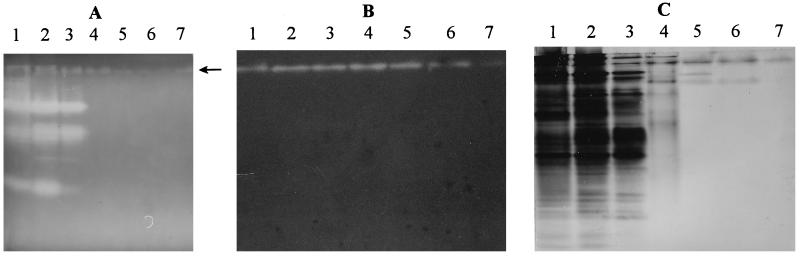
FIG. 2.
Zymograms and protein staining of a native PAGE gel for ArfI after each purification step. (A) Oat spelt xylan zymogram (stained with Congo red) of ENDOX activity. (B) MU-AF zymogram showing ARAF activity as UV-fluorescent bands. (C) Native PAGE (silver-stained protein) gel. Lane 1 contained Triton extract, and lanes 2 through 7 contained the fractions obtained after purification steps 1 through 6 listed in Table 1, respectively; lanes 1 through 7 contained 105, 123, 49, 9.1, 1.1, 0.9, and 1.2 μg of protein, respectively (B and C), and the gel for panel A was loaded with three times as much protein per lane. The arrow in panel A indicates a small zone of Congo red nonbinding induced by the presence of ArfI (see text). Note that panels B and C are magnified more than panel A.
Enzyme assays.
ARAF activity was determined semiquantitatively by a modification of the Bachmann-McCarthy method (3), as follows. Portions (1.0 μl) of enzyme (e.g., column chromatography fractions) were added to separate wells of 96-well microtiter plates that also contained 90 μl of a 4-methylumbelliferyl-α-l-arabinofuranoside (MU-AF) solution (10 μg · ml−1) per well. After various periods of incubation (from 10 min to 24 h at RT, depending on the relative activity), the microtiter plates were placed on a UV transilluminator, and active fractions were identified by the intense fluorescence of liberated methylumbelliferone.
Quantitative assays for ARAF activity were performed as described by Greve et al. (11) with 1-ml (total volume) reaction mixtures containing 1 mM para-nitrophenyl-α-l-arabinofuranoside (pNP-AF) in 50 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer (pH 6.0) plus enzyme. Reaction mixtures lacking enzyme were prewarmed to 45°C, and reactions were started by adding 20 μl of appropriately diluted enzyme (0.24 to 42 mg of protein), also prewarmed in the same buffer (the actual pH of the reaction mixture at 45°C was determined to be 5.8). After 1 min, the reaction was terminated by adding 2 ml of 1 M NH4OH, and the A405 nm of the resulting solution was determined with a Bausch & Lomb Spectronic 20 colorimeter. Absorbance readings were converted to micromoles of p-nitrophenol by comparison to a standard curve. One unit of ARAF activity was defined as the amount of enzyme that produced 1 μmol of p-nitrophenol per min under the assay conditions used. Catalytic constants of purified ArfI were determined in a similar way, but triplicate reaction mixtures were used; these reaction mixtures had a total volume of 4.5 ml and contained 50 mM MES (pH 6.0), pNP-AF (concentration range, 25 μM to 5 mM), and 0.06 U of enzyme in 50 mM MES (pH 6.0). Periodically during incubation, 1-ml samples were removed and added to separate tubes containing 2 ml of 1 M NH4OH, after which the A405 nm was determined as described above. Michaelis-Menten kinetic parameters were determined by using the method of Wilkinson (48).
Glycosidase activities other than ARAF activity, as well as acetyl esterase activities, were determined by using other p-nitrophenyl and 4-methylumbelliferyl derivatives as substrates (at concentrations of 2.5 and 5.0 mM, respectively); the assays were performed either quantitatively, as described above for pNP-AF, or semiquantitatively by the microtiter plate method, as described above for MU-AF.
The ability of purified ArfI to release arabinose from hemicellulose substrates was tested by incubating (at 40°C for 48 h) 0.52 U of enzyme in 1-ml MOPS (10 mM; pH 6.5)-buffered reaction mixtures containing (at a concentration of 10 to 25 mg per reaction mixture) Lenzing beechwood, rye, wheat, and oat spelt (arabino)xylans; three corn cob arabinoxylan fractions (CCXA, containing 5.6% arabinose, 87.7% xylose, 3% glucose, 0.1% galactose, and 3.6% other; CCXB, containing 15.3% arabinose, 77.1% xylose, 2.7% galactose, and 4.9% glucose; and CCX, containing 34.8% arabinose, 57.3% xylose, 7.2% galactose, 0.5% glucose, and 0.2% other [all corn cob arabinoxylan fractions were prepared and analyzed at the USDA Agricultural Research Service, Peoria, Ill.]); arabinogalactan; and sugar beet arabinan. At the end of incubation, the reaction mixtures were centrifuged at 11,000 × g for 15 min to sediment the particulate material. The resulting supernatant fluids were then brought to 70% ethanol by adding 100% absolute ethanol and recentrifuged. The resulting second supernatants were removed, placed in fresh microcentrifuge tubes, and lyophilized. When dry, the samples were redissolved in 50 μl of H2O, and 1 to 5 μl of each sample was spotted onto a thin-layer chromatography (TLC) plate (20 by 20 cm) precoated with a 250-μm layer of Whatman silica gel (K5) 150A. The plate was placed in a glass-enclosed tank and chromatographed with a solvent mixture consisting of n-butanol, acetic acid, and H2O (2:1:1, vol/vol/vol). Lanes containing authentic arabinose, xylose, xylobiose, xylotriose, xylotetraose, and xylopentaose (10 μg of each) were included as standards. After chromatography, the plate was air dried and then sprayed uniformly with ca. 4 ml of aniline-diphenylamine reagent (Sigma Chemical Co., St. Louis, Mo.) and developed by heating it at 85°C for 20 min. Sugars gave blue-green or brown spots.
ENDOX activity (i.e., any glycosidase activity capable of hydrolyzing xylan or portions of xylan) was assayed by measuring the release of reducing sugar from Lenzing beechwood, oat spelt, and rye (arabino)xylans (final concentration, 0.5 mg · ml−1). Reducing sugar was quantified by the method of Nelson (33), as modified by Somogyi (42), and xylose was used as the standard. Samples were boiled for 30 min with the copper reagent for optimal detection of reducing sugars. One unit of activity was defined as the amount of enzyme that liberated 1 μmol of reducing sugar equivalent (as xylose) per min under the assay conditions. Synergy between the ArfI and ENDOX pools was determined by using reaction mixtures containing rye arabinoxylan (2 mg · ml−1 in 10 mM HEPES buffer [pH 6.8] containing 0.5 mM CaCl2) and various amounts of each enzyme (or enzyme pool). The assays were run for 90 min, and during the assays aliquots were removed for determinations of reducing sugar as described above.
Glyceraldehyde-3-phosphate dehydrogenase (GPD) activity was determined by the method of Velick (45), as modified by Hespell and Canale-Parola (15), by using 1-ml reaction mixtures containing 150 mM HEPES (pH 8.4), 2 mM NAD, 10 mM dl-glyceraldehyde 3-phosphate, 50 mM KH2AsO4, 10 μM dithiothreitol, and appropriately diluted enzyme. The reaction (at 25°C) was started by adding enzyme and was followed continuously for 2 min at 340 nm with a Lambda 14 spectrophotometer (Perkin-Elmer, Norwalk, Conn.) equipped with a thermal jacketed cuvette holder. The amount of NADH was estimated by using a molar extinction coefficient (1-cm light path) of 6.22 × 103 at 340 nm (17).
Determination of pH and temperature optima and stability.
The pH optimum for ArfI activity was determined by using reaction mixtures buffered with the following compounds (pHs were adjusted in 0.5-pH unit increments): sodium citrate, pH 3 to 6; MES, pH 5.5 to 6.5; and MOPS, pH 6.5 to 8.0. Prewarmed (45°C) reaction mixtures (final volume, 4 ml) contained 3.575 ml of 50 mM buffer and 400 μl of 10 mM pNP-AF, and the reactions were initiated by adding 25 μl of ArfI (0.026 U in 2 mM MES [pH 6.0]). Then 1-ml aliquots were removed at 2-min intervals for the p-nitrophenol assay (see above). A similar procedure was used to determine the temperature optimum for ArfI.
To determine the stability of ArfI when it was exposed to various pHs, 50-μl portions of purified ArfI (0.26 U in 10 mM MES [pH 6.0]) were added to 50-μl portions of the following buffers (all at a concentration of 50 mM and adjusted in 0.5-pH unit increments): acetate, pH 4.0 to 5.5; MES, pH 5.5 to 6.5; MOPS, pH 6.5 to 8.0; N-tris(hydroxymethyl)methyl-3-aminopropanesulfonic acid, pH 8.0 to 9.0; and 2-(N-cyclohexylamino)ethanesulfonic acid, pH 9.0 to 10.0. Control tubes containing 100 μl of 10 mM MES buffer (pH 6.0) and 100 μl of each test buffer at a concentration of 50 mM were checked to ensure that ArfI was being exposed to the intended pH. The tubes were incubated at RT, and after 24 and 72 h the residual activity was determined. To determine the stability of ArfI to various temperatures, 400-μl samples of purified ArfI (0.52 U in 10 mM MES [pH 6.0]) were incubated for 1 h at temperatures ranging from 25 to 65°C in 5 ± 0.2°C increments, after which the residual enzyme activity was determined.
Electrophoresis and zymograms.
Sodium dodecyl sulfate (SDS)-PAGE was performed by using 4% (wt/vol) polyacrylamide stacking gels and 12.5% (wt/vol) polyacrylamide resolving gels (22). Native PAGE (4% [wt/vol] polyacrylamide stacking gel; 10, 12, or 7.5 to 18% [wt/vol] polyacrylamide gradient resolving gel) was performed without SDS and 2-mercaptoethanol and without preexposure of sample proteins to 100°C (3). Unless otherwise noted, all native and SDS-PAGE gels were stained with a Silver Stain Plus kit (Bio-Rad) according to the manufacturer’s instructions. Isoelectric focusing (IEF) of proteins was performed with a model 111 mini IEF cell (0.4-mm-thick minigels; Bio-Rad) and pH 5 to 7 ampholytes according to the manufacturer’s instructions. Unless otherwise noted, all electrophoresis gels were prepared with GelBond PAG films (for native PAGE and SDS-PAGE; FMC BioProducts, Rockland, Maine) or Gel Support films (for IEF; Bio-Rad) to facilitate handling. The gels used for detection of nucleic acids were prepared and electrophoresed by using standard methods (40).
Zymograms were prepared by overlaying native PAGE or IEF gels with 7.5% polyacrylamide gels containing the test substrate (5, 38). The zymogram gels for detecting ENDOX activity contained 1% (wt/vol) ethanol-extracted oat spelt xylan in 50 mM HEPES (pH 6.8) containing 0.5 mM CaCl2 and were poured with GelBond film. After an oat spelt xylan zymogram gel was placed on a native PAGE gel, the two gels were wrapped together in Sealwrap (Borden Chemical, North Andover, Mass.) and incubated overnight at 40°C. After incubation, the zymogram gel was stained with Congo red (10 mg · ml−1) for 2 h and destained for ca. 30 min with several changes of 1 M NaCl until bands of hydrolysis were visible and the destaining solution was fairly clear. The zymogram was then treated with 0.1% acetic acid, which converted Congo red-stained xylan to a dark purple color that enhanced the contrast of hydrolysis zones for photography. Zymogram gels for detecting ARAF were prepared in a similar manner, but contained MU-AF (200 μg · ml−1) instead of xylan and were incubated for only 20 min at RT before photography on a UV transilluminator.
Fractionation of cells for ArfI localization.
Cells from a 2-liter culture were harvested by centrifugation at 10,000 × g as described above, and the supernatant fluid (secreted enzyme fraction) was pooled and concentrated (10,000-molecular-weight cutoff membrane). One-half of the cell pellets (containing cell-associated enzymes) were pooled by resuspending them in 15 ml of 10 mM Tris buffer (pH 7.6) and subjected to sonication (see above), followed by centrifugation (30,000 × g, 30 min, 4°C), which yielded a particulate enzyme fraction and a supernatant (i.e., soluble cytoplasmic-periplasmic) enzyme fraction. The other half of the cell pellets were subjected to an osmotic shock procedure similar to that described by Godchaux and Leadbetter (10). First, the pellets were pooled by resuspending them in 5 ml of ice-cold 10 mM Tris buffer (pH 7.6) containing 0.3 M NaCl and then rapidly warmed to 25°C in a 40°C water bath and incubated at 25°C for 5 min. The suspension was next chilled on ice, and 0.75 mg of lysozyme (75 μl of a 10-mg/ml stock solution) was added with rapid stirring. The lysozyme-treated cells were then subjected to a slow addition of 10 ml of ice-cold 10 mM Tris buffer (pH 7.6) and incubated for 2.5 min on ice. The suspension was then warmed as described above to 25°C and kept at that temperature until more than 90% of the cells were converted into spheroplasts (as determined by phase-contrast light microscopy). The spheroplasts were centrifuged (30,000 × g, 30 min, 4°C), and the supernatant (containing periplasmic enzymes) was removed and saved. The spheroplast pellet was then resuspended in 10 ml of 10 mM Tris buffer (pH 7.6), sonicated (see above), and recentrifuged to yield a second supernatant containing soluble cytoplasmic enzymes. A portion of the remaining pellet was treated with 0.2% Triton X-100 for 2 h at room temperature and then recentrifuged, which yielded a third supernatant containing Triton X-100-extractable enzymes associated with particulate spheroplast material.
Antibody production and Western blotting.
Polyclonal anti-ArfI antiserum was produced by injecting a female New Zealand White rabbit with 100 μg of purified enzyme emulsified in TiterMax (CytRx Corp., Norcross, Ga.) by using the method of Harlow and Lane (14). A booster injection (100 μg, prepared as described above) was administered after 28 days, and serum was collected 14 days later. The anti-ArfI antiserum titer was determined by using 1 μg of purified ArfI per well in a dot blot apparatus (Bio-Rad) containing an Immobilon PSQ nylon membrane (Millipore Corp., Bedford, Mass.). The preparation was developed with a goat anti-rabbit immunoglobulin G (H + L) alkaline phosphatase immun-blot assay kit (Bio-Rad) according to the manufacturer’s instructions. Western blotting was performed by using 10% polyacrylamide native PAGE gels without GelBond film, and the blots were blotted onto Immobilon PSQ nylon membranes with a Trans-blot Cell apparatus (Bio-Rad) by using the method of Matsudaira (29). ArfI was detected as described above with anti-ArfI antiserum diluted 1:20,000. Preimmune rabbit serum (1:1,000 dilution) was used to screen a duplicate blot for nonspecific binding.
Other analytical procedures.
The native molecular masses of purified ArfI and each partially purified ENDOX pool were determined by size exclusion chromatography with a Pharmacia fast protein liquid chromatography (FPLC) system equipped with either a Superose 6 or Superose 12 HR 10/30 column (Pharmacia Biotech, Uppsala, Sweden). The columns were preequilibrated with 20 mM MOPS (pH 6.5) containing 50 mM NaCl and were run at a flow rate of 0.5 ml · min−1. The column effluents were monitored for A280, as well as for enzyme activity, by using the MU-AF plate assay (for ArfI) or the reducing sugar assay and native PAGE zymograms (for ENDOX pools). The size exclusion chromatography standards used for the Superose 6 column included blue dextran (2,000 kDa), bovine thyroglobulin (670 kDa), ferritin (440 kDa), catalase (232 kDa), and aldolase (158 kDa). The size exclusion chromatography standards used for Superose 12 columns included thyroglobulin (670 kDa), gamma globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1.35 kDa).
Glycosylation of purified ArfI was examined after SDS-PAGE of 24- and 48-μg samples as described above, but without GelBond PAG film. The subunits were then electroblotted onto an Immobilon PSQ membrane (Millipore) (29) and tested for glycosylation by using a digoxigenin glycan detection kit according to the instructions of the manufacturer (Boehringer Mannheim, Indianapolis, Ind.). Positive (transferrin) and negative (creatinase) controls were included and were supplied by the manufacturer.
Protein was measured as described by Smith et al. (41) by using a micro bicinchoninic acid protein assay reagent kit (Pierce, Rockford, Ill.) or by performing the Bradford assay (6) with bovine serum albumin as the standard.
Chemicals and other materials.
Oat spelt xylan, arabinogalactan, xylose, p-nitrophenyl derivatives, 4-methylumbelliferyl derivatives, DEAE-Sephadex A-50, NAD, dl-glyceraldehyde 3-phosphate, KH2AsO4, dithiothreitol, and bovine thyroglobulin were obtained from Sigma Chemical Co. Phenyl-Sepharose CL-4B, blue dextran 2000, ferritin, catalase, and aldolase were obtained from Pharmacia Biotech. Low-molecular-weight SDS-PAGE standards, IEF standards, FPLC size exclusion standards, pH 5 to 7 ampholytes, hydroxylapatite, and Macro Prep 50Q were obtained from Bio-Rad Laboratories. DEAE-cellulose was obtained from Whatman Specialty Products Inc. (Fairfield, N.J.). Xylobiose, xylotriose, xylotetraose, xylopentaose, rye and wheat arabinoxylans, and sugar beet arabinan were obtained from Megazyme, Inc. (Sydney, Australia). SPECTRA/POR molecularporous membrane tubing (width, 45 mm; 3,500-molecular-weight cutoff) was obtained from Spectrum Medical Industries, Inc. (Los Angeles, Calif.). Lenzing beechwood xylan and the three corn cob arabinoxylan fractions were gifts from Robert B. Hespell (Agricultural Research Service, U. S. Department of Agriculture, Peoria, Ill.). The other chemicals were reagent grade and were obtained from commercial sources. All H2O used was double distilled (Millipore Corp.).
RESULTS
Enzyme activities in crude extracts.
Triton extracts of C. xylanolytica contained a variety of enzymatic activities important for hydrolysis of xylans and other saccharides, including ARAF (but not α-l-arabinopyranosidase), β-d-xylosidase, α-d- and β-d-glucosidases, α-d- and β-d-galactosidases, and acetylesterase (Fig. 1). Likewise, native PAGE of Triton extracts revealed an array of electrophoretically separable proteins, including approximately 15 proteins with ENDOX activity distributed in the top (slowest migrating), middle, and bottom (fastest migrating) zones of the gel and detectable by zymograms with oat spelt xylan (Fig. 2A and C, lane 1). By contrast, analogous zymograms with MU-AF as the substrate revealed that ARAF activity was associated with only a single band, and this band was referred to as ArfI (Fig. 2B, lane 1). No apparent β-d-glucuronidase or β-d-cellobiosidase activity was observed when preparations were assayed by using 4-methylumbelliferyl-linked substrates in microtiter plate wells (Fig. 1).
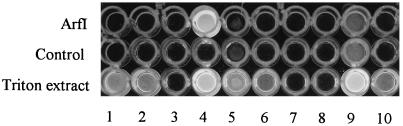
FIG. 1.
Microtiter plate assay comparing the activity of purified ArfI (1st row) to activities present in a crude Triton extract of cells (3rd row) with the following 4-methylumbelliferyl derivatives: column 1, β-d-galactoside; column 2, α-d-galactoside; column 3, α-l-arabinopyranoside; column 4, α-l-arabinofuranoside; column 5, α-d-glucoside; column 6, β-d-glucoside; column 7, β-d-glucuronide; column 8, β-d-cellobioside; column 9, acetate; column 10, β-d-xyloside. Each well containing purified ArfI contained 48 ng of protein (equivalent to 10.6 U of ARAF activity). Each well containing Triton extract contained 4.2 μg of protein (equivalent to 0.011 U of ARAF activity). The control (2nd row) contained test substrate, but lacked enzyme. Incubation was for 12 h at RT.
Purification and physicochemical properties of ArfI.
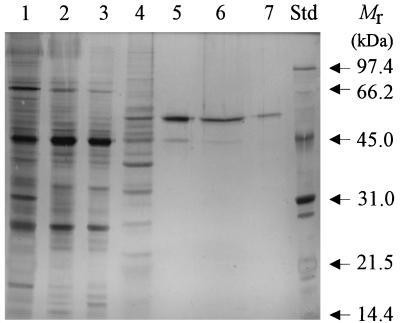
ArfI was purified 85-fold to homogeneity from Triton extracts by using anion-exchange and hydrophobic interaction column chromatography (Table 1; Fig. 2B and C and 3). With these procedures, 23.6% of the original activity in Triton extracts was recovered. A major step in the purification procedure was chromatography on hydroxylapatite, which alone yielded an 8-fold increase in purity. Although a slight decrease in specific activity was seen after Macro Prep 50Q chromatography (presumably because of some inactivation of ArfI), this step eliminated a significant amount of contaminating protein (compare lanes 5 and 6 of Fig. 2C and 3).
TABLE 1.
Summary of purification of ArfI from C. xylanolytica
| Purification stepa
|
Total amt of protein (mg) | Total activity (U)b | Sp act (U · mg−1) | Purifi- cation (fold) | Yield (%) | |
|---|---|---|---|---|---|---|
| No. | Column | |||||
| None (Triton extract) | 429.9 | 1,118 | 2.6 | 1.0 | 100.0 | |
| 1 | DEAE-cellulose | 183.9 | 1,067 | 5.8 | 2.2 | 95.4 |
| 2 | DEAE-Sephadex | 73.1 | 951 | 13.0 | 5.0 | 85.1 |
| 3 | Hydroxylapatite | 7.7 | 808 | 104.9 | 40.3 | 72.3 |
| 4 | Phenyl-Sepharose | 1.7 | 347 | 204.1 | 78.5 | 31.0 |
| 5 | Macro Prep 50Q | 1.6 | 291 | 181.9 | 70.0 | 26.0 |
| 6 | Phenyl-Sepharose | 1.2 | 264 | 220.0 | 84.6 | 23.6 |
See Materials and Methods for details.
Measured with pNP-AF as the substrate.
FIG. 3.
SDS-PAGE (silver-stained) gel of fractions obtained during purification of ArfI. Lane 1 contained Triton extract, and lanes 2 through 7 contained the fractions obtained after purification steps 1 through 6 listed in Table 1, respectively; lanes 1 through 7 contained 10.5, 9.8, 4.9, 3.6, 2.3, 1.7, and 2.4 μg of protein, respectively. Lane Std contained the following molecular weight markers: rabbit muscle phosphorylase B (molecular weight, 97,400), bovine serum albumin (66,200), hen egg white ovalbumin (45,000), bovine carbonic anhydrase (31,000), soybean trypsin inhibitor (21,500), and hen egg white lysozyme (14,400).
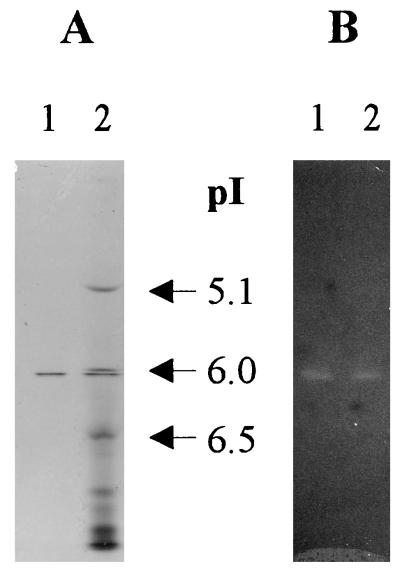
Size-exclusion FPLC of purified ArfI yielded a single, symmetrical peak corresponding to native molecular masses of 210 kDa with Superose 6 and 160 kDa with Superose 12 (data not shown). However, SDS-PAGE revealed that ArfI consisted of 56-kDa subunits (Fig. 3, lane 7), implying that the native enzyme was a trimer or tetramer. IEF of purified ArfI yielded a single protein band with a pI of 6.1, which corresponded to the only ARAF activity in the gel detectable by zymogram analysis (Fig. 4).
FIG. 4.
IEF gel of purified ArfI (1 μg of protein per lane). (A) Gel stained with crocein scarlet. (B) Gel overlaid with an MU-AF zymogram gel. Lane 1, purified ArfI; lane 2, purified ArfI plus pI standards (β-lactoglobulin B [pI 5.1], bovine carbonic anhydrase [pI 6.0], and human carbonic anhydrase [pI 6.5]).
At no time during the purification of ArfI was more than a single peak of activity observed to elute from chromatographic columns or was more than a single band of activity observed on zymograms of native PAGE gels (Fig. 2B).
Thus, the major physical properties of ArfI are as follows: Mr of native protein, 160,000 to 210,000; Mr of subunits, 56,000; and pI 6.1.
Enzymatic activity and stability of ArfI.
With pNP-AF as the substrate, ArfI had optimum activity at pH 5.8 (at 45°C) and 45 to 50°C and exhibited a Km of 0.504 ± 0.034 mM and a Vmax of 319 ± 6.6 μmol · min−1 · mg of protein−1. ArfI was highly specific for the α-l-arabinofuranoside linkage, as no hydrolytic activity was seen with the following p-nitrophenyl derivatives: α-l- and β-l-arabinopyranosides, α-d- and β-d-glucopyranosides, α-d- and β-d-galactopyranosides, α-l-rhamnopyranoside, α-l-fucopyranoside, β-d-lactopyranoside, β-d-xylopyranoside, β-d-cellobioside, α-d-mannopyranoside, β-d-glucuronide, and acetate. This specificity was also observed when 4-methylumbelliferyl derivatives of many of these same compounds were tested (Fig. 1).
Small amounts of reducing sugar were liberated when ArfI was incubated with oat spelt xylan, and this was due primarily to the release of arabinose residues (see below). However, bands of electrophoretically separated ArfI also coincided with a faint zone of Congo red nonbinding in zymogram gels designed to screen for ENDOX activity (Fig. 2A). The basis for such Congo red nonbinding is not known, but this phenomenon does not appear to represent ENDOX activity associated with ArfI (see below).
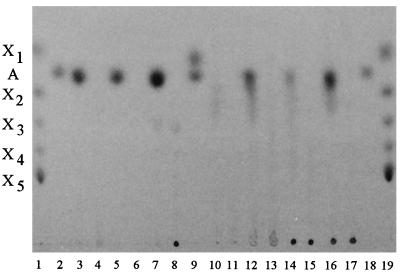
Purified ArfI also liberated arabinose from rye, wheat, corn cob, and oat spelt arabinoxylans and sugar beet arabinan (Fig. 5). Although the spot on TLC plates corresponding to arabinose was usually the most intense spot in all such digests, there was also a region of less intensely stained material below the arabinose spot in digests of corn cob and oat spelt arabinoxylans (Fig. 5, lanes 10, 12, 14, and 16). The nature of this material is unknown, but it migrated between the xylobiose and xylotriose standards, and it may be a branched arabinose-containing oligosaccharide. The barely detectable arabinose spot obtained with ArfI digests of CCXA (Fig. 5, lane 10) was not surprising, inasmuch as this substrate contained only 5.6% arabinose by weight. ArfI displayed no apparent hydrolytic activity on arabinogalactan or Lenzing beechwood xylan (data not shown).
FIG. 5.
Thin-layer chromatogram of various arabinoxylans and an arabinan before (controls) and after exposure to purified ArfI. Lanes 1 and 19, xylose, xylobiose, xylotriose, xylotetraose, and xylopentaose xylooligosaccharide standards; lanes 2 and 18, arabinose standard; lane 3, rye arabinoxylan plus ArfI; lane 4, rye arabinoxylan control; lane 5, wheat arabinoxylan plus ArfI; lane 6, wheat arabinoxylan control; lane 7, sugar beet arabinan plus ArfI; lane 8, sugar beet arabinan control; lane 9, arabinose and xylose standards; lane 10, CCXA plus ArfI; lane 11, CCXA control; lane 12, CCXB plus ArfI; lane 13, CCXB control; lane 14, CCX plus ArfI; lane 15, CCX control; lane 16, oat spelt arabinoxylan plus ArfI; lane 17, oat spelt arabinoxylan control. A, arabinose; X1, xylose; X2, xylobiose; X3, xylotriose; X4, xylotetraose; X5, xylopentaose.
Whether present in Triton extracts or in purified form, ArfI was quite stable in solution. Essentially full ARAF activity was retained in Triton extracts when they were stored at 4°C for periods as long as 24 months. In fact, zymograms of native PAGE gels of Triton extracts stored at 4°C for up to 48 months had ArfI and ENDOX activities that were only slightly less than those obtained with fresh extracts (data not shown). Likewise, purified ArfI retained ≥80% of its activity when it was exposed at RT to pH 4 to 10 for 24 h or to pH 6 to 10 for 72 h. Full activity was retained after exposure to temperatures up to 45°C for 1 h; however, activity began to decline sharply after exposure to temperatures of more than 50°C and was completely lost at 65°C.
Interaction of ArfI with ENDOXs.
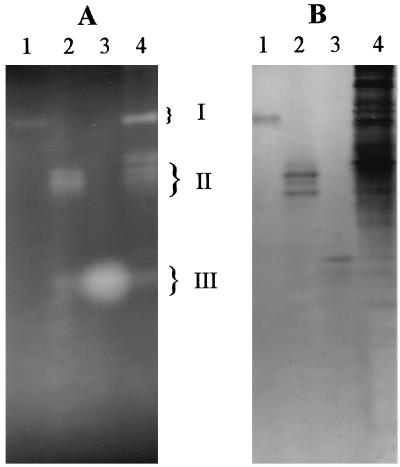
The documented ability of many ARAFs to interact synergistically with ENDOXs (3, 11, 21, 23, 28, 34, 46, 50) prompted us to explore this phenomenon with the analogous enzymes of C. xylanolytica. ENDOX components from three main zones separable by native PAGE of Triton extracts (Fig. 2A) were partially purified and designated ENDOX I, ENDOX II, and ENDOX III (Fig. 6). The ENDOX II pool consisted of at least four ENDOX enzymes, including a small but significant amount of a component(s) present in ENDOX III. In contrast, the ENDOX I and ENDOX III pools each contained only one or perhaps two electrophoretically resolvable ENDOX components. Each ENDOX pool, however, contained far less non-ENDOX, Coomassie blue-stainable material than crude Triton extracts (Fig. 6, lane 4). Size-exclusion FPLC of each ENDOX pool yielded a single activity peak that eluted at a position corresponding to 130 kDa for ENDOX I, 63 kDa for ENDOX II, and 43 kDa for ENDOX III.
FIG. 6.
Oat spelt xylan zymogram (A) and Coomassie blue protein staining (B) of a native PAGE gel of partially purified ENDOX pools. Lane 1, ENDOX I; lane 2, ENDOX II; lane 3, ENDOX III; lane 4, Triton extract. Lanes 1 through 4 contained 0.24, 1.03, 1.30, and 110.9 μg of protein, respectively.
When ArfI was mixed with the ENDOX I, II, or III pool, the rate of hydrolysis of rye arabinoxylan was 22 to 46% greater than the sum of the rates for each component acting alone (Table 2). The increased rate of hydrolysis was apparently due to catalytic synergy, as opposed to enhanced stability of one enzyme component conferred by the presence of the other, because inclusion of an equivalent amount of bovine serum albumin as a protein surrogate for any one of the enzyme components in the two-component reaction mixtures did not affect the rate of hydrolysis catalyzed by the remaining component acting alone (data not shown).
TABLE 2.
Synergy between ArfI and ENDOX pools during hydrolysis of rye arabinoxylan
| Enzyme(s) | Amt of reducing sugar released (μmol of xylose equivalent · min−1 · ml−1) | % Enhance- menta |
|---|---|---|
| ArfI | 2.41 | |
| ENDOX I | 0.30 | |
| ArfI + ENDOX I | 3.42 | 26.2 |
| ENDOX II | 2.02 | |
| ArfI + ENDOX II | 6.45 | 45.6 |
| ENDOX III | 2.33 | |
| ArfI + ENDOX III | 5.79 | 22.2 |
Increase in the amount of reducing sugar released compared with the amount expected from the action of ArfI alone or an ENDOX fraction alone.
Cellular distribution of ArfI and ENDOX activities.
The relatively high molecular weight of most arabinoxylans (1, 32) requires that they undergo hydrolysis to fragments small enough to pass through the cell membrane before further dissimilation can take place by cytoplasmic enzymes. Inasmuch as most of the enzymes of the xylanase system of C. xylanolytica were cell associated, it seemed likely that some or all of them, including ArfI, might be on or in the outer membrane or periplasm of this gram-negative bacterium, where they would have access to the native substrate or to large fragments released from it. This notion was supported by the ready extractability of the xylanase system from whole cells with a low concentration (0.2%) of the detergent Triton X-100, a treatment which caused no obvious disruption of cells, as assessed previously by phase-contrast and electron microscopy (13). However, Triton extracts were recently found to exhibit GPD activity (a typically cytoplasmic enzyme), and 0.8% agarose gel electrophoresis of Triton extracts revealed the presence of ethidium bromide-stainable material that migrated to a position typical of that of RNA (data not shown). These observations suggested that Triton X-100 did in fact disrupt the cytoplasmic membrane enough to liberate cytoplasmic components. Therefore, a cell fractionation procedure was used to determine more precisely the cellular location of ArfI and other enzymes of the xylanase system.
Sonication of xylan-grown cells, followed by centrifugation, yielded a soluble supernatant fraction that contained almost all of the ArfI and GPD activities (Table 3). This indicated that ArfI resided in the cytoplasm and/or periplasm and was not an integral membrane protein. When cells were subjected to an osmotic shock, little or no ArfI or GPD activity was released into the shock fluid (Table 3), suggesting that each activity was located primarily, if not entirely, in the cytoplasm. Unfortunately, a legitimate positive control could not be included in this experiment, as no authentic periplasmic enzymes have been identified yet for C. xylanolytica and alkaline phosphatase (a typically periplasmic enzyme in many gram-negative bacteria) was almost undetectable in this bacterium. However, the ability of exogenously added lysozyme to induce spheroplast formation during the osmotic shock procedure implied that the outer membrane was disrupted enough to allow proteins such as lysozyme to pass through it (spheroplasts were not formed in the absence of lysozyme). Thus, if ArfI was located in the periplasm, a significant amount of it should have been released into the extracellular shock fluid, but that was not the case. Rather, most of the ArfI and GPD activities from the spheroplasts were liberated into the soluble fraction following sonic disruption, a result entirely consistent with these enzymes being largely, if not entirely, cytoplasmic. The relatively high proportion of GPD associated with the particulate fraction of spheroplasts (18.5%) and the somewhat low overall recovery of this activity (75.7%) were curious, but reproducible, and may reflect some change in this enzyme during osmotic shock, since virtually all of the GPD was routinely recovered in the soluble fraction of nonosmotically shocked cells.
TABLE 3.
Distribution of ArfI and ENDOX activities in cells of C. xylanolytica
| Treatment | Cellular fraction | % of total activity or proteina
|
|||
|---|---|---|---|---|---|
| ArfI | ENDOX | GPD | Protein | ||
| Sonication | Soluble fraction | 96.6 | 60.3 | 99.9 | 79.7 |
| Particulate fraction | 3.4 | 39.7 | <0.1 | 20.3 | |
| Osmotic shock | Extracellular shock fluid | 1.8 | 8.7 | <0.1 | 2.3 |
| Soluble (cytoplasmic) fraction of spheroplasts | 87.0 | 72.5 | 57.2 | 77.4 | |
| Particulate fraction of spheroplasts | 2.6 | 19.7 | 18.5 | 13.1 | |
| Total recoveryb | 91.4 | 100.9 | 75.7 | 92.8 | |
The total amounts in 1010 sonicated C. xylanolytica cells were as follows: ArfI, 0.36 U; ENDOX, 0.36 U; GPD, 0.66 U; and protein, 1.33 mg.
Based on values given in footnote a.
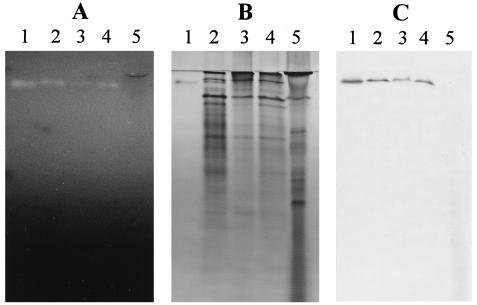
In each cell fraction in which ArfI activity was detected, the activity could be attributed to the ArfI protein, based on MU-AF zymograms prepared from native PAGE of homologous fractions (Fig. 7A and C, lanes 2 through 4). In this experiment, no ArfI (or any other ARAF activity) was detected in concentrated samples of spent, cell-free growth medium (Fig. 7A and C, lane 5), although such samples contained a variety of proteins (Fig. 7B, lane 5), including some ENDOX components (see below). However, a trace amount of ARAF activity observed in culture fluids in some experiments eluted in the same position as ArfI during column chromatography and had a similar pI in IEF gels (data not shown).
FIG. 7.
Specificity of anti-ArfI antiserum. (A) MU-AF zymogram of native PAGE gel. (B) Native PAGE (silver-stained) gel. (C) Western blot with anti-ArfI antiserum. Lane 1, purified ArfI; lane 2, soluble cell extract (obtained by sonication and centrifugation); lane 3, Triton extract of particulate cell material (obtained by sonication and centrifugation); lane 4, Triton extract of untreated whole cells; lane 5, concentrated cell-free growth medium. Lanes 1 through 5 contained 0.6, 75.0, 72.5, 60.0, and 72.0 μg of protein, respectively.
Attempts to confirm the cytoplasmic location of ArfI by immunoelectron microscopy were unsuccessful, presumably because the absolute amount of ArfI protein in the cytoplasm was too low to be detected in thin sections. However, the polyclonal anti-ArfI antiserum was used for screening other strains of C. xylanolytica for cross-reactive proteins in Western immunoblots (see below).
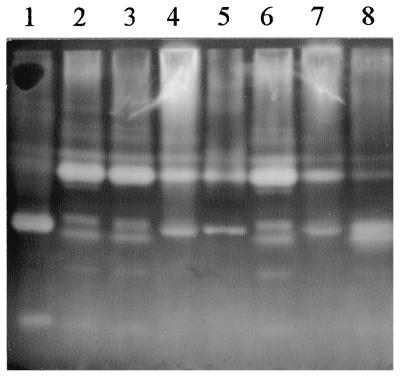
In contrast to ArfI, ENDOX activities were more widely distributed in C. xylanolytica, a result not surprising given the diversity of electrophoretically separable ENDOX proteins produced by the cells (Fig. 2A, lane 1, and Fig. 6A, lane 4). After sonication of whole cells, about 60% of the ENDOX activity was recovered in the soluble fraction (Table 3), which was rich in a variety of ENDOX I components, as well as three fairly discrete components representing the ENDOX II pool (Fig. 8, lanes 2 and 3). Triton X-100 extraction of the particulate fraction revealed that it also contained components of ENDOX I, as well as a band of activity from the ENDOX II region (Fig. 8, lane 4) that appeared to be different from the bands present in the soluble fraction of sonicated cells.
FIG. 8.
Oat spelt xylan zymogram of a native PAGE gel showing the distribution of ENDOX components in cells and culture fluids of C. xylanolytica. Lane 1, concentrated cell-free growth medium; lane 2, soluble cell extract; lane 3, soluble cell extract plus Triton X-100; lane 4, Triton extract of particulate cell material; lane 5, osmotic shock fluid (= cell periplasmic fraction); lane 6, soluble (cytoplasmic) fraction of spheroplasts; lane 7, Triton extract of particulate fraction of spheroplasts; lane 8, Triton extract of untreated whole cells. Lanes 1 through 8 contained 0.014, 3.7, 3.7, 2.8, 0.094, 5.1, 1.8, and 4.4 mg of protein, respectively.
When cells were subjected to osmotic shock, about 9% of the total ENDOX activity was released into the shock fluid (Table 3), and this activity presumably represented ENDOX components present in the periplasm. The zymograms of this material (Fig. 8, lane 5) were similar to those of Triton X-100-extracted particulate material obtained after sonication of whole cells (Fig. 8, lane 4), suggesting that some of the ENDOX activity associated with the latter (Table 3) was due to periplasmic ENDOXs that were not completely liberated by sonication of cells and remained trapped within the particulate fraction. Most of the ENDOX activity remaining in the lysozyme-induced spheroplasts formed during the osmotic shock procedure (ca. 73% of the total) appeared to be of cytoplasmic origin, as it was released as soluble activity following sonication of the spheroplasts (Table 3). As expected, the zymogram profile of this activity (Fig. 8, lane 6) was virtually identical to that of the soluble fraction of sonicated whole cells (Fig. 8, lane 2). Triton X-100 extraction of the particulate fraction of spheroplasts contained about 20% of the total activity and yielded a zymogram pattern (Fig. 8, lane 7) similar to that of material extracted with Triton X-100 from the particulate fraction of whole cells and that released by osmotic shock (Fig. 8, lanes 4 and 5, respectively).
Although cell-free spent growth medium generally contained ≤5% of the total ENDOX activity of cultures, zymograms of this material after native PAGE (Fig. 8, lane 1) revealed activities with electrophoretic mobilities similar to some of those seen in cell-associated ENDOXs—in particular, an ENDOX II component released by osmotic shock (Fig. 8, lane 5) and by Triton X-100 extraction of particulate cell material (Fig. 8, lanes 4 and 7) and an ENDOX III component that was present in cytoplasmic contents (Fig. 8, lanes 2, 3, and 6).
ArfI-like proteins in other strains of C. xylanolytica.
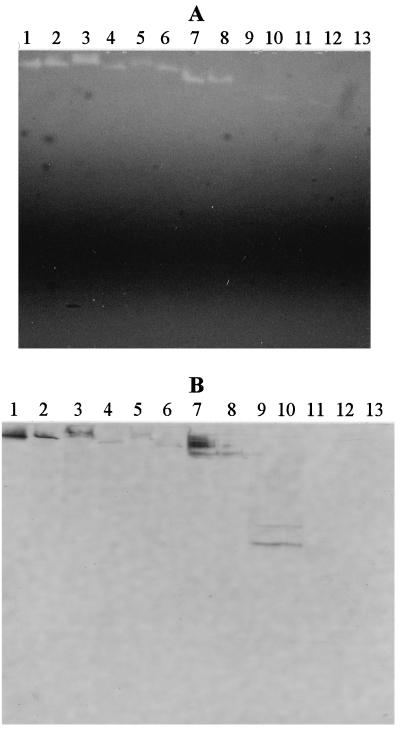
Polyclonal antiserum produced against purified ArfI was specific for ArfI in C. xylanolytica XM3, as judged by a Western immunoblot analysis of native PAGE gels containing proteins from various cell fractions (Fig. 7C). Like zymograms, Western immunoblots failed to demonstrate that ArfI was present in cell-free spent culture fluids (Fig. 7A and C, lane 5). This anti-ArfI antiserum was then used to determine whether ArfI-like proteins were present in soluble cell-free extracts of other freshwater and marine strains of C. xylanolytica isolated previously (12). Zymograms of native PAGE gels revealed that most of the strains possessed ARAF activity, which appeared as a single band migrating to a position that was similar (but not necessarily identical) to the position of ArfI (Fig. 9A). However, three marine strains (OP2E, OP2F, and PR2L) (Fig. 9A, lanes 11 through 13, respectively) did not appear to express any ARAF activity, and the ARAF activity bands for two other marine strains (EPA and EPB) (Fig. 9A, lanes 9 and 10, respectively) were faint.
FIG. 9.
ArfI-like proteins and ARAF activity of various freshwater isolates (strains XM3, SL1, MA3, and EW1) and marine isolates (strains EPFW, EPA, EPB, OP2E, OP2F, and PR2L) of C. xylanolytica after growth in freshwater medium (FW medium) or marine medium (MM medium). (A) MU-AF zymogram of native PAGE gel showing ARAF activity (viewed by UV light). (B) Western immunoblot of a native PAGE gel with polyclonal anti-ArfI antiserum. Lane 1, purified ArfI; lane 2, strain XM3 (FW medium); lane 3, strain XM3 (MM medium); lane 4, strain MA3 (FW medium); lane 5, strain EW1 (FW medium); lane 6, strain SL1 (FW medium); lane 7, strain EPFW (FW medium); lane 8, strain EPFW (MM medium); lane 9, strain EPA (MM medium); lane 10, strain EPB (MM medium); lane 11, strain OP2E (MM medium); lane 12, strain OP2F (MM medium); lane 13, strain PR2L (MM medium). Lanes 1 through 13 contained 0.6, 91.0, 142.0, 138.0, 121.0, 166.0, 146.0, 68.0, 226.0, 201.0, 85.0, 130.0, and 59.0 μg of protein, respectively. Lanes 2 through 13 contained soluble cell extract produced after sonification and centrifugation. Note that panel B is magnified more than panel A.
Zymograms prepared from Triton extracts of the particulate fraction of sonicated cells yielded patterns that were essentially identical to those seen for the soluble cell fraction (data not shown). Western immunoblots revealed that ARAFs produced by various freshwater strains appeared to cross-react with the anti-ArfI antiserum. Although these ARAFs cross-reacted weakly, they were nevertheless the only apparent proteins in cell extracts to do so (Fig. 9B, lanes 4 through 6). The pattern exhibited by marine strains was more varied. Strain EPFW had an ARAF that appeared to cross-react with anti-ArfI antiserum (Fig. 9, lanes 7 and 8), but it also possessed a cross-reactive protein(s) that migrated more slowly than ARAF and that had no ARAF activity. The latter was more abundant in extracts of cells grown in freshwater medium (Fig. 9, lane 7). In contrast, strains EPA and EPB had cross-reactive proteins that did not correspond to ARAF (Fig. 9, lanes 9 and 10). Strains OP2F and PR2L had neither ARAF activity nor cross-reactive material (Fig. 9, lanes 12 and 13).
DISCUSSION
A cytoplasmic arabinofuranosidase (ArfI) from C. xylanolytica was purified 85-fold by column chromatography and was judged to be a single protein species on the basis of SDS-PAGE, native PAGE, and IEF analyses, all of which yielded a single protein band (the latter two preparations retained ARAF activity in zymograms), as well as on the basis of FPLC (during which ArfI eluted as a single, sharp symmetrical peak) and on the basis of a Western immunoblot analysis with anti-ArfI antiserum (which revealed only one cross-reactive protein, ArfI, in cell extracts of strain XM3). We obtained no evidence which suggested that ArfI existed in multiple forms, as has been observed with some ARAFs and ENDOXs produced by other organisms (20, 30, 44, 46).
ArfI accounted for virtually all of the ARAF activity in cultures of C. xylanolytica XM3, and it appeared to be the only ARAF produced by this bacterium under our growth conditions. Likewise, single ARAFs were produced by certain other freshwater and marine strains of C. xylanolytica isolated from geographically distant sites, and most of these cross-reacted with anti-ArfI antiserum, suggesting that they are structurally similar to ArfI. A recent study (44) suggested that some ARAFs may go undetected because of their inability to cleave pNP-AF, the substrate commonly used to assay for ARAF activity. However, MU-AF was also used as a substrate in the present study, and again the only MU-AF-hydrolyzing activity observed was that attributable to ArfI and present as a single electrophoretically separable band throughout the purification procedure (Fig. 2B). The trace amount of extracellular ARAF activity occasionally observed in cell-free culture fluids of xylan- and xylose-grown cells (13) was probably the result of lysis of some cells in the population, since the protein responsible for such activity has properties similar to those of ArfI during column chromatography and IEF. However, although ArfI was the only ARAF detected in the present study, it may not be the only ARAF that cells are capable of producing. Indeed, in another paper (19) Kim et al. describe the cloning and sequencing of the following two different ARAF-encoding genes from C. xylanolytica: arfI, which encodes ArfI; and arfII, which encodes an ARAF (ArfII) expressed by Escherichia coli, but which has not yet been observed in cells or culture fluids of C. xylanolytica. In any case, the single protein (ArfI) accounting for ARAF activity was in marked contrast to the multiple ENDOX activities which segregated into three major zones (slow, moderate, and fast migrating) during native PAGE and which were associated with ca. 15 individual bands. Such a multiplicity of endoxylanases is not uncommon in xylanolytic systems (8, 49).
Some of the properties of ArfI from C. xylanolytica (see above) were typical of the properties of ARAFs from various other organisms; these properties include subunit molecular mass (which is usually 30 to 95 kDa), an acidic pH optimum (typically between pH 2.5 and 6.9), and the ability to release arabinose from pNP-AF, MU-AF, arabinoxylans, and arabinan (but otherwise the substrate specificity is narrow). However, the apparent native molecular mass of 160 to 210 kDa, which was consistent with the molecular mass of a trimer or tetramer composed of 56-kDa subunits, was higher than that of many other ARAFs. Among purified ARAFs, ArfI bears perhaps the closest resemblance to the analogous enzyme from Butyrivibrio fibrisolvens GS113 in terms of physical parameters (similar pIs, identical subunits [56 and 31 kDa, respectively], and location [both cytoplasmic] [see below]), catalytic properties (similar Km values and temperature and pH optima), and narrow substrate specificity (i.e., they are specific for the furanoside configuration, with no activity on arabinogalactan [16]).
One curious property of the ArfI of C. xylanolytica was its ability to suppress Congo red binding to oat spelt arabinoxylan zymogram gels (Fig. 2A). However, we are reluctant to attribute true endoxylanase activity (hence bifunctionality) to this enzyme because (i) little or no reducing sugar was liberated (above the amount expected from release of arabinose residues alone) when ArfI acted on oat spelt arabinoxylan, (ii) neither a significant amount of reducing sugar nor spots on TLC plates corresponding to xylooligomers were liberated by ArfI from essentially arabinose-free Lenzing beechwood xylan (data not shown), and (iii) no apparent xylooligomers accompanied the release of arabinose from rye or wheat arabinoxylan (Fig. 5). Nevertheless, it is conceivable that a small amount of xylan-depolymerizing activity might be associated with ArfI and that this activity might be akin to that seen with certain ENDOXs of Clostridium thermocellum (which liberate too little reducing sugar to be detected by conventional colorimetric assays, but can effect enough depolymerization of the substrate to be detected in Congo red-stained zymograms [26]) or the ARAF of Streptomyces lividans (which hydrolyzes the xylan backbone after prolonged incubation [46]). Such activity might also be responsible for the small amount of saccharide trailing the arabinose spot after TLC of ArfI digests of corn cob and oat spelt arabinoxylans (Fig. 5). However, the ArfI of C. xylanolytica bears little resemblance to the arabinose-releasing ENDOXs that release arabinose side groups before attacking the xylan backbone (30). Such debranching ENDOXs do not act on MU-AF and pNP-AF, whereas the ArfI of C. xylanolytica had significant activity on these substrates.
An osmotic shock procedure, originally developed during the isolation of inner and outer membrane components from several Cytophaga species (10), was part of a cell fractionation study that implied that the location of ArfI is almost entirely cytoplasmic. In contrast, this same study revealed a broad cellular distribution of ENDOX components, including at least one component whose location was primarily periplasmic (Fig. 8, lane 5), and it also revealed several ENDOX components that appear to be secreted from cells and comprise the small, but significant, fraction of total ENDOX activity that is not cell associated (Fig. 8, lane 1). However, we regard this assessment of the cellular location of ENDOX components as a first approximation. Other workers have shown that the distribution of xylanase components can be affected by the amount and type of growth substrate, the culture conditions employed, and the growth phase of cells at the time of harvest (24, 25, 31, 37).
In light of the largely (if not entirely) cytoplasmic nature of ArfI, it seems reasonable to assume that the major role of this enzyme in C. xylanolytica is to remove α-l-arabinofuranosyl residues from xylooligosaccharide fragments that are small enough to pass through (or be actively transported through) the cytoplasmic membrane and whose presence might otherwise impede the action of cytoplasmic ENDOXs and/or β-xylosidases. Such xylooligosaccharide fragments are undoubtedly liberated from arabinoxylans by ENDOX components external to the cell membrane. This interpretation is consistent with the ability of ArfI to release arabinose from various arabinoxylans (Fig. 5) and to interact synergistically with various ENDOX components (Table 2), including some components present in the cytoplasmic fraction of cells (Fig. 8, lanes 2 and 6). Such synergy is not unusual and has been reported previously for the xylanolytic enzyme systems of other organisms (3, 30). Although one might assume that xylooligosaccharides capable of passing through the cytoplasmic membrane are restricted to short oligomers, preliminary evidence that uptake of large oligosaccharides occurs has been obtained for Bacteroides thetaiotaomicron (39), a member of the same phylogenetic group as C. xylanolytica (i.e., the Bacteroides group in the Flexibacter-Cytophaga-Bacteroides phylum [27]).
Given the ability of C. xylanolytica to compete so well for the degradation of insoluble xylan particles in anoxic enrichment cultures, further studies on the nature and cellular location of specific ENDOXs and other debranching enzymes, as well as on the mechanisms and limits of saccharide uptake by cells, should undoubtedly refine our concept of arabinoxylan degradation by cells. Hence, we consider this study only one step in dissecting the complex xylanase system of this fascinating gliding bacterium.
ACKNOWLEDGMENTS
We are grateful to Robert B. Hespell for his generous gift of Lenzing beechwood xylan and corn cob arabinoxylan fractions and for many helpful discussions. We are also grateful to E. R. Leadbetter and W. Godchaux III for advice on suitable osmotic shock procedures and to R. P. Hausinger for use of equipment and helpful advice.
This research was supported by grant DE-FG02-94ER20141 from the U. S. Department of Energy and by grant BIR91-20006 from the National Science Foundation to the Michigan State University Center for Microbial Ecology.
Footnotes
Dedicated to the memory of Richard M. Behmlander, whose love of science and good nature inspired us every day.
REFERENCES
- 1.Aspinall G O. Chemistry of cell wall polysaccharides. In: Preiss J, editor. The biochemistry of plants. Vol. 3. New York, N.Y: Academic Press; 1980. pp. 473–500. [Google Scholar]
- 2.Aspinall G O. Structural chemistry of the hemicelluloses. Adv Carbohydr Chem. 1959;14:429–468. doi: 10.1016/s0096-5332(08)60228-3. [DOI] [PubMed] [Google Scholar]
- 3.Bachmann S L, McCarthy A J. Purification and cooperative activity of enzymes constituting the xylan-degrading system of Thermomonospora fusca. Appl Environ Microbiol. 1991;57:2121–2130. doi: 10.1128/aem.57.8.2121-2130.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biely P. Microbial xylanolytic systems. Trends Biotechnol. 1985;3:286–290. [Google Scholar]
- 5.Biely P. Sensitive detection of endo-1,4-β-glucanases and endo-1,4-β-xylanases in gels. Anal Biochem. 1985;144:147–151. doi: 10.1016/0003-2697(85)90096-x. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for quantitation of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Davis B J. Disk electrophoresis. II. Method and application to human serum proteins. Ann N Y Acad Sci. 1964;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- 8.Dekker R F H. Biodegradation of the hemicelluloses. Orlando, Fla: Academic Press, Inc.; 1985. [Google Scholar]
- 9.Dekker R F H, Richards G N. Hemicellulases: their occurrence, purification, properties and mode of action. Adv Carbohydr Chem Biochem. 1976;32:277–352. doi: 10.1016/s0065-2318(08)60339-x. [DOI] [PubMed] [Google Scholar]
- 10.Godchaux W, III, Leadbetter E R. Sulfonolipids are localized in the outer membrane of the gliding bacterium Cytophaga johnsonae. Arch Microbiol. 1988;150:42–47. [Google Scholar]
- 11.Greve L C, Labavitch J M, Hungate R E. α-l-Arabinofuranosidase from Ruminococcus albus 8: purification and possible role in hydrolysis of alfalfa cell wall. Appl Environ Microbiol. 1984;47:1135–1140. doi: 10.1128/aem.47.5.1135-1140.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haack S K, Breznak J A. Cytophaga xylanolytica sp. nov., a xylan-degrading, anaerobic gliding bacterium. Arch Microbiol. 1993;159:6–15. [Google Scholar]
- 13.Haack S K, Breznak J A. Xylan-degrading enzyme system of a new, anaerobic Cytophaga. In: Visser J, Beldman G, Kusters-van Someren M A, Voragen A G J, editors. Xylans and xylanases. Vol. 7. Amsterdam, The Netherlands: Elsevier; 1992. pp. 491–492. [Google Scholar]
- 14.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 15.Hespell R B, Canale-Parola E. Carbohydrate metabolism in Spirochaeta stenostrepta. J Bacteriol. 1970;103:216–226. doi: 10.1128/jb.103.1.216-226.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hespell R B, O’Bryan P J. Purification and characterization of an α-l-arabinofuranosidase from Butyrivibrio fibrisolvens GS113. Appl Environ Microbiol. 1992;58:1082–1088. doi: 10.1128/aem.58.4.1082-1088.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horecker B L, Kornberg A. The extinction coefficients of the reduced band of pyridine nucleotides. J Biol Chem. 1948;175:385–390. [PubMed] [Google Scholar]
- 18.Kaji A. l-Arabinosidases. Adv Carbohydr Chem Biochem. 1984;42:383–394. [Google Scholar]
- 19.Kim, K. S., T. G. Lilburn, M. J. Renner, and J. A. Breznak.arfI and arfII: two genes encoding α-l-arabinofuranosidases in Cytophaga xylanolytica. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 20.Komae K, Kaji A, Sato M. An α-l-arabinofuranosidase from Streptomyces purpurascens IFO 3389. Agric Biol Chem. 1982;46:1899–1905. [Google Scholar]
- 21.Kormelink F J M, Searle-Van Leeuwen M J F, Wood T M, Voragen A G J. Purification and characterization of a β-1,4-arabinoxylan arabinofuranohydrolase from Aspergillus awamori. Appl Microbiol Biotechnol. 1991;35:753–758. [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Lee S F, Forsberg C W. Purification and characterization of an α-l-arabinofuranosidase from Clostridium acetobutylicum ATCC 824. Can J Microbiol. 1987;33:1011–1016. doi: 10.1128/aem.53.4.644-650.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee Y-E, Lowe S E, Zeikus J G. Regulation and characterization of xylanolytic enzymes of Thermoanaerobacterium saccharolyticum B6A-RI. Appl Environ Microbiol. 1993;59:763–771. doi: 10.1128/aem.59.3.763-771.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee Y-E, Zeikus J G. Genetic organization, sequence and biochemical characterization of recombinant β-xylosidase from Thermoanaerobacterium saccharolyticum strain B6A-RI. J Gen Microbiol. 1993;139:1235–1243. doi: 10.1099/00221287-139-6-1235. [DOI] [PubMed] [Google Scholar]
- 26.MacKenzie C R, Yang R C A, Patel G B, Bilous D, Narang S A. Identification of three distinct Clostridium thermocellum xylanase genes by molecular cloning. Arch Microbiol. 1989;152:377–381. doi: 10.1007/BF00425176. [DOI] [PubMed] [Google Scholar]
- 27.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manin C, Shareek F, Morosoli R, Kluepfel D. Purification and characterization of an α-l-arabinofuranosidase from Streptomyces lividans 66 and DNA sequence of the gene (abfA) Biochem J. 1994;302:443–449. doi: 10.1042/bj3020443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene diflouride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 30.Matte A, Forsberg C W. Purification, characterization, and mode of action of endoxylanases 1 and 2 from Fibrobacter succinogenes S85. Appl Environ Microbiol. 1992;58:157–168. doi: 10.1128/aem.58.1.157-168.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDermid K P, MacKenzie C R, Forsberg C W. Esterase activities of Fibrobacter succinogenes subsp. succinogenes S85. Appl Environ Microbiol. 1990;56:127–132. doi: 10.1128/aem.56.1.127-132.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McNeil M, Darvill A G, Fry S C, Albersheim P. Structure and function of the primary cell walls of plants. Annu Rev Biochem. 1984;53:625–663. doi: 10.1146/annurev.bi.53.070184.003205. [DOI] [PubMed] [Google Scholar]
- 33.Nelson N. A photometric adaption of the Somogyi method for the determination of glucose. J Biol Chem. 1944;153:375–380. [Google Scholar]
- 34.Poutanen K. An α-l-arabinofuranosidase of Trichoderma reesei. J Biotechnol. 1988;7:271–282. [Google Scholar]
- 35.Puls J, Poutanen K. Mechanisms of enzymic hydrolysis of hemicelluloses (xylans) and procedures for determination of the enzyme activities involved. In: Coughlan M P, editor. Enzyme systems for lignocellulose degradation. London, United Kingdom: Elsevier; 1989. pp. 151–219. [Google Scholar]
- 36.Renner M J, Breznak J A. Purification and properties of the α-l-arabinofuranosidase component of the xylanase system of Cytophaga xylanolytica. In: van Dijken J P, Scheffers W A, editors. Book of abstracts of the Beijerinck centennial. Delft, The Netherlands: Delft University Press; 1995. pp. 272–273. [Google Scholar]
- 37.Rodriguez H, Enriquez A, Volfova O. The localization and activity of Cellulomonas xylanase on sugarcane bagasse pith. Can J Microbiol. 1985;31:754–756. [Google Scholar]
- 38.Royer J C, Nakas J P. Simple, sensitive zymogram technique for detection of xylanase activity in polyacrylamide gels. Appl Environ Microbiol. 1990;56:1516–1517. doi: 10.1128/aem.56.6.1516-1517.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salyers A A, Reeves A, D’Elia J. Solving the problem of how to eat something as big as yourself: diverse bacterial strategies for degrading polysaccharides. J Ind Microbiol. 1996;17:470–476. [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 42.Somogyi M. Notes on sugar determination. J Biol Chem. 1952;195:19–23. [PubMed] [Google Scholar]
- 43.Timell T E. Recent progress in the chemistry of wood hemicelluloses. Wood Sci Technol. 1967;1:45–70. [Google Scholar]
- 44.Van Laere K M, Beldman G, Voragen A G. A new arabinofuranohydrolase from Bifidobacterium adolescentis able to remove arabinosyl residues from double-substituted xylose units in arabinoxylan. Appl Microbiol Biotechnol. 1997;47:231–235. doi: 10.1007/s002530050918. [DOI] [PubMed] [Google Scholar]
- 45.Velick S F. Glyceraldehyde-3-phosphate dehydrogenase from muscle. Methods Enzymol. 1955;1:401–406. [Google Scholar]
- 46.Vincent P, Shareck F, Dupont C, Morosoli R, Kluepfel D. New α-l-arabinofuranosidase produced by Streptomyces lividans: cloning and DNA sequence of the abfB gene and characterization of the enzyme. Biochem J. 1997;322:845–852. doi: 10.1042/bj3220845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilkie K C B. Hemicellulose. Chemtech. 1983;13:306–319. [Google Scholar]
- 48.Wilkinson G N. Statistical estimations in enzyme kinetics. Biochem J. 1961;80:324–332. doi: 10.1042/bj0800324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong K K Y, Tan L U L, Saddler J N. Multiplicity of β-1,4-xylanase in microorganisms: functions and applications. Microbiol Rev. 1988;52:305–317. doi: 10.1128/mr.52.3.305-317.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wood T M, McCrae S I. Arabinoxylan-degrading enzyme system of the fungus Aspergillus awamori: purification and properties of an α-l-arabinofuranosidase. Appl Microbiol Biotechnol. 1996;45:538–545. doi: 10.1007/BF00578468. [DOI] [PubMed] [Google Scholar]