Abstract
Insertion element ISD1, discovered when its transposition caused the insertional inactivation of an introduced sacB gene, is present in two copies in the genome of Desulfovibrio vulgaris Hildenborough. Southern blot analysis indicated at least two insertion sites in the sacB gene. Cloning and sequencing of a transposed copy of ISD1 indicated a length of 1,200 bp with a pair of 44-bp imperfect inverted repeats at the ends, flanked by a direct repeat of the 4-bp target sequence. AAGG and AATT were found to function as target sequences. ISD1 encodes a transposase from two overlapping open reading frames by programmed translational frameshifting at an A6G shifty codon motif. Sequence comparison showed that ISD1 belongs to the IS3 family. Isolation and analysis of the chromosomal copies, ISD1-A and ISD1-B, by PCR and sequencing indicated that these are not flanked by direct repeats. ISD1-A is inserted in a region of the chromosome containing the gapdh-pgk genes (encoding glyceraldehyde-3-phosphate dehydrogenase and phosphoglycerate kinase). Active transposition to other loci in the genome was demonstrated, offering the potential of a new tool for gene cloning and mutagenesis. ISD1 is the first transposable element described for the sulfate reducers, a large and environmentally important group of bacteria. The distribution of ISD1 in genomes of sulfate-reducing bacteria is limited. A single copy is present in the genome of D. desulfuricans Norway.
Bacterial insertion sequences (ISs) are mobile genetic elements of 0.7 to 2 kb that code only for functions necessary for their transposition (11, 14). The majority contains imperfect inverted repeats (IRs) of up to 46 bp at the ends and produce target site duplication upon insertion. The transposition of an IS element can have different genetic consequences, including insertional mutation of a gene and activation or inactivation of nearby genes (1, 14). IS elements are found either alone or at the ends of composite transposable elements (transposons). In spite of their diversity, sequence analysis has revealed the existence of several large families, e.g., IS3 (9, 32) and IS4 (26). The IS3 family includes members isolated from both gram-negative and gram-positive bacteria. All are organized similarly, with two overlapping open reading frames (orfA and orfB) and a string of purines (A6G or similar motif containing a run of adenines) in the overlap region, usually followed by a potential stem-loop structure (3, 24). This unusual arrangement has been shown to produce an OrfA-OrfB fusion protein with transposase activity by a programmed −1 translational frameshift at the A6G-like site in IS911 (21), IS150 (39), and IS3 (34). OrfA, which is not highly conserved, contains a potential helix-turn-helix motif, possibly involved in binding to the terminal IRs of the cognate IS element (24). OrfB is more conserved among the family and contains a D-(1)-G-(33)-E or D-(35)-E motif which is also shared by the retroviral/retrotransposon integrases (7, 9, 18).
Bacteria of the genus Desulfovibrio are gram-negative sulfate-reducing anaerobes, for which the genetics and molecular biology have been relatively well studied (29, 41–44). In the course of a gene replacement mutagenesis study of dcrA of Desulfovibrio vulgaris Hildenborough, encoding an oxygen-sensing protein, using the Bacillus subtilis sacB gene as a counterselection marker, we obtained mutants which were sucrose resistant by insertion of a 1.2-kb DNA element into sacB (10). The cloning and characterization of this element, which we named ISD1, its homology to the IS3 family, and its distribution among Desulfovibrio spp. are reported here.
MATERIALS AND METHODS
Bacterial strains, phages, plasmids, and growth conditions.
Bacteria, phages, and plasmids used in this study are listed in Table 1. D. vulgaris Hildenborough and its derivative strains and Escherichia coli TG2 were grown in medium C and TY medium, respectively, as described previously (10). Chromosomal DNA samples from the following bacteria were available in the laboratory for screening of the distribution of ISD1 by Southern blot analysis: D. vulgaris NCIMB8399, D. vulgaris Miyazaki, D. vulgaris subsp. oxamicus, D. desulfuricans Norway, D. desulfuricans NCIMB8407, D. gigas, D. africanus, D. salexigens NCIMB8365, D. baarsii DSM2075, Desulfobotulus sapovorans, Desulfobulbus propionicus, Desulfococcus multivorans, Desulfosarcina variabilis, Desulfotomaculum ruminis NCIMB8452, Desulfotomaculum ruminis ATCC 23192, Shewanella putrefaciens, and Clostridium sp. These are not listed in Table 1.
TABLE 1.
Bacterial strains, plasmids, and phages used for ISD1 characterization
| Bacterial strain, plasmid, or phage | Genotype, comments | Reference or source |
|---|---|---|
| D. vulgaris | ||
| Hildenborough | NCIMB 8303; wild type, source of the ISD1 element | 23 |
| F1 | Derivative of Hildenborough with pΔDcrA2CTB integrated into the chromosome; Kmr Cmr Sucs | 10 |
| F1SR | All sucrose-resistant derivatives of F1 with sacB gene mutated; Kmr Cmr Sucr | This study |
| F1SR12 | One of the F1SR strains containing an ISD1 insertion within the sacB gene | This study |
| F1SR11 | One of the F1SR strains containing an ISD1 insertion within the sacB gene and two additional newly transposed copies on the chromosome | This study |
| E. coli TG2 | Δ(lac-pro) supE thi hsdM hsdR recA F′ (traD36 proAB+ lacZΔM15 Iq) | 30 |
| Plasmids and phages | ||
| pNOT19 | Cloning vector derived from pUC19; Apr | 33 |
| pNEB193 | Cloning vector derived from pUC19; Apr | New England Biolabs |
| pHIS1 | pNOT19 with a 3.8-kb PstI fragment from D. vulgaris F1SR12 containing the sacB gene with an ISD1 insertion; Apr | This study |
| DvH λ library | Ordered genomic λ library of D. vulgaris Hildenborough | 5, 40 |
| λ-G724 | Clone from the D. vulgaris λ library, with a 18-kb insert containing ISD1 |
Southern blot analysis of D. vulgaris F1SR strains.
Chromosomal DNAs of D. vulgaris F1SR strains were isolated from 5-ml cultures by a minipreparation protocol (10). The DNAs were restricted with BamHI and HindIII, and the digests were fractionated on an agarose gel and blotted onto a Hybond-N nylon membrane (Amersham). The blot was probed with 32P-labeled sacB DNA, obtained as a 2.4-kb XbaI fragment from pMOB2 (33) to characterize the insertion mutation.
Cloning of ISD1.
Chromosomal DNA from D. vulgaris F1SR12, containing a putative IS element inserted into the sacB gene, was restricted with PstI. Restriction fragments of 3 to 4 kb were isolated from a low-melting-temperature agarose gel and ligated into the PstI site of plasmid pNOT19. The ligation mix was used to transform E. coli TG2. Ampicillin-resistant colonies were screened by hybridization with the sacB probe. Plasmids isolated from positive clones were mapped to verify the presence of the putative insertion element. One of these was named pHIS1. Fragments of the sacB gene containing the insertion element were gel isolated, 32P labeled, and used to probe a Southern blot of chromosomal DNAs to verify the D. vulgaris origin of the insertion element. The same probe was also used to probe an ordered D. vulgaris Hildenborough genomic λ library (5, 40) to identify λ clones containing the native ISD1 element.
Nucleotide sequence determination.
The insertion element DNA cloned in plasmid pHIS1 was mapped and subcloned to facilitate nucleotide sequence determination. The recombinant plasmids were used directly for double-stranded DNA sequencing by the dideoxy-chain termination method using a T7 sequencing kit (Pharmacia). A cycling sequencing kit (Pharmacia) was used for sequencing the flanking regions of the native ISD1 elements obtained by PCR amplification of wild-type D. vulgaris DNA, using the PCR primers described below. The sequencing gel autoradiograms were read manually, and the sequence data were assembled into a contiguous sequence by using the Fragment Assembly system of the Genetics Computer Group (GCG) package (version 8.0.1-UNIX).
Cloning of the flanking regions of native ISD1 by PCR.
After determination of the complete nucleotide sequence of the cloned insertion element, two outward-pointing oligonucleotide primers, GCACTCCATGAGGCAATC (P101) and AGTACAACGAGGAACGAC (P102), complementary to sequences near the two ends of the element were synthesized. Chromosomal DNA from wild-type D. vulgaris was digested with HincII or ClaI, and the fragments produced were circularized with T4 DNA ligase and then used as templates for PCRs to amplify the flanking sequences. These PCRs were conducted in 100 μl of buffer (50 mM KCl, 1.5 mM MgCl2, 10 mM Tris-HCl [pH 9.0]) containing 2 mM deoxynucleoside triphosphates 0.4 μM primers, and 2.5 U of Taq DNA polymerase, with 5 min of preheating at 94°C followed by 30 cycles of 45 s at 94°C, 30 s at 50°C, and 4 min at 72°C. The last polymerization at 72°C was extended to 10 min. The PCR product was cloned into the PmeI site of plasmid pNEB193, or used directly, for sequence determination with a cycling sequencing kit (Pharmacia) as described above.
Nucleotide sequence analysis.
The nucleotide sequence of ISD1 and amino acid sequences deduced from open reading frames and longer than 50 amino acids (aa) were used to search for homologous sequences in the GenBank and protein databases, using the BLAST program in the network server at blast@ncbi.nlm.nih.gov. Homologous sequences were aligned by using the multiple-sequence alignment program PILEUP of the GCG package by varying parameters for gap creation and extension penalties until a satisfactory alignment was achieved. The output file of aligned multiple sequences was used to deduce a matrix of the pairwise evolutionary distances with the DISTANCES program, which uses the Kimura protein distance correction method. Phylogenetic trees were constructed from the distance matrix by using the GROWTREE program of the GCG package (12).
Distribution of ISD1-like elements.
Chromosomal DNAs from 18 different bacteria were restricted with PvuII, which has no site in the cloned ISD1, and blotted onto Hybond-N membranes. The blots were probed with 32P-labeled ISD1 DNA under highly stringent conditions. The final posthybridization washing step was in 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate at 68°C for 30 min.
Nucleotide sequence accession number.
The complete nucleotide sequence of ISD1 described in this report has been deposited in GenBank under accession no. AF034211.
RESULTS
Cloning of ISD1 from an insertionally inactivated sacB gene.
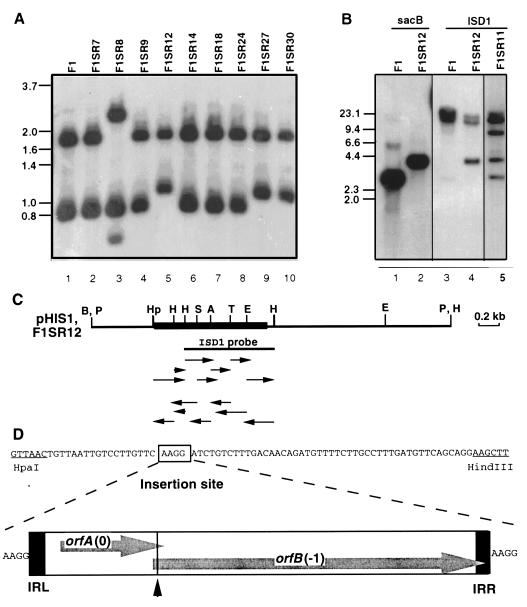
In earlier work, Southern analysis using the sacB gene as a probe had shown that 6 of 16 D. vulgaris F1SR strains contain a 1.2-kb insertion in the sacB gene (10). We found two insertion patterns, one represented by F1SR8 (Fig. 1A, lane 3) and one represented by F1SR12, F1SR27, and F1SR30 (Fig. 1A, lanes 5, 9, and 10). Other sucrose-resistant mutants of D. vulgaris F1 (Fig. 1A, F1SR7, F1SR9, F1SR14, F1SR18, and F1SR24) had a hybridization pattern unchanged from that of D. vulgaris F1, indicating that sucrose resistance was not caused by sacB inactivation through IS element insertion.
FIG. 1.
Cloning and sequencing of ISD1 from a D. vulgaris F1SR strain. (A) Southern blot analysis of D. vulgaris FISR strains. Chromosomal DNAs were restricted with BamHI and HindIII, and the blot was hybridized with the sacB probe. (B) Southern hybridization of F1, F1SR11, and F1SR12 strains restricted with PstI, using the sacB probe (lanes 1 and 2) and the ISD1 probe (lanes 3 to 5). In both panels, the size markers are denoted in kilobases on the left. (C) Restriction map of a 3.8-kb fragment of the sacBR genes (thin line) with the ISD1 insertion (thick line) as cloned in plasmid pHIS1. Restriction sites: A, AccI; B, BamHI; E, EcoRI; H, HindIII; Hp, HpaI; P, PstI; S, SacI; T, TaqI. The 0.7-kb HindIII fragment used as the ISD1 probe in panel B is indicated by the solid line below the map. The arrows indicate the sequencing strategy. (D) Nucleotide sequence of the HpaI-HindIII region of the sacB gene from strain F1SR12 indicating the site of ISD1 insertion. A survey of the structure of ISD1, which is flanked by a duplicated target sequence (AAGG) and contains IRL and IRR, is also shown. OrfA and OrfB in reading frames 0 and −1, respectively, are separated by a shifty codon motif (▴) as discussed in the text.
A 3.8-kb PstI fragment containing the 1.2-kb IS element was identified in D. vulgaris F1SR12 (Fig. 1B, lane 2) and was cloned into the PstI site of pNOT19 to give plasmid pHIS1 (see Materials and Methods). A 0.7-kb HindIII fragment located mostly within the element (Fig. 1C, ISD1 probe) was isolated. Use in probing a Southern blot of PstI-restricted chromosomal DNA showed a 3.8-kb fragment, corresponding to the IS element-inactivated sacB gene, as well as two additional PstI-fragments of 15 and 23 kb for D. vulgaris F1SR12 (Fig. 1B, lane 4). The latter two were also found in D. vulgaris F1 (Fig. 1B, lane 3) and in wild-type D. vulgaris Hildenborough (not shown), indicating that ISD1 is present in two identical or very similar copies in the D. vulgaris genome.
Nucleotide sequence of ISD1.
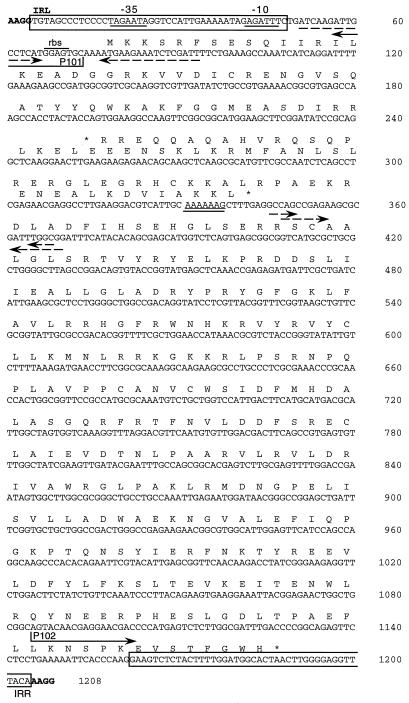
The nucleotide sequence of the 3.8-kb PstI insert of pHIS1 was determined by the strategy shown in Fig. 1C. Parts of the sequence, determined for both strands, are shown in Fig. 1D and 2. The actual ISD1 element is 1,200 bp long (Fig. 2, nucleotides [nt] 5 to 1204) with a GC content of 52.2%, considerably lower than that of the D. vulgaris genome (62 to 65%). The sequence has a pair of 44-bp imperfect IRs with 11 mismatches at the left (IRL) and right (IRR) ends. These are flanked by a pair of 4-bp direct repeats of the target site sequence (AAGG), indicating that transposition of ISD1 causes a 4-bp target site duplication. ISD1 carries two overlapping open reading frames, orfA and orfB, on its coding strand. The presence of only one putative ς70-like promoter within the IRL region (Fig. 2, −35 [TAGAAT at nt 18 to 23] and −10 [GAGATT at nt 41 to 46]) indicates that orfA and orfB are cotranscribed. A BLAST search of the nonredundant protein database revealed that orfA and orfB encode a transposase of the IS3 family (24). A multiple sequence alignment with transposases of this family, generated by using the PILEUP program of the GCG package, is shown in Fig. 3.
FIG. 2.
Nucleotide sequence of ISD1. The flanking duplicated target site (AAGG) is indicated in bold. IRL and IRR are boxed. The two open reading frames that encode the cognate transposase (orfA [nt 77 to 343] and orfB [nt 256 to 1188]) are indicated by translation into protein. orfA gene is preceded by a ribosome-binding site (rbs). As in other IS elements, the exact translational start codon of orfB is unclear. A shifty codon motif (nt 331 to 337), a putative promoter (35 [TAGAAT, nt 18 to 23] and −10 [GAGATT, nt 41 to 46]), several IRs (dashed arrows), and the primers P101 and P102 are indicated.
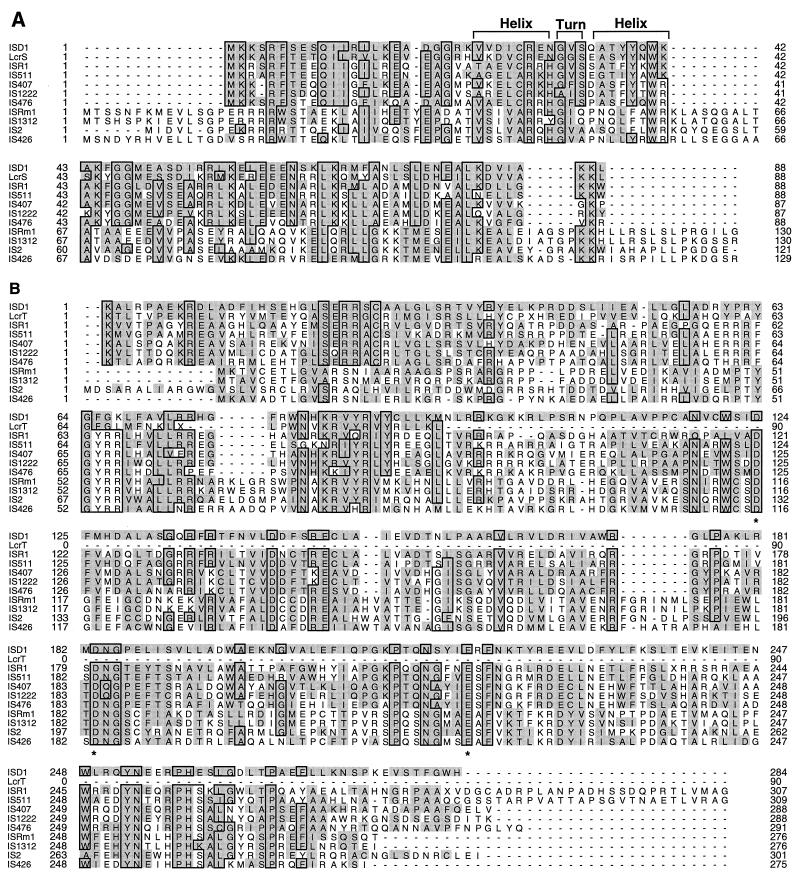
FIG. 3.
Amino acid sequence comparisons of IS transposases. The putative transposase encoded by orfA and orfB of ISD1 is compared with its counterparts in the IS3 family and with proteins encoded by the lcr operon. (A) Sequence alignment of OrfA sequences and LcrS. (B) Sequence alignment of OrfB sequences and LcrT. Identical residues in 6 or more of the 11 sequences are boxed, and residues homologous to those in ISD1 are shaded. The DNA binding helix-turn-helix motif in OrfA and the D,G-(33)-E motif in OrfB are also indicated by overlining and asterisks, respectively.
Sequence of an additional target site.
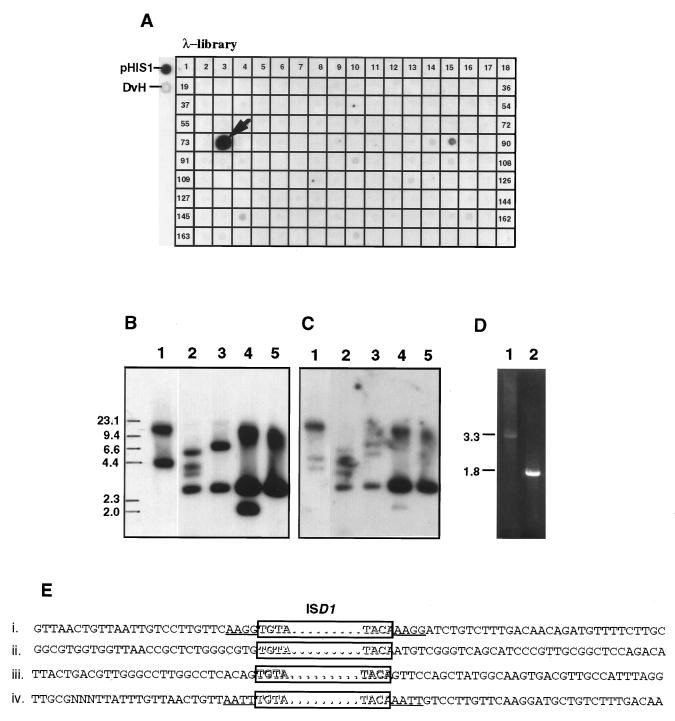
ISD1 is present in two copies in the D. vulgaris chromosome, to which we will refer as ISD1-A and ISD1-B. One of the F1SR strains, F1SR11, was unusual in having three additional ISD1 copies on the chromosome; one was inserted in the sacB gene to give the 3.8-kb PstI fragment, and the other two were present in 3- and 7-kb PstI fragments, respectively (Fig. 1B, lane 5). PstI fragments of 3 kb were isolated by agarose gel electrophoresis and circularized by ligation. PCR of the product of the ligation reaction with the pair of outward primers (Fig. 2, P101 and P102) gave a 1.9-kb PCR product. Sequence analysis of this PCR product indicated that the target site sequence of the ISD1 transposition was AATT in this case (Fig. 4E, line iv).
FIG. 4.
Cloning and sequencing of the flanking regions of native ISD1 from the D. vulgaris Hildenborough genome. (A) Dot blot hybridization of an ordered genomic library with the 0.7-kb ISD1 probe indicated in Fig. 1C. pHIS1 DNA (pHIS1) and D. vulgaris Hildenborough chromosomal DNA (DvH) served as positive controls. Positive clone 75 (λ-G724) is indicated with an arrow. (B) A Southern blot of wild-type D. vulgaris chromosomal DNA restricted with ClaI (lane 1), AccI (lane 2), and HincII (lane 3) and λ-G724 restricted with AccI (lane 4) and HincII (lane 5) was hybridized with the 0.7-kb ISD1 probe. The size markers are indicated in kilobases on the left. (C) Southern blot as in panel B but probed with the 1.8-kb PCR product containing the flanking regions of ISD1-A (Fig. 1D, lane 2). (D) PCR products amplified from ClaI (lane 1)- or HincII (lane 2)-restricted and circularized chromosomal DNA of wild-type D. vulgaris Hildenborough, using the P101-P102 primer pair. These contain the flanking regions of ISD1-A and ISD1-B, respectively. (E) Comparison of flanking region sequences for ISD1 inserted into the sacB gene in D. vulgaris F1SR12 (i), of the two native copies ISD1-A (ii) and ISD1-B (iii), and of the one transposed in D. vulgaris F1SR11 (iv). The ISD1 element is boxed. The duplicated target sites flanking transposed ISD1 are underlined.
Characterization of chromosomal flanking regions.
Digestion of the genome with HincII, Southern blotting, and hybridization of the blot with the 0.7-kb ISD1 probe indicated hybridizing fragments of 3 and 7 kb, corresponding to ISD1-A and ISD1-B, respectively (Fig. 4B, lane 3). Screening an ordered λ library, estimated to contain ca. 70% of the D. vulgaris genome (5), with the 0.7-kb ISD1 probe gave one positive λ clone (Fig. 4A, λ-G724). Detection of the 3-kb HincII fragment in this clone (Fig. 4B, lane 5) indicated it to contain ISD1-A. To retrieve ISD1-B, D. vulgaris DNA was digested with HincII and circularized by ligation, and the product of the ligation reaction was used for PCR with the pair of outward primers. Agarose gel electrophoresis indicated that only a 1.8-kb PCR product, derived from the 3-kb HincII fragment, was obtained (Fig. 4D, lane 2). This PCR product hybridized to the 3-kb HincII fragments obtained from both genomic DNA and λ-G724 samples (Fig. 4C, lanes 3 and 5). The ISD1-B containing 7-kb HincII fragment was expected to give rise to a 5.9-kb PCR product. However, this was not observed, indicating bias toward amplification of the smaller product. Southern blot analysis of a ClaI digest of D. vulgaris DNA indicated that 4.4- and 15-kb fragments hybridized with the ISD1 probe (Fig. 4B, lane 1). Hybridization with the 1.8-kb PCR product indicated that the 15-kb ClaI fragment contained ISD1-A (Fig. 4C, lane 1). PCR of a ClaI-restricted, circularized chromosomal DNA sample gave a 3.3-kb PCR product (Fig. 4D, lane 1), derived from the 4.4-kb fragment containing ISD1-B. The flanking sequences of ISD1-A and ISD1-B were determined by cycled sequencing of the two PCR products using primers P101 and P102, and the results are shown in Fig. 4E. No direct repeat sequences are flanking ISD1-A and ISD1-B in the D. vulgaris chromosome, implying that these flanking regions may have undergone secondary mutations following ISD1 insertion.
ISD1-A is located downstream from gap.
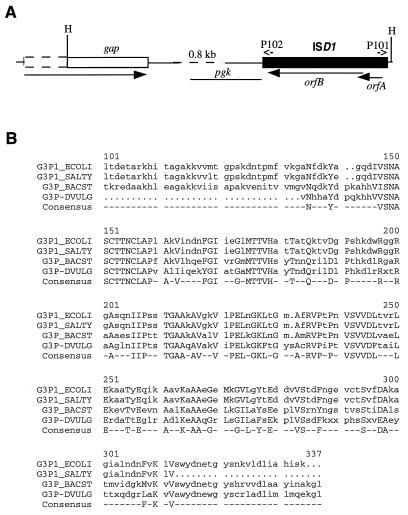
Sequence analysis of the 1.8-kb PCR product flanking ISD1-A and cloned in pNEB193 revealed the presence of a HincII site 16 nt upstream from IRL (Fig. 5A). The sequence data beyond this point therefore represented the sequence downstream from ISD1-A. Translation gave a truncated open reading frame coding for a polypeptide with homology to the C-terminal parts of glyceraldehyde-3-phosphate dehydrogenases (GAPDHs) from eubacteria, archaebacteria, and eukaryotes (Fig. 5B). The identity to the enzyme of Bacillus stearothermophilus (2, 35) is 58.5% in a 204-aa overlap. This result indicates that ISD1-A is located ca. 0.8 kb downstream from a gap gene, encoding GAPDH of D. vulgaris in a reverse orientation, as indicated in Fig. 5A.
FIG. 5.
Position of ISD1-A relative to the gap gene of D. vulgaris Hildenborough. (A) Physical map showing the 3′ end of gap (open box) and ISD1-A (filled box). The gene orientation (arrows), HincII sites (H), and locations of primers P101 and P102, used for PCR and sequencing, are indicated. The sequence of the region represented by the dashed line was not determined. The location of a putative pgk gene is also indicated. (B) Alignment of the amino acid sequence of D. vulgaris GAPDH (G3P_DVULG) with sequences from B. stearothermophilus (G3P_BACST), E. coli (G3P_ECOLI), and Salmonella typhimurium (G3P_SALTY). The numbering refers to the E. coli sequence. Residues conserved in all four species are indicated as the consensus sequence.
Distribution of ISD1 in other bacteria.
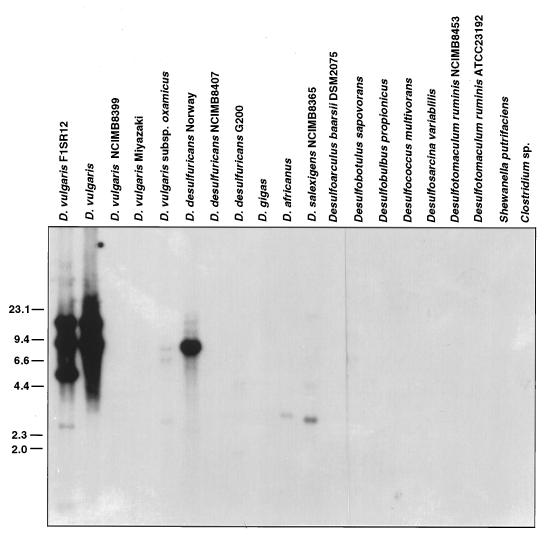
Probing of a Southern blot of PvuII-restricted genomic DNAs of 18 different bacteria with the 0.7-kb ISD1 fragment gave the results shown in Fig. 6. Wild-type D. vulgaris and D. vulgaris F1 showed two hybridizing bands, while D. vulgaris F1SR12 showed an expected extra band. Among 10 other Desulfovibrio spp., only D. desulfuricans Norway showed a strongly hybridizing band, indicating the presence of an ISD1-like element in this organism. Weakly hybridizing bands were observed for D. vulgaris subsp. oxamicus, D. africanus, and D. salexigens, indicating the presence of distantly related IS elements in these organisms. Under these same conditions, no hybridization was found with DNA from sulfate-reducing bacteria other than Desulfovibrio spp. and with DNA from S. putrefacieus, Clostridium sp., and E. coli TG2.
FIG. 6.
Distribution of ISD1 among different bacteria. Genomic DNA samples of Desulfovibrio spp. and of other bacteria as indicated above each lane were restricted with PvuII, Southern blotted onto Hybond-N nylon membranes, and probed with the 0.7-kb ISD1 probe indicated in Fig. 1C. Size markers are indicated in kilobases on the left.
DISCUSSION
Transposition of ISD1.
ISD1 was discovered when cells of D. vulgaris F1, which contains the B. subtilis sacB gene integrated into the chromosome at a site immediately downstream from the dcrA gene, were subjected to selection pressure by adding sucrose (10). Insertion of ISD1 was in an AAGG target site of the sacB gene (Fig. 2). The second insertion site in the sacB gene (Fig. 1A, lane 3) has not been characterized. Sucrose selection did also cause ISD1 to jump to other sites in the chromosome, and characterization of one of these indicated an AATT target site. Thus, ISD1 transposase may have AAXX as recognition sequence, although more experiments are needed to completely define the insertion specificity. The occurrence of several simultaneous transposition events in the same cell subjected to sacB counterselection suggests that the transposition activity may have been induced by the sacB lethality in the presence of sucrose. This induction increased the frequency of random mutagenesis under stress conditions and thereby the probability of surviving the harsh selection pressures. The fact that ISD1 can insert at more than one target site may make it a powerful natural mutagenesis tool for D. vulgaris. ISD1 is not widely distributed in sulfate-reducing bacteria and may have been acquired by D. vulgaris Hildenborough sufficiently long ago to allow mutation of the direct repeats of the initial target sites (Fig. 5E).
Analysis and comparison of the ISD1 sequence.
The orfA gene of ISD1 (nt 77 to 340) encodes a polypeptide of 88 aa, starting from an ATG codon preceded by a ribosome-binding site (Fig. 2, nt 67 to 70). The orfB gene overlaps with the 3′ end of orfA. Within the overlapping region, a putative translational frameshifting motif A6G (nt 331 to 337) is located close to the 3′ end of orfA, followed by two potential stem-loop structures (Fig. 2, nt 345 to 370). This A6G motif and the downstream secondary structures have been shown to promote the programmed translational frameshifting between orfA and orfB to produce an OrfAB fusion. This OrfAB fusion protein functions as the transposase in IS911, IS150, and IS3 members of the IS3 family (21, 22, 34, 39). The sequence homology suggests that this same mechanism is used by ISD1 for regulation of transposase production and transposition of the element. An additional IR (Fig. 2, nt 50 to 93) overlapping the ribosome-binding site and start codon of orfA could provide additional translational regulation by forming a stable stem-loop structure at the 5′ end of the mRNA. This would sequester the orfA translational start codon, especially when transcription is initiated from outside the IS element after its transposition into a transcriptionally active region, as in the case of IS10 and IS50 (4, 16) as reviewed by Chandler and Fayet (3). Otherwise, the transcript is expected to start at nt 57 and the stem-loop structure is not expected to be stable.
Multiple sequence alignment of transposases and retroviral polyproteins of the IS3 family with OrfA and OrfB of ISD1 revealed that IS1222, IS511, ISR1, IS407, IS476, IS1313, ISRm1, IS2, and IS426 are most closely related to ISD1 (Fig. 3). OrfA of ISD1 also showed high homology with LcrS encoded by the lcrS gene of plasmid pIB1 from Yersinia pseudotuberculosis (27), while the N terminus of OrfB of ISD1 showed homology with LcrT, which is only 55 aa long and is encoded by the lcrT gene located downstream from lcrS. Comparison of ISD1 and the lcr operon sequence (accession no. M83986) revealed the presence of an ISD1-like sequence at the 3′ end of the available sequence data (nt 3925 to 4521). The presence of an IRL-like sequence upstream of the lcrS gene and an A7 shifty codon motif-like structure (3) between the lcrS and lcrT genes raises the possibility that these two genes originated in Y. pseudotuberculosis from an IS element that resembled ISD1. Both OrfA and OrfB of ISD1 show highest homology to LcrS and LcrT (27). Within the IS3 family, IS1222 (36), IS511 (19), ISR1 (25), IS407 (46), and IS476 (15) have distinctly higher homology to ISD1 than do IS1312 (6), ISRm1 (45), IS2 (28), IS426 (45), and other IS3 family members (Fig. 3). The helix-turn-helix motif suggested for OrfA of IS2, IS476, and ISR1, which could be involved in recognition of and binding to the terminal IRs (24), was also found in ISD1 OrfA (Fig. 3A, aa 23 to 43). A D,D-(35)-E motif which is conserved among all IS3 family elements identified so far as well as in retroviral integrases (18, 20) and which has been proposed to be involved directly in catalysis (20) is present in ISD1 OrfB (Fig. 3B, aa 124, 183, and 219).
The aligned sequences for OrfA and OrfB were used to construct phylogenetic trees (not shown). The two trees implied similar phylogenetic relationships in which ISD1 is first grouped with the lcrS and lcrT genes and then grouped with ISR1, IS511, IS407, IS1222, and IS476. The phylogenetic relationship of these IS elements is different from that of the hosts as derived by aligning 16S rRNA sequences. This finding implies horizontal transfer of these IS elements, which is also confirmed by the significantly different GC content of ISD1 (52%) compared to the rest of the D. vulgaris genome (64%).
Insertion site of ISD1-A.
GAPDH is one of the crucial enzymes in the Embden-Meyerhof-Parmas pathway of glycolysis. Desulfovibrio spp., with the exception of D. fructosovorans (13, 29), cannot derive energy for growth from the fermentation of hexoses, indicating the absence of a complete glycolytic pathway. However, D. vulgaris Hildenborough and many other Desulfovibrio spp. can oxidize glycerol to acetate and CO2 through the action of a glycerol kinase, a membrane-bound glycerol-3-phosphate dehydrogenase, and glycolytic pathway enzymes to convert dihydroxyacetone phosphate to pyruvate (13). These include GAPDH, phosphoglycerate kinase (PGK), and triosephosphate isomerase (TPI). This represents the only route in Desulfovibrio spp. known to result in NADH production (37). The pgk and tpi genes of several bacteria, including Bacillus megaterium (31) and Corynebacterium glutamicum (8), are located downstream from the gap gene in the same operon. Assuming that this same arrangement of genes is present in the gap operon in D. vulgaris, the presence of an ISD1 element downstream from the gap gene suggest that the pgk and tpi genes have been inactivated by ISD1 insertion. This idea is supported by the observation that the limited stretch of nucleotide sequence determined on the 5′ side of ISD1-A (Fig. 5A, pgk), encodes a peptide that aligns with the central part of the sequence of known PGKs (not shown). The genes surrounding ISD1-A may thus not encode enzymes that are currently involved in glycerol metabolism in D. vulgaris Hildenborough. An intriguing hypothesis is that these genes formerly provided the organism with the capacity to derive energy for growth from the glycolysis of hexoses, but that this capacity has been lost through gene inactivation (as documented here) and deletion allowing only products of glycolysis (pyruvate, lactate) supplied externally to be used as electron donors for sulfate reduction.
Potential of ISD1 as a tool for mutagenesis of D. vulgaris.
Wall et al. (44) have recently shown that a modified Tn7 can be used for random mutagenesis of D. desulfuricans G20 at a frequency of 10−6 per donor. Tn5 derivatives were also found to transpose at similar frequency following conjugation from E. coli. These transposons may also be useful for random mutagenesis of D. vulgaris Hildenborough, although initial experiments in this regard have been unsuccessful. The fact that ISD1 is actively transposing to a frequently occurring site (AAXX) offers another route toward random mutagenesis by providing a starting point for construction of artificial transposons (38) for random mutagenesis of genes from D. vulgaris Hildenborough and other Desulfovibrio spp.
ACKNOWLEDGMENTS
This work was supported by a research grant from the Natural Science and Engineering Research Council of Canada to G.V. and by a postgraduate studentship from the Alberta Heritage Foundation for Medical Research to R.F.
We thank M. Jiang for technical assistance.
REFERENCES
- 1.Bennett P M. Transposable elements and transposition in bacteria. In: Streips U N, Yasbin R E, editors. Modern microbial genetics. New York, N.Y: Wiley-Liss; 1991. pp. 323–364. [Google Scholar]
- 2.Biesecker G, Harris J I, Thierry J C, Walker J E, Wonacott A J. Sequence and structure of d-glyceraldehyde 3-phosphate dehydrogenase from Bacillus stearothermophilus. Nature (London) 1977;266:328–333. doi: 10.1038/266328a0. [DOI] [PubMed] [Google Scholar]
- 3.Chandler M, Fayet O. Translational frameshifting in the control of transposition in bacteria. Mol Microbiol. 1993;7:497–503. doi: 10.1111/j.1365-2958.1993.tb01140.x. [DOI] [PubMed] [Google Scholar]
- 4.Davis M A, Simons R W, Kleckner N. Tn10 protects itself at two levels from fortuitous activation by external promoters. Cell. 1985;43:379–387. doi: 10.1016/0092-8674(85)90043-1. [DOI] [PubMed] [Google Scholar]
- 5.Deckers H M, Voordouw G. Identification of a large family of genes for putative chemoreceptor proteins in an ordered library of the Desulfovibrio vulgaris Hildenborough genome. J Bacteriol. 1994;176:351–358. doi: 10.1128/jb.176.2.351-358.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng W, Gordon M P, Nester E W. Sequence and distribution of IS1312: evidence for horizontal DNA transfer from Rhizobium meliloti to Agrobacterium tumefaciens. J Bacteriol. 1995;177:2554–2559. doi: 10.1128/jb.177.9.2554-2559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doak T G, Doerder F P, Jahn C L, Herrick G. A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common “D35E” motif. Proc Natl Acad Sci USA. 1994;91:942–946. doi: 10.1073/pnas.91.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eikmanns B J. Identification, sequence analysis, and expression of a Corynebacterium glutamicum gene cluster encoding the three glycolytic enzymes glyceraldehyde-3-phosphate dehydrogenase, 3-phosphoglycerate kinase, and triosephosphate isomerase. J Bacteriol. 1992;174:6076–6086. doi: 10.1128/jb.174.19.6076-6086.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fayet O, Ramond P, Polard P, Prere M F, Chandler M. Functional similarities between retroviruses and the IS3 family of bacterial insertion sequences? Mol Microbiol. 1990;4:1771–1777. doi: 10.1111/j.1365-2958.1990.tb00555.x. [DOI] [PubMed] [Google Scholar]
- 10.Fu R, Voordouw G. Targeted gene-replacement mutagenesis of dcrA, encoding an oxygen sensor of the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Microbiology. 1997;143:1815–1826. doi: 10.1099/00221287-143-6-1815. [DOI] [PubMed] [Google Scholar]
- 11.Galas D J, Chandler M. Bacterial insertion sequences. In: Berg D M, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 109–162. [Google Scholar]
- 12.Genetics Computer Group. Program manual for the Wisconsin package, version 8. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 13.Hansen T A. Carbon metabolism of sulfate-reducing bacteria. In: Odom J M, Singleton R Jr, editors. The sulfate-reducing bacteria: contemporary perspectives. New York, N.Y: Springer-Verlag; 1993. pp. 21–40. [Google Scholar]
- 14.Iida S, Meyer J, Arber W. Prokaryotic IS elements. In: Shapiro J A, editor. Mobile genetic elements. New York, N.Y: Academic Press; 1983. pp. 159–221. [Google Scholar]
- 15.Kearney B, Staskawicz B J. Characterization of IS476 and its role in bacterial spot disease of tomato and pepper. J Bacteriol. 1990;172:143–148. doi: 10.1128/jb.172.1.143-148.1990. . (Erratum, 172:2199.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krebs M P, Reznikoff W S. Transcriptional and translational initiation sites of IS50. Control of transposase and inhibitor expression. J Mol Biol. 1986;192:781–791. doi: 10.1016/0022-2836(86)90028-8. [DOI] [PubMed] [Google Scholar]
- 17.Kremer D A, Hansen T A. Glycerol and dihydroxyacetone dissimilation in Desulfovibrio strains. Arch Microbiol. 1987;147:249–256. [Google Scholar]
- 18.Kulkosky J, Jones K S, Katz R A, Mack J P G, Skalka A M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohta N, Mullin D A, Tarleton J, Ely B, Newton A. Identification, distribution, and sequence analysis of new insertion elements in Caulobacter crescentus. J Bacteriol. 1990;172:236–242. doi: 10.1128/jb.172.1.236-242.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polard P, Chandler M. Bacterial transposases and retroviral integrases. Mol Microbiol. 1995;15:13–23. doi: 10.1111/j.1365-2958.1995.tb02217.x. [DOI] [PubMed] [Google Scholar]
- 21.Polard P, Prere M F, Chandler M, Fayet O. Programmed translational frameshifting and initiation at an AUU codon in gene expression of bacterial insertion sequence IS911. J Mol Biol. 1991;222:465–477. doi: 10.1016/0022-2836(91)90490-w. [DOI] [PubMed] [Google Scholar]
- 22.Polard P, Prere M F, Fayet O, Chandler M. Transposase-induced excision and circularization of the bacterial insertion sequence IS911. EMBO J. 1992;11:5079–5090. doi: 10.1002/j.1460-2075.1992.tb05615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Postgate J R. The sulfate-reducing bacteria. 2nd ed. Cambridge, United Kingdom: Cambridge University Press; 1984. [Google Scholar]
- 24.Prère M-F, Chandler M, Fayet O. Transposition in Shigella dysenterae: isolation and analysis of IS911, a new member of the IS3 group of insertion sequences. J Bacteriol. 1990;172:4090–4099. doi: 10.1128/jb.172.7.4090-4099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Priefer U B, Kalinowski J, Rüger B, Heumann W, Pühler A. ISR1, a transposable DNA sequence resident in Rhizobium class IV strains, shows structural characteristics of classical insertion elements. Plasmid. 1989;21:120–128. doi: 10.1016/0147-619x(89)90055-3. [DOI] [PubMed] [Google Scholar]
- 26.Rezsohazy R, Hallet B, Delcour J, Mahillon J. The IS4 family of insertion sequences: evidence for a conserved transposase motif. Mol Microbiol. 1993;9:1283–1295. doi: 10.1111/j.1365-2958.1993.tb01258.x. [DOI] [PubMed] [Google Scholar]
- 27.Rimpiläinen M, Forsberg Å, Wolf-Watz H. A novel protein, LcrQ, involved in the low-calcium response of Yersinia pseudotuberculosis shows extensive homology to YopH. J Bacteriol. 1992;174:3355–3363. doi: 10.1128/jb.174.10.3355-3363.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ronecker H J, Rak B. Genetic organization of insertion element IS2 based on a revised nucleotide sequence. Gene. 1987;59:291–296. doi: 10.1016/0378-1119(87)90337-4. [DOI] [PubMed] [Google Scholar]
- 29.Rousset M, Dermoun Z, Chippaux M, Belaich J P. Marker exchange mutagenesis of the hydN genes in Desulfovibrio fructosovorans. Mol Microbiol. 1991;5:1735–1740. doi: 10.1111/j.1365-2958.1991.tb01922.x. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Schlapfer B S, Zuber H. Cloning and sequencing of the genes encoding glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase and triosephosphate isomerase (gap operon) from mesophilic Bacillus megaturium: comparison with corresponding genes from thermophilic Bacillus stearothermophilus. Gene. 1992;122:53–62. doi: 10.1016/0378-1119(92)90031-j. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz E, Kroger M, Rak B. IS150: distribution, nucleotide sequence and phylogenetic relationships of a new E. coli insertion element. Nucleic Acids Res. 1988;16:6789–6802. doi: 10.1093/nar/16.14.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schweizer H P. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol. 1992;6:1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 34.Sekine Y, Eisaki N, Ohtsubo E. Translational control in production of transposase and in transposition of insertion sequence IS3. J Mol Biol. 1994;235:1406–1420. doi: 10.1006/jmbi.1994.1097. [DOI] [PubMed] [Google Scholar]
- 35.Skarzynsky T, Moody P C E, Wonacott A J. Structure of holo-glyceraldehyde-3-phosphate dehydrogenase from Bacillus stearothermophilus at 1.8 Å resolution. J Mol Biol. 1987;193:171–187. doi: 10.1016/0022-2836(87)90635-8. [DOI] [PubMed] [Google Scholar]
- 36.Steibl H D, Lewecke F M. IS1222: analysis and distribution of a new insertion sequence in Enterobacter agglomerans 339. Gene. 1995;156:37–42. doi: 10.1016/0378-1119(95)00003-o. [DOI] [PubMed] [Google Scholar]
- 37.Stokkermans J P. Molecular studies on iron-sulfur proteins in Desulfovibrio. Ph.D. thesis. Wageningen, The Netherlands: Agricultural University; 1993. [Google Scholar]
- 38.Vertes A A, Asai Y, Kobayashi M, Kurusu Y, Yukawa H. Transposon mutagenesis of coryneform bacteria. Mol Gen Genet. 1994;245:397–405. doi: 10.1007/BF00302251. [DOI] [PubMed] [Google Scholar]
- 39.Vögele K, Schwartz E, Welz C, Schiltz E, Rak B. High-level ribosomal frameshifting directs the synthesis of IS150 gene products. Nucleic Acids Res. 1991;19:4377–4385. doi: 10.1093/nar/19.16.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voordouw G. Cloning of genes encoding redox proteins of known amino acid sequence from a library of the Desulfovibrio vulgaris (Hildenborough) genome. Gene. 1988;69:75–83. doi: 10.1016/0378-1119(88)90010-8. [DOI] [PubMed] [Google Scholar]
- 41.Voordouw G. Molecular biology of the sulfate-reducing bacteria. In: Odom J M, Singleton R Jr, editors. The sulfate-reducing bacteria: contemporary perspectives. New York, N.Y: Springer-Verlag; 1993. pp. 88–130. [Google Scholar]
- 42.Voordouw G. The genus Desulfovibrio: the centennial. Appl Environ Microbiol. 1995;61:2813–2819. doi: 10.1128/aem.61.8.2813-2819.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wall J D. Genetics of the sulfate-reducing bacteria. In: Odom J M, Singleton R Jr, editors. The sulfate-reducing bacteria: contemporary perspectives. New York, N.Y: Springer-Verlag; 1993. pp. 77–87. [Google Scholar]
- 44.Wall J D, Murnan T, Argyle J, English R S, Rapp-Giles B J. Transposon mutagenesis in Desulfovibrio desulfuricans: development of a random mutagenesis tool from Tn7. Appl Environ Microbiol. 1996;62:3762–3767. doi: 10.1128/aem.62.10.3762-3767.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watson R J, Wheatcroft R. Nucleotide sequence of Rhizobium meliloti insertion sequence ISRm1: homology to IS2 from Escherichia coli and IS426 from Agrobacterium tumefaciens. DNA Seq. 1991;2:163–172. doi: 10.3109/10425179109039686. [DOI] [PubMed] [Google Scholar]
- 46.Wood M S, Byrne A, Lessie T G. IS406 and IS407, two gene-activating insertion sequences from Pseudomonas cepacia. Gene. 1991;105:101–105. doi: 10.1016/0378-1119(91)90519-h. [DOI] [PubMed] [Google Scholar]