Abstract
In Pichia stipitis, fermentative and pyruvate decarboxylase (PDC) activities increase with diminished oxygen rather than in response to fermentable sugars. To better characterize PDC expression and regulation, two genes for PDC (PsPDC1 and PsPDC2) were cloned and sequenced from P. stipitis CBS 6054. Aside from Saccharomyces cerevisiae, from which three PDC genes have been characterized, P. stipitis is the only organism from which multiple genes for PDC have been identified and characterized. PsPDC1 and PsPDC2 have diverged almost as far from one another as they have from the next most closely related known yeast gene. PsPDC1 contains an open reading frame of 1,791 nucleotides encoding 597 amino acids. PsPDC2 contains a reading frame of 1,710 nucleotides encoding 570 amino acids. An 81-nucleotide segment in the middle of the β domain of PsPDC1 codes for a unique segment of 27 amino acids, which may play a role in allosteric regulation. The 5′ regions of both P. stipitis genes include two putative TATA elements that make them similar to the PDC genes from S. cerevisiae, Kluyveromyces marxianus, and Hanseniaspora uvarum.
Pichia stipitis is one of the best-known xylose-fermenting yeasts (5). Besides metabolizing all common monosaccharides, it uses xylan (21) and spent sulfite waste liquors (3) as carbon sources for ethanol production. Wild-type strains of P. stipitis, however, will not ferment xylose at rates or with yields that enable commercial ethanol production from hemicellulosic sugars (17, 24). Xylose fermentation is essential for the economic production of ethanol from angiosperm residue (9). Thus, to better understand xylose fermentation, identify rate-limiting steps, and improve overall ethanol production, we are isolating and altering expression of key genes involved in xylose metabolism (31).
Pyruvate decarboxylase (PDC; EC 4.1.1.1.) is one of the key enzymes involved in the fermentative process. It converts pyruvate to acetaldehyde, which is then reduced to ethanol by alcohol dehydrogenase (for a recent review, see reference 22). PDC is found in microorganisms whose predominant fermentation product is ethanol. Classically, pyruvate is viewed as partitioning between acetyl coenzyme A (leading to respiration) and acetaldehyde (leading to fermentation) through the activities of pyruvate dehydrogenase and PDC, respectively (16). Genetic and allosteric regulation of PDC activity seem to be instrumental in directing metabolite flow. During continuous cultivation of Saccharomyces cerevisiae, fermentative activity is induced in response to glucose, whereas activities of alcohol dehydrogenase, acetaldehyde dehydrogenase, and acetyl coenzyme A synthetase remain unchanged (30). In S. cerevisiae, PDC activity is induced by growth on glucose (28), and its appearance coincides with ethanol production (19). PDC1 appears to be essential for fermentative growth of S. cerevisiae on glucose. Beyond fermentation, however, disruption of all three known PDC genes renders S. cerevisiae unable to grow on glucose in a defined minimal medium even though it can grow on a complex medium (7). This finding indicates that PDC may also serve some essential role for growth on glucose.
PDC genes have been cloned and sequenced from the yeasts S. cerevisiae (10, 12, 13, 19), Kluyveromyces marxianus (14), and Hanseniaspora uvarum (15). In S. cerevisiae, three PDC structural genes, PDC1 (12, 19), PDC5 (13, 29), and PDC6, have been characterized (10, 11). These genes are differently expressed at the transcriptional level (7, 11), and they appear to be under autoregulation because PDC5 is expressed only in PDC1 deletion mutants (13). The PDC gene of K. marxianus (YskPDC1a) and the gene from H. uvarum are very similar to those of S. cerevisiae (14, 15). In P. stipitis, PDC activity is induced as oxygen is restricted (23).
Our objective in this study was to determine the number and nature of the PDC genes in P. stipitis. The results show that P. stipitis PDC1 (PsPDC1) is substantially different in structure from other yeast PDC genes. The two P. stipitis genes have also diverged significantly from one another.
MATERIALS AND METHODS
Strains and plasmids.
P. stipitis CBS 6054 (NRRL Y-11545, ATCC 58785) was the source of all DNA. Escherichia coli DH5α (F− recA1 endA1 hsdR17 [rK− mK+] supE44 thi-1 gyrA relA1) (Gibco BRL, Gaithersburg, Md.) was used for routine recombinant DNA experiments that required a bacterial host. E. coli XL-1 Blue MRF′ (recA mcrA mcrB mrr) and SOLR (Stratagene, La Jolla, Calif.) were used in conjunction with the λ-ZAP genomic DNA library. The PDC1 and PDC5 genes from S. cerevisiae (ScPDC1 and ScPDC5) were kindly provided by S. Hohmann.
Media.
E. coli was routinely cultivated in Luria broth. Yeast strains were routinely cultivated in YPD medium (1% yeast extract, 2% peptone, 2% glucose). Selective medium contained 0.17% yeast nitrogen base without amino acids, with 0.5% ammonium sulfate. Fermentation medium consisted of 0.17% yeast nitrogen base without amino acids and without ammonium sulfate (Difco, Detroit, Mich.), 0.23% urea, 0.66% peptone, and 8% glucose or xylose.
DNA and RNA isolation.
Plasmid DNA was isolated and purified by using a QIAprep Spin Plasmid kit (Qiagen Inc., Chatsworth, Calif.). Yeast genomic DNA was isolated and purified as described previously (25).
Genomic DNA library.
Genomic DNA was purified from P. stipitis CBS 6054 (wild type), partially digested with Tsp509I, and fractionated by electrophoresis. The 5- to 10-kb DNA fragments were ligated into λ-ZAP (Stratagene) digested with EcoRI. The resultant library has approximately 106 individual, recombinant phages with an average insert size of 5 kb. Assuming that P. stipitis has a genome equivalent to that of S. cerevisiae (14,000 kb/haploid genome), this library has a complexity of 23 genome equivalents.
DNA sequencing.
Nucleotide sequences of PsPDC1 and PsPDC2 were determined by the dideoxy method of Sanger et al. (27), using a Sequenase kit (United States Biochemicals, Cleveland, Ohio).
Enzymes and chemicals.
Restriction enzymes and other DNA modification enzymes were obtained from New England Biolabs (Beverly, Mass.), Stratagene, or Promega Corp. (Madison, Wis.). Reaction conditions were those recommended by the suppliers. SeaKem GTG and SeaPlaque GTG agarose were obtained from FMC BioProducts (Rockville, Md.). Gelase (Epicenter Technology, Madison, Wis.) was used to purify DNA from low-melting-point agarose gels. RNase inhibitor (RNasin) was obtained from Promega.
Southern blot analysis.
Southern transfer by capillary blotting was performed as described by Sambrook et al. (26). Both radioactive (32P) and nonradioactive (Genius nonradioactive system; Boehringer Mannheim Biochemicals, Indianapolis, Ind.) probes were used for hybridizations under moderate conditions (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]–25% formamide at 37°C) and washes (2× SSC at 25°C and 0.5× SSC at 37 to 50°C).
RT-PCR.
Reverse transcription (RT) and subsequent PCR amplifications were performed as described by Kawasaki (18), with the following modifications. Fifty units of reverse transcriptase in 100 μM MgCl2, 20 μM each deoxynucleotide, 50 mM KCl, 10 mM Tris (pH 9.0), 0.1% Triton, 21.8 U of RNasin, 20 mM deoxynucleoside triphosphate, 21 pmol of oligo (dT), and 1 μg of total RNA (DNase treated) in a 20-μl final reaction volume were incubated at 23°C for 10 min and then at 42°C for 45 min. Reaction tubes were heated at 95°C for 10 min and then kept on ice for further use. To start the PCR, 10 μl of 10× reaction buffer, 2 U of Taq DNA polymerase, and 21 pmol of each primer were added to each tube in a final reaction volume of 100 μl, and then reaction mixtures were carried through a standard thermal cycle profile. The first cycle consisted of 94°C for 6 min, 54°C for 2 min, and 72°C for 40 s. This cycle was followed by 35 cycles of 94°C for 1 min, 54°C for 2 min, 72°C for 5 min, and finally 72°C for 15 min.
Sequence analysis and deposition.
BLAST searches (1) were performed on the National Center for Biotechnology Information server. All sequence assembly, alignment, and analysis were performed with the Genetics Computer Group sequence analysis software package (4). Distances were calculated as substitutions per 100 amino acids by using the Kimura method (20) following deletion of gapped regions. The phylogenetic tree was drawn by using the neighbor-joining method.
Nucleotide sequence accession numbers.
The sequences of PsPDC1 and PsPDC2 were deposited in GenBank, and the accession numbers are U75310 and U75311, respectively.
RESULTS
PsPDC1 and PsPDC2 clones.
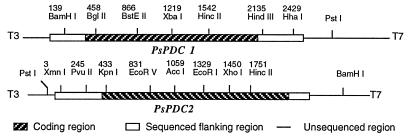
Since other yeast genes for PDC had proven similar to ScPDC1 and ScPDC5 (14, 15), we reasoned that the P. stipitis PDC genes could be cloned through cross-hybridization with the coding sequences of homologous genes from S. cerevisiae. Southern hybridizations with either ScPDC1 or ScPDC5 resulted in the same banding patterns in blots of BamHI-digested genomic DNA of P. stipitis. Only two bands were apparent (data not shown), which suggested that no more than two genes for PDC were closely homologous to the S. cerevisiae genes. ScPDC1 was used to screen a total of 200,000 phage plaques from a λ library of P. stipitis CBS 6054. We identified five individual plaques that strongly cross-hybridized to ScPDC1. Restriction enzyme digestion and Southern hybridization showed that these plaques belonged to two distinct classes. Clones 17 and 18 overlapped to form PsPDC1, while clones 4, 5, and 20 overlapped to form PsPDC2 (Fig. 1). These results likewise suggest that only two close PDC homologs were present.
FIG. 1.
Restriction map of PDC clones from P. stipitis. Restriction sites and approximate lengths of each clone (in kilobases) are indicated. T3 and T7 are adjacent regions of pBluescript KS II. Numbers indicate positions of restriction sites in the sequenced region.
Sequences of PsPDC1 and PsPDC2.
PsPDC1 was sequenced by primer walking from the universal primer T3 located next to the multiple cloning site of clone 17. The 2,534-bp nucleotide sequence of PsPDC1 contains an open reading frame of 1,791 nucleotides (nt) encoding a polypeptide of 597 amino acids. Two putative TATA elements, upstream of the putative AUG start codon, are located at −64 (TAAATATA) and −228 (TATATAAA). The 4.0-kb XhoI fragment containing the 3′-flanking region and partial coding sequence of PsPDC2 was deleted from clone 5, and the religated plasmid was sequenced by primer walking from the universal primer T7. The 2,305-bp sequence of PsPDC2 contains an open reading frame of 1,710 nt encoding a polypeptide of 570 amino acids. Two putative TATA elements are located at −126 (TATAAAT) and at −177 (TATAAT) relative to the start of translation. No obvious cis-acting sequences are apparently shared between the PsPDC genes or with the ScPDC genes. The thiamine binding structural motif which is characteristic of thiamine pyrophosphate binding proteins (8) was present in both of the predicted PsPDC structures.
Characteristics of PsPDC1.
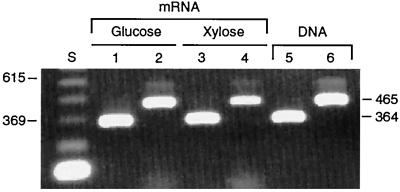
PsPDC1 has an 81-nt segment (coding for 27 amino acids) that is not found in other known PDC genes (Fig. 2). This sequence in P. stipitis corresponds to and replaces amino acid residues 255 to 257 in S. cerevisiae. To determine whether this sequence is actually encoded within the mature mRNA—and is not an intron or cloning artifact—we amplified this segment from mRNA and DNA by using RT-PCR primers specific for each of the two PsPDC genes. We observed bands corresponding to PsPDC1 and PsPDC2 in mRNA from cells grown on both glucose and xylose, and these bands matched those observed from genomic DNA (Fig. 3). These results indicate that the 81-nt segment is indeed transcribed within the PsPDC1 mRNA. BLAST searches (1) against all published sequences in all data banks provided no good matches or clues to its function.
FIG. 2.
Alignment of predicted protein sequences of yeast PDC genes where PsPDC1 contains an additional segment. Accession numbers (from top to bottom): X85968, L09727, X04675, X15668, X55905, U13635, U75301, U75311. Position numbering is based on PsPDC2.
FIG. 3.
Amplification of PsPDC1 mRNA (lanes 1, 3, and 5) and PsPDC2 mRNA (lanes 2, 4, and 6) from cells grown in glucose (lanes 1 and 2) or xylose (lanes 3 and 4) under aerobic conditions and of genomic DNA (lanes 5 and 6) yielded bands of the same size. Lane S is a standard DNA ladder with 123-bp increments.
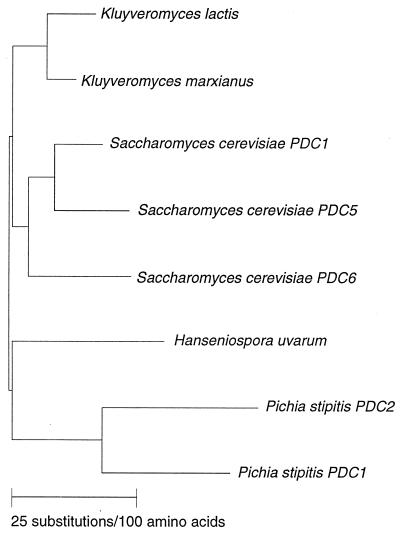
At the amino acid and nucleotide levels, the PsPDC1 gene is 70 and 63%, respectively, identical to the ScPDC1 gene; the PsPDC2 gene is slightly less conserved (68 and 62% identity to ScPDC1), and the PsPDC genes are 72.5 and 70% identical to each other. A phylogenetic analysis of PsPDC1 and PsPDC2 with other yeast PDC genes showed that the two Pichia genes have diverged farther from one another than have the three PDC genes in S. cerevisiae (Fig. 4).
FIG. 4.
Phylogenetic tree showing similarity among yeast PDC genes. Sequences of eight different PDC genes available from GenBank and EMBL data banks were compared by using Genetics Computer Group PileUp, Distances, and GrowTree programs. Source names are shown. Accession numbers are the same as those for Fig. 2.
DISCUSSION
Southern blot analysis indicated that at least two genes in P. stipitis were homologous to the PDC1 and PDC5 genes of S. cerevisiae. Plaque hybridization against a λ library of P. stipitis genomic DNA identified five clones that strongly hybridized to the S. cerevisiae sequence. Restriction analysis and southern blot hybridizations showed that these clones belonged to two overlapping groups, which we designated PsPDC1 and PsPDC2. Examination of the 5′ flanking region revealed no obvious cis-acting sequences that could indicate trans-acting factors or environmental conditions that control their expression. We found that the predicted protein of PsPDC1 contains a unique 27-amino-acid insertion relative to other known PDC proteins.
The fact that the two P. stipitis proteins diverged almost as far from one another as they did from other known yeast PDC sequences suggests that they may play different roles in P. stipitis metabolism. Aside from the three structural PDC genes from S. cerevisiae, this is the only organism from which multiple genes for PDC have been identified and characterized. The 5′ regions of PsPDC1 and PsPDC2 include two putative TATA elements, which make them similar to the PDC genes from S. cerevisiae (19), K. marxianus, and H. uvarum (15). Determination of the significance of potential cis-acting sequences in the 5′ region must await further sequence analysis and regulatory studies.
The predicted primary amino acid sequence of PsPDC1 differs substantially from ScPDC1 in only three places. The most intriguing change is in the β domain with the previously mentioned 27-amino-acid insertion (amino acids 264 to 290) at amino acid residue 255 in ScPDC1p. Based on the three-dimensional structure of PDC from S. cerevisiae (2) and Saccharomyces uvarum (6), this loop could form an amphipathic α helix, based on hydrophobicity and probability, on the surface of the molecule. The new structure is very close in space to Cys221, which is known to be important for substrate activation in the tertiary structure (32). It is reasonable to suspect that the 27-amino-acid extension may be important for allosteric regulation of the molecule, especially in light of the different kinetic regulation of P. stipitis and S. cerevisiae PDC activities (23). This hypothesis could be directly tested by deletion of the 27-amino-acid insert by site-specific mutagenesis and expression, by either targeted gene replacement in P. stipitis or heterologous expression in PDC mutants of S. cerevisiae.
ACKNOWLEDGMENTS
P.L. and B.P.D. were supported by National Renewable Energy Laboratory subcontract XAU-4-1193-02 and by USDA NRICGP grant 96-35500-3172.
We thank U. Klinner, Reinisch-Westfälisch Technische Hochschule, for useful comments. The ScPDC1 and ScPDC5 clones from S. Hohmann were greatly appreciated.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Arjunan P, Umland T, Dyda F, Swaminathan S, Furey W, Sax M, Farrenkopf B, Gao Y, Zhang D, Jordon F. Crystal structure of the thiamin diphosphate-dependent enzyme pyruvate decarboxylase from the yeast Saccharomyces cerevisiae at 2.3 Å resolution. J Mol Biol. 1996;256:590–600. doi: 10.1006/jmbi.1996.0111. [DOI] [PubMed] [Google Scholar]
- 3.Bjorling T, Lindman B. Evaluation of xylose-fermenting yeasts for ethanol production from spent sulfite liquor. Enzyme Microb Technol. 1989;11:240–246. [Google Scholar]
- 4.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du Preez J C, Bosch M, Prior B A. Xylose fermentation by Candida shehatae and Pichia stipitis: effects of pH, temperature and substrate concentration. Enzyme Microb Technol. 1986;8:360–364. [Google Scholar]
- 6.Dyda F, Furey W, Swaminathan S, Sax M, Farrenkopf B, Jordon F. Catalytic centers in the thiamin diphosphate-dependent enzyme pyruvate decarboxylase at 2.4 Å resolution. Biochemistry. 1993;32:6165–6170. doi: 10.1021/bi00075a008. [DOI] [PubMed] [Google Scholar]
- 7.Flikweert M T, van der Zanden L, Janssen W M T M, Steensma H Y, van Dijken J P, Pronk J T. Pyruvate decarboxylase: an indispensible enzyme for growth of Saccharomyces cerevisiae on glucose. Yeast. 1996;12:247–257. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C247::AID-YEA911%3E3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 8.Hawkins C F, Borges A, Perham R N. A common structure motif in thiamin pyrophosphate-binding enzymes. FEBS Lett. 1989;255:77–82. doi: 10.1016/0014-5793(89)81064-6. [DOI] [PubMed] [Google Scholar]
- 9.Hinman N D, Wright D J, Hoagland W, Wyman C E. Xylose fermentation, an economic analysis. Appl Biochem Biotechnol. 1989;28/29:369–375. [Google Scholar]
- 10.Hohmann S. Characterization of PDC6, a third structural gene for pyruvate decarboxylase in Saccharomyces cerevisiae. J Bacteriol. 1991;173:7963–7969. doi: 10.1128/jb.173.24.7963-7969.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hohmann S. PDC6, a weakly expressed pyruvate decarboxylase gene from yeast, is activated when placed spontaneously under the control of the PDC1 promoter. Curr Genet. 1991;20:373–378. doi: 10.1007/BF00317064. [DOI] [PubMed] [Google Scholar]
- 12.Hohmann S. Characterisation of PDC2, a gene necessary for high level expression of pyruvate decarboxylase structural genes PDC1 and PDC5. Eur J Biochem. 1993;241:657–666. doi: 10.1007/BF00279908. [DOI] [PubMed] [Google Scholar]
- 13.Hohmann S, Cederberg H. Autoregulation may control the expression of yeast pyruvate decarboxylase structural genes PDC1 and PDC5. Eur J Biochem. 1990;188:615–621. doi: 10.1111/j.1432-1033.1990.tb15442.x. [DOI] [PubMed] [Google Scholar]
- 14.Holloway P, Subden R E. The isolation and nucleotide sequence of the pyruvate decarboxylase gene from Kluyveromyces marxianus. Curr Genet. 1993;24:274–277. doi: 10.1007/BF00351804. [DOI] [PubMed] [Google Scholar]
- 15.Holloway P, Subden R E. The nucleotide sequence and initial characterization of pyruvate decarboxylase from the yeast Hanseniaspora uvarum. Yeast. 1994;10:1581–1589. doi: 10.1002/yea.320101207. [DOI] [PubMed] [Google Scholar]
- 16.Holzer H. Regulation of carbohydrate metabolism by enzyme competition. Cold Spring Harbor Q Symp Biol. 1961;26:277–288. doi: 10.1101/sqb.1961.026.01.034. [DOI] [PubMed] [Google Scholar]
- 17.Jeffries T W. Emerging technology for fermenting d-xylose. Trends Biotechnol. 1985;3:208–212. [Google Scholar]
- 18.Kawasaki E S. Amplification of RNA protocols: a guide to methods and applications. New York, N.Y: Academic Press, Inc.; 1990. pp. 21–27. [Google Scholar]
- 19.Kellermann E, Seeboth P G, Hollenberg C P. Analysis of the primary structure and promoter function of a pyruvate decarboxylase gene (PDC1) from Saccharomyces cerevisiae. Nucleic Acids Res. 1986;14:8963–8977. doi: 10.1093/nar/14.22.8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimura M. The neutral theory of molecular evolution. Cambridge, England: Cambridge University Press; 1983. [Google Scholar]
- 21.Lee H, Biely P, Latta R K, Barbosa M F S, Schneider H. Utilization of xylan by yeasts and its conversion to ethanol by a Pichia stipitis strain. Appl Environ Microbiol. 1986;52:320–324. doi: 10.1128/aem.52.2.320-324.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liesen T, Cornelis P, Hollenberg C P, Jeinisch J J. ERA, a novel cis-acting element required for autoregulation and ethanol repression of PDC1 transcription in Saccharomyces cerevisiae. Mol Microbiol. 1996;21:621–632. doi: 10.1111/j.1365-2958.1996.tb02570.x. [DOI] [PubMed] [Google Scholar]
- 23.Passoth V, Zimmermann M, Klinner U. Peculiarities of the regulation of fermentation and respiration in the Crabtree-negative, xylose fermenting yeast Pichia stipitis. Appl Biochem Biotechnol. 1996;57/58:201–212. doi: 10.1007/BF02941701. [DOI] [PubMed] [Google Scholar]
- 24.Prior B A, Kilian S G, Du Preez J C. Fermentation of d-xylose by the yeast Candida shehatae and Pichia stipitis; prospects and problems. Proc Biochem. 1989;24:21–32. [Google Scholar]
- 25.Rose M D, Winston F, Hieter P. Methods in yeast genetics. A laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmitt H D, Ciriacy M, Zimmermann F F. The synthesis of yeast pyruvate decarboxylase is regulated by large variations in messenger RNA level. Mol Gen Genet. 1983;192:247–252. doi: 10.1007/BF00327674. [DOI] [PubMed] [Google Scholar]
- 29.Seeboth P G, Bohnsach K, Hollenberg C P. pdc1 mutants of Saccharomyces cerevisiae give evidence for an additional structural PDC gene; cloning of PDC5, a gene homologous to PDC1. J Bacteriol. 1990;172:678–685. doi: 10.1128/jb.172.2.678-685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Urk H, Voll W S L, Scheffers W A, Van Dijken V P. Transient-state analysis of metabolic fluxes in crabtree-positive and crabtree-negative yeast. Appl Microbiol Biotechnol. 1990;56:281–287. doi: 10.1128/aem.56.1.281-287.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang V W, Marks J A, Davis B P, Jeffries T W. High-efficiency transformation of Pichia stipitis based on its URA3 gene and a homologous autonomous replication sequence, ARS2. Appl Environ Microbiol. 1994;60:4245–4254. doi: 10.1128/aem.60.12.4245-4254.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng X, Farrenkopf B, Hohmann S, Jordon F, Dyda F, Furey W. Role of cysteines in the activation and inactivation of brewer’s yeast pyruvate decarboxylase investigated with a PDC1-PDC6 fusion protein. Biochemistry. 1993;32:2704–2709. doi: 10.1021/bi00061a031. [DOI] [PubMed] [Google Scholar]