Abstract
Green mold disease (causal agent, Trichoderma) has resulted in severe crop losses on mushroom farms worldwide in recent years. We analyzed 160 isolates of Trichoderma from mushroom farms for morphological, cultural, and molecular characteristics and classified these isolates into phenotypic groups. The most common group comprised approximately 40% of the isolates and was identified as a strain of Trichoderma harzianum. This group was consistently recovered from farms with severe green mold disease but not from farms with little or no problem. In addition, the strain identified as the major cause of green mold disease in Ireland and the United Kingdom grouped with these North American isolates in having very similar randomly amplified polymorphic DNA patterns.
Trichoderma species are common contaminants of spawn, compost, and wood in commercial mushroom-growing facilities. Most species are considered indicators of compost quality or horticultural practices (22). In the mid-1980s, extensive crop losses attributed to Trichoderma harzianum occurred on mushroom farms in the British Isles (18). Mushroom compost or casing infected with an aggressive strain of T. harzianum did not produce mushrooms, and the crop loss was proportional to the area infected. Since then, Trichoderma has caused severe problems in many areas in North America, including Ontario, British Columbia, and Pennsylvania (16). This infection has come to be known as green mold disease. Economic losses incurred by growers in North America have not been calculated accurately but have been estimated to be in the tens of millions of dollars.
Aggressive and nonaggressive forms of T. harzianum are found on mushroom farms, often in the same cultivation area (9, 10). The Trichoderma species colonizing mushroom cultivation media are difficult to distinguish from each other. Therefore, a rapid, accurate, and sensitive method for identification of the aggressive strains of T. harzianum is essential to disease management and is a prerequisite for studies of environmental and cultural factors related to disease occurrence. These factors might include the mechanism and site of Trichoderma introduction to a farm, modes of dispersal once the infection is established, horticultural practices which allow or even promote disease, and efficacy of sanitation procedures.
Muthumeenakshi et al. (12) developed randomly amplified polymorphic DNAs (RAPDs) and mitochondrial restriction fragment length polymorphisms (RFLPs) as molecular markers for the identification of T. harzianum isolates from mushroom farms in the British Isles. These markers confirmed the separation of isolates into three groups; the aggressive group was designated TH2. Our objective was to develop molecular identification procedures suitable for North American strains of T. harzianum and to determine the genetic variation in Trichoderma isolates from British Columbia, Alberta, Ontario, and Pennsylvania.
MATERIALS AND METHODS
Sampling and growth.
Samples were collected during farm visits or were received from mushroom growers from May 1994 to March 1995. Most samples were compost or casing materials that were visibly colonized by Trichoderma. These were used as inocula on malt extract agar plates (MEA) (30 g of Difco malt extract and 15 g of agar per liter) containing 0.78 g of streptomycin sulfate (Fisher) per liter. The plates were incubated at room temperature (21). Axenic cultures were established by transferring hyphae which grew out from the inoculum to fresh MEA or MEA plus streptomycin. Representative isolates from the study were deposited in the CCFC culture collection (Agriculture and Agrifood Canada, Ottawa, Ontario). The CCFC accession numbers are 222156 for RAPD group 1 (Tables 1 and 2); 222136 for group 2; 222121 for group 3; 222115 for group 4 T. citrinoviride (Table 3); 222124 for group 4 T. koningii; 222105 for group 5; 222096 for group 6 (both primers); 222123 for group 7; 222144 for group 8 (primer 1) and group 6 (primer 2); and 222110 for group 9. Strains TH1, TH2, and TH3, recovered from farms in Ireland (18), were obtained from the International Mycological Institute for comparison to the North American isolates.
TABLE 1.
Primer 1 RAPD patterns of Trichoderma isolates
| RAPD fragment size (kbp)a for:
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Group 1 | Group 2 | Group 3 | Group 4 | Group 5 | Group 6 | Group 7 | Group 8 | Group 9 |
| 2.80* | 2.70f | 2.10* | 2.40f | 2.10v | 2.50v | 2.50vf | 2.60f | 4.40* |
| 2.20f | 2.40f | 1.80* | 1.80* | 1.30f | 1.80* | 2.10* | 2.40* | 3.60* |
| 1.90f | 2.20v | 1.60vf | 1.40v | 0.74f | 1.60* | 1.70f | 1.70vf | 3.00* |
| 1.20* | 1.60* | 1.50* | 0.91* | 0.44* | 1.30* | 1.30f | 1.10v | 1.70v |
| 1.00* | 1.10v | 1.30* | 0.77vf | 0.26* | 1.10v | 1.10* | 0.91* | 1.50* |
| 0.86* | 1.00* | 1.10vf | 0.68f | 0.86* | 0.86v | 0.84v | 1.30* | |
| 0.68f | 0.68vf | 1.00* | 0.51* | 0.73* | 0.85f | 0.77v | 1.10v | |
| 0.57f | 0.48* | 0.88vf | 0.17v | 0.36* | 0.60f | 0.63f | 0.82* | |
| 0.10f | 0.73vf | 0.42f | 0.15* | 0.66v | ||||
| 0.52* | 0.10* | 0.59v | ||||||
| 0.53v | ||||||||
| 0.47v | ||||||||
| 0.38f | ||||||||
| 0.31f | ||||||||
| 0.28f | ||||||||
Asterisks denote amplification products which were used to define a group. All isolates in the group produced these products. Faint (f) and variable (v) amplification products are noted.
TABLE 2.
Primer 2 RAPD patterns of Trichoderma isolates
| RAPD fragment size (kbp)a for:
| |||||||
|---|---|---|---|---|---|---|---|
| Group 1 | Group 2/4 | Group 3 | Group 5 | Group 6 | Group 7 | Group 8 | Group 9 |
| 2.70* | 1.80vf | 2.50f | 2.50f | 3.80v | 1.90* | 3.00vf | 3.30f |
| 2.00f | 1.70vf | 2.20f | 2.00* | 3.20v | 1.70v | 2.50vf | 2.70f |
| 1.80f | 1.20v | 1.60* | 1.80* | 2.70* | 1.40* | 2.20* | 2.40vf |
| 1.40* | 0.93* | 1.30* | 1.60* | 2.10* | 1.10v | 1.80* | 2.30f |
| 0.92* | 0.81* | 1.20v | 1.50* | 1.80v | 1.00v | 1.60* | 1.90v |
| 0.41* | 0.70vf | 1.00* | 1.30* | 1.70v | 0.68* | 1.00* | 1.70f* |
| 0.64vf | 0.91vf | 1.20* | 1.50v | 0.83* | 1.50* | ||
| 0.82vf | 1.10* | 1.00v | 0.72* | 1.40* | |||
| 0.71f | 0.91* | 0.95v | 0.65* | 1.20f | |||
| 0.47f | 0.66* | 0.61v | 0.46* | 1.10* | |||
| 0.57* | 0.91f | ||||||
| 0.79f | |||||||
| 0.68f | |||||||
| 0.53* | |||||||
| 0.46f | |||||||
Asterisks denote amplification products which were used to define a group. Faint (f) and variable (v) amplification products are noted.
TABLE 3.
Characteristics of Trichoderma isolates from North American mushroom farmsa
| Species | T. harzianum morphologyb | RAPD groupc
|
ITSd type | Plasmid typee | RFLP size (kbp)f | |
|---|---|---|---|---|---|---|
| Primer 1 | Primer 2 | |||||
| T. harzianum | A (2/49) | 1 | 1 (62/62) | 1 (45/45) | L, S (6/62) | 6.5 (32/34) |
| B (46/49) | L (4/62) | 7.0 (1/34) | ||||
| C (1/49) | S (26/62) | NH (1/34) | ||||
| ND (26/62) | ||||||
| T. harzianum | A (19/21) | 2 | 2/4 (24/24) | 1 (21/21) | L, S (2/24) | 5.0 (3/4) |
| C (2/21) | S (1/24) | NH (1/4) | ||||
| ND (21/24) | ||||||
| T. harzianum | A (5/6) | 3 | 3 (12/12) | 1 (7/7) | S (5/12) | 7.0 (2/4) |
| C (1/6) | ND (7/12) | 8.0 (1/4) | ||||
| 6.5 (1/4) | ||||||
| T. citrinoviride | 4 | 2/4 (5/5) | 2 (5/5) | ND (5/5) | NT | |
| T. koningii | 4 | 2/4 (10/10) | 3 (9/9) | S (1/10) | NH (1/1) | |
| ND (1/10) | ||||||
| T. koningii | 5 | 5 (8/8) | 3 (7/7) | ND (8/8) | NH (2/2) | |
| T. atroviride | 6 or 8 | 6 or 8 (15/15) | 3 (13/13) | ND (15/15) | NH (1/1) | |
| T. longibrachiatum | 7 | 7 (6/6) | 2 (6/6) | ND (6/6) | NH (2/2) | |
| T. longibrachiatum | 9 | 9 (4/4) | 2 (4/4) | ND (4/4) | NT | |
| Other | A (1/2) | Many | Many (14/14) | 1 (10/14) | ND (14/14) | NT |
| C (1/2) | 2 (2/14) | |||||
| 3 (2/14) | ||||||
Information pertinent to each Trichoderma species or species subgroup is summarized under the column headings. The number of isolates with the reported phenotype out of the total number examined for each characteristic is in parentheses. For example, 46 of 49 isolates in the first T. harzianum subgroup showed morphological phenotype A; the rest showed phenotypes B and C. All 62 isolates were grouped by RAPD phenotypes with primers 1 and 2. All 45 tested isolates showed ITS1–4 phenotype 1. Plasmids were observed in 36 of 62 isolates with some variation in the type of plasmid present. The cloned DNA fragment hybridized to a 6.5-kbp fragment in 32 of 34 isolates.
Three distinct morphologies termed phenotypes A, B and C were noted for T. harzianum isolates. Characteristics are given in Results.
BamHI digestion of the ITS1–4 amplification product produced three phenotypes: type 1 did not digest and was observed as a 700-bp fragment, type 2 was characterized by 560- and 140-bp fragments, and type 3 was characterized by 500- and 200-bp fragments.
Plasmids were characterized as large (L, 7.5 to 8 kbp of supercoiled DNA) or small (S, 2.7 kbp of double-stranded RNA). ND, no plasmids detected.
The size of the DNA fragment hybridizing to an anonymous cloned fragment from T. harzianum isolate T97 is given in kilobase pairs. NH, no hybridization; NT, not tested.
Morphological observations and identification.
Morphological observations were made from cultures grown on 2% MEA at about 24°C under ambient laboratory conditions of diffuse daylight. The microscopic characteristics were observed for the more complex conidiophores developing from the characteristic tufted or pustulate areas of conidiation, usually 3 to 5 days after inoculation. Identifications were performed by using the identification keys provided by Rifai (15) and Bissett (1, 3, 4) and confirmed by comparison with cultures of ex type strains from the CCFC.
DNA isolation.
Fungal cultures were grown at room temperature in 2% malt extract broth for 2 to 4 days. Hyphae were collected on cheesecloth in a Buchner funnel and washed with 25 mM EDTA followed by distilled water. The samples were frozen in liquid nitrogen and lyophilized.
For most analytical procedures, 50 to 100 μg of lyophilized material was placed in a microcentrifuge tube with 500 μl of 10 mM EDTA–10 mM Tris (pH 8)–0.5% sodium dodecyl sulfate (SDS)–pronase (0.5 mg/ml) (modified from reference 17). Samples were mixed by vortexing and incubated at 37°C for 30 to 45 min. Proteins were extracted by centrifugation (12,500 × g for 1 min) in 25:24:1 (by volume) phenol-chloroform-isoamyl alcohol. Nucleic acids in the aqueous phase were precipitated with 0.25 volume of 7.5 M ammonium acetate and separately with 2.5 volumes of ice-cold ethanol. The samples were centrifuged (12,500 × g for 10 min), and the pellets were washed with 70% ethanol, air dried, and suspended in 100 μl of TE (10 mM Tris [pH 8], 1 mM EDTA). RNase A was added to a final concentration of 10 μg/ml. The samples were incubated at 37°C for 30 min, and the deproteinizing and alcohol precipitation steps were repeated as above. DNA was dissolved in 50 μl of TE and quantified by spectrophotometry.
DNA cloning.
DNA used for cloning was extracted by the procedure of Murray and Thompson (11) and purified by bis-benzimide CsCl gradient centrifugation (6). Approximately 30 mg of lyophilized hyphae from T. harzianum isolate T97 was ground to a fine powder and mixed with 1.5 ml of 1% hexadecyltrimethylammonium bromide (CTAB)–0.7 M NaCl–50 mM Tris (pH 8)–10 mM EDTA–1% 2-mercaptoethanol. The sample was incubated at 65°C for 30 min and extracted with 24:1 (vol/vol) chloroform-isoamyl alcohol. After centrifugation, the aqueous phase was mixed with an equal volume of 1% CTAB–50 mM Tris (pH 8)–10 mM EDTA, and the nucleic acid was allowed to precipitate at room temperature for 30 min. The precipitate was collected by centrifugation, and the pellet was suspended in 2 ml of TE. The DNA-CTAB complex was dissolved by the addition of 0.2 g of CsCl, and the solution was brought to a density of 1.69 g/ml by the addition of 2.44 g of CsCl. Bis-benzimide was added to a final concentration of 100 μg/ml. The sample was centrifuged overnight at 200,000 × g. Nuclear DNA was dialyzed against TE, ethanol precipitated, and resuspended in TE.
Nuclear DNA was digested with EcoRI. Random fragments were ligated to plasmid vector pUC19 and transformed into Escherichia coli DH5α. Transformants were selected on Luria-Bertani plates containing 50 μg of ampicillin per ml and 50 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml.
PCR analysis.
RAPD characters (20) were developed with primer 1 (CAGCACCCAG) and primer 2 (AGTCAGCCAC) (Operon Technologies primer OPA03). The samples contained 12.5 ng of Trichoderma DNA, 100 μM each deoxynucleoside triphosphate, 1.25 U of Taq polymerase, 1.5 mM MgCl2, 0.4 μM primer, 10 mM Tris (pH 8.3), 50 mM KCl, and 0.001% gelatin in a total reaction volume of 25 μl. The samples were heated to 94°C for 5 min and then subjected to 40 cycles of annealing at 34°C for 1 min, extension at 72°C for 2 min, and denaturation at 94°C for 1 min followed by a final annealing and extension for 10 min.
Ribosomal internal transcribed spacer (ITS) DNA was amplified by similar procedures, except that the annealing temperature was 52°C and 30 cycles were used. The primers were ITS1 and ITS4 (19). Amplified DNAs from 16 isolates were digested with seven restriction enzymes in a search for site polymorphisms.
The DNA samples were separated by electrophoresis on 0.8 to 1.2% agarose gels in 40 mM Tris–20 mM acetic acid–1 mM EDTA (pH 8) (1× TAE). The fragments were visualized by staining with 1 μg of ethidium bromide per ml and UV illumination.
RFLPs.
Genomic DNA samples were digested with EcoRI, and the fragments were separated on 0.8% agarose gels in 1× TAE. After visualization and photography, the gels were treated with 250 mM HCl for 10 min and rinsed briefly with distilled water and then for 30 min with 0.4 N NaOH. The gels were blotted onto a GeneScreen Plus membrane (DuPont, NEN) in 0.4 N NaOH for 16 h (14).
Prior to hybridization, the membrane was incubated with 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–2× Denhardt’s solution–10% dextran sulfate–1% SDS at 65°C for 1 h. An anonymous EcoRI nuclear DNA fragment from T. harzianum isolate T97 was randomly primed and biotin labelled with a kit from New England Biolabs and was hybridized to the membrane. The hybridization conditions were the same as those for the prehybridization, except that denatured probe and sheared salmon sperm DNA (100 μg/ml) were added and incubated with the membrane for 16 h. The membrane was washed once with 2× SSC for 5 min at room temperature and then twice with 2× SSC–1% SDS for 20 min at 65°C. A final wash with 0.1× SSC was carried out at room temperature for 20 min. Chemiluminescence-bound probe was detected with a NEBlot phototope kit (New England Biolabs) as specified by the manufacturer.
RESULTS
Morphological observations.
Eight species were represented in 160 isolates from mushroom farms. T. harzianum, T. koningii, T. atroviride, T. citrinoviride, and T. longibrachiatum were the most common species in this sample. Four isolates were classified as T. crassum, two were classified as T. hamatum, and one was classified as T. spirale. Since the isolates were collected in a nonrandom fashion, emphasizing Trichoderma strains growing on compost and casing material, the sample may not accurately represent all species found on mushroom farms or their relative abundance.
Subtle morphological variation was noted among isolates of some species. Three morphologically discernible phenotypes (designated A, B, and C) were noted for T. harzianum. The first phenotype (A) was characterized by having entirely effuse conidiation after 3 to 4 days of growth on 2% MEA. Compact aggregations of conidiophores and flat conidiogenous pustules developed later. In these respects and in microscopic observations, these strains were morphologically indistinguishable from common strains of T. harzianum isolated from a variety of habitats and from isolates of Hypocrea nigricans, which is a purported teleomorph of T. harzianum. The other two morphological phenotypes had conidiophores aggregated in small tufts after 3 to 4 days. Group B had a less regular branching pattern, with branching being flexuous and frequently dichotomous, whereas group C had a regular verticillate branching pattern with phialides arranged mostly in crowded verticils. Multiple morphological phenotypes were also observed for T. atroviride, T. koningii, and T. longibrachiatum. These morphological differences will be described in more detail elsewhere (4a).
Genetic analysis.
DNA amplification with primer 1 defined nine phenotypic groups (Table 1) representing 146 isolates (see Table 3). Variation within a group was observed as absence of one of the amplified fragments or presence of an extra fragment. The remaining isolates either produced unique RAPD patterns or fit into very small groups of two or three isolates. These isolates are grouped into the last category, “other,” in Table 3.
Amplification with primer 2 defined eight major groups of isolates (Table 2). The categorization of the isolates was largely consistent with the primer 1 grouping, with two exceptions. First, the primer 2 amplification patterns observed with isolates from the second and fourth groups, as defined from primer 1 RAPDs, were too similar to be distinguished with any reliability. Second, different combinations of patterns characteristic of groups 6 and 8 were observed in T. atroviride. For example, some isolates were categorized as group 6 with both of the primers whereas others had the group 6 pattern with primer 1 and the group 8 pattern with primer 2. Three of the four possible combinations were evident.
T. harzianum isolates were divided into three groups on the basis of the RAPD results. Morphological phenotype A corresponded to RAPD groups 2 and 3, and morphological phenotype B corresponded to RAPD group 1. Morphological phenotype C was not clearly defined in the RAPD results, since one isolate showed the RAPD group 1 phenotype, two were RAPD group 2, one was RAPD group 3, and one was classified into the “other” RAPD group. Similarly, two groups of T. koningii and T. longibrachiatum were evident. T. citrinoviride and one of the T. koningii groups had common patterns for the two primers. T. atroviride isolates formed a single group. Other species, including the four T. crassum isolates, each produced unique RAPD patterns.
Amplification of the ITS with primers ITS1 and ITS4 yielded a product estimated by gel electrophoresis to be approximately 700 bp for all isolates. Amplified DNAs from 16 isolates representing the eight groups defined by RAPD patterns were digested with seven restriction enzymes. No digestion was apparent with HindIII, KpnI, BglII, XbaI, or PstI. Single SmaI and EcoRI sites were conserved in all isolates. SmaI digestion produced two fragments of 550 and 125 bp, and two 350-bp fragments were recovered after EcoRI digestion. Digestion with BamHI produced three phenotypes, and this characteristic was used subsequently to analyze a larger sample of isolates (Fig. 1). Some ITS amplification products were not digested (phenotype 1). This phenotype was observed with T. harzianum, T. crassum, T. hamatum, and T. spirale isolates. Isolates of T. citrinoviride and T. longibrachiatum displayed phenotype 2 (two fragments with sizes of approximately 560 and 140 bp). Phenotype 3, with two fragments of 480 and 220 bp, was found with isolates of T. koningii and T. atroviride (see Table 3).
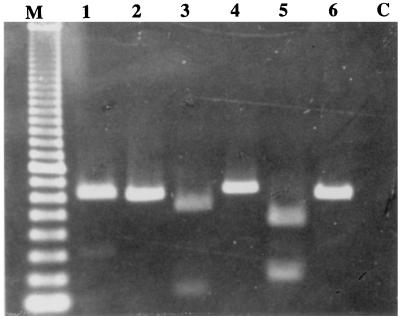
FIG. 1.
Amplification of the ITS region of Trichoderma isolates. DNA sequences flanked by ITS1 and ITS4 primers were amplified by PCR. The products were digested with BamHI, and the fragments were separated by agarose gel electrophoresis. Lane M contains a 100-bp size standard (Pharmacia). Results characteristic of phenotype 1 are observed in lanes 1 (BamHI-digested amplification product) and 2 (undigested). Lanes 3 (digested) and 4 (undigested) show phenotype 2, due to the presence of a single restriction site which resulted in 560- and 140-bp digestion products. Phenotype 3 is seen in lanes 5 (digested) and 6 (undigested). Two fragments, 480 and 220 bp in length, were produced by BamHI digestion. Lane C contains products from a water control reaction in which a DNA template was omitted.
Two plasmids were seen in some isolates when undigested DNA was analyzed by gel electrophoresis. One, with a size of approximately 7.5 to 8 kbp, showed a ladder characteristic of supercoiled DNA. The second type appeared as a single band of about 2.7 kbp. This plasmid was DNase I resistant and RNase A sensitive in 0.03 M NaCl but not in 0.3 M NaCl and therefore is probably a double-stranded RNA (5). Both plasmids were observed in numerous isolates of T. harzianum, and the small plasmid was found in one isolate of T. koningii (see Table 3).
The cloned DNA fragment hybridized to single DNA fragments from 40 of 42 tested T. harzianum isolates from all three RAPD groups (Table 3). Polymorphisms were evident within two of the three T. harzianum groups, and T. harzianum isolates comprising RAPD group 2 were distinguished from those in the other two groups by the presence of a 5-kbp fragment. These polymorphisms were observed in two (in some instances three) repeated hybridization trials. No hybridization was evident with DNAs from six isolates (three T. koningii isolates, two T. longibrachiatum isolates, and one T. atroviride isolate).
Relationship of Irish isolates to North American isolates.
TH2, reported to be the major cause of green mold disease on farms in Ireland and the United Kingdom (12, 18), produced RAPD patterns characteristic of group 1 with both primers and showed ITS1–4 phenotype 1. TH1 yielded RAPD pattern 3 with both primers and showed ITS1–4 phenotype 1. TH1 therefore appeared to be a common T. harzianum isolate similar to those recovered from a variety of habitats. TH3 matched North American RAPD group 6 with both primers and showed ITS1–4 phenotype 3. Based on these criteria, TH3 is an isolate of T. atroviride (data not shown).
Association of isolates with green mold disease.
Green mold disease problems were correlated strongly with the presence of T. harzianum isolates in RAPD group 1. Isolates which fit into this group were consistently found on farms with severe crop damage but not on farms without Trichoderma disease problems. In addition, Trichoderma isolated from large diseased areas on mushroom beds was invariably associated with this group. The match between the Irish aggressive isolate TH2 and RAPD group 1 substantiated the conclusion that North American aggressive green mold strains are in this group.
Genetic variants of aggressive green mold strains.
No variation in the RAPD phenotype was observed among isolates in RAPD group 1. However, differences in the presence of a plasmid were noted; some isolates contained plasmids, whereas others did not. There were also differences in the type of plasmid present and in the RFLP phenotype (Table 3). Five distinct genotypes, as defined by at least one difference, were noted. There appeared to be no correlation between genotype and geographic location. Different genotypes were recovered from a common area, even the same farm, and four of the five genotypes were isolated from widely separated regions, e.g., Pennsylvania and British Columbia.
DISCUSSION
Bissett (2) proposed the organization of the genus Trichoderma into five sections, containing groups of species with similar morphological characteristics. ITS1-4 phenotypes were consistent with this sectional classification. Phenotype 1 was observed in isolates representative of species in the section Pachybasium. These species included T. harzianum, T. crassum, T. spirale, and T. hamatum. The last three species are included in the “other” category in Table 2. Species in the section Longibrachiatum, including T. longibrachiatum and T. citrinoviride, had ITS1-4 phenotype 2. All isolates of T. koningii and T. atroviride, representative of the section Trichoderma, showed phenotype 3. Thus, this molecular characteristic should provide a useful diagnostic tool for placement of a novel isolate into one of these three sections. Representatives of the sections Hypocreanum and Saturnisporum were not recovered in this sample.
The plasmids can be used to separate T. harzianum from other species, since they were observed almost exclusively in T. harzianum. However, numerous T. harzianum isolates lacked plasmids, and isolates in different RAPD groups had similar plasmids. These results indicate that these plasmids are not useful for the identification of T. harzianum RAPD groups. Comparable results were obtained by Meyer (7) on mitochondrial plasmids in group I and group II T. viride isolates. Similarly, the RFLPs did not reliably separate T. harzianum RAPD groups.
Muthumeenakshi et al. (12) developed RAPDs as molecular markers for T. harzianum, including those observed with the A3 primer (primer 2 in this study). Our results confirm their findings. Distinct groups within T. harzianum can be defined with these markers. At the interspecific level, each species was distinguished, with the exception of one group of T. koningii and one of T. citrinoviride, which had common RAPD phenotypes with both primers. This exception emphasizes the need to examine isolates with a diverse array of characteristics, including molecular and morphological attributes.
The characteristics reviewed differentiated T. harzianum isolates that cause green mold disease from the other isolates. Five different genotypes, as defined by reproducible differences in at least one characteristic, were detected in the sample of 62 isolates. No nuclear DNA differences were found in 10 isolates of TH2 (12), the disease-causing form of T. harzianum in Ireland and the United Kingdom. However, in a sample of 36 isolates, five mitochondrial DNA types were observed in this group. In addition, TH2 is distinct from North American isolates of aggressive T. harzianum (13), even though some similarities do exist. Thus, green mold disease is not caused by a single strain.
Seaby (18) proposed that disease-causing T. harzianum in the British Isles was derived from a mutant particularly suited for growth on mushroom compost. Green mold disease could have radiated throughout Ireland and the United Kingdom from this single source. If so, the genetic variation in TH2 isolates, as seen with the mitochondrial polymorphisms, must have arisen through multiple mutational events after the initial mutation promoting compost colonization. A similar series of events could account for the diversity of the North American isolates. An alternate hypothesis is that the narrow range of environmental conditions used to optimize mushroom production provided strong selection for a specific type of T. harzianum from preexisting populations. In other words, genetically distinct forms of aggressive T. harzianum arose from multiple independent sources. This latter hypothesis almost certainly accounts for the differences between the North American and the Irish and British isolates. These two groups arose from at least two different sources. The North American isolates are now referred as TH4 according to the practice developed by Muthumeenakshi and Mills (13).
Whichever hypothesis is correct, the source of aggressive T. harzianum in North America is not the common, cosmopolitan forms observed in this study as RAPD groups 2 and 3. Very similar conclusions were presented by Muthumeenakshi et al. (12); TH2 was not derived from the nonaggressive or less aggressive group, TH1. Searches for possible environmental sources of the aggressive forms of T. harzianum are under way, but at present the origin of these strains is unknown.
These studies, along with the work of Muthumeenakshi et al. (12) and Meyer et al. (8), provide a basis for the quick identification of aggressive forms of T. harzianum. The sensitivity and rapidity of DNA-based identification procedures facilitate the development of diagnostic methods which would indicate the presence of aggressive forms of T. harzianum: (i) on mushroom production surfaces prior to the appearance of disease symptoms, (ii) on possible vectors such as insects or in air samples, and (iii) in sites which might serve as reservoirs between disease outbreaks. This information would allow intervention aimed at controlling green mold disease, thereby reducing crop loss.
ACKNOWLEDGMENTS
We thank Carol Ann Lunam and Ann Pouliot for technical assistance with the morphological studies at ECORC. Hank Taylor and the Canadian Mushroom Growers Association (CMGA) facilitated the collaboration between J.B. and colleagues at HRIO and Brock University. Biolog Inc. (Hayward, Calif.) provided resources for activities at ECORC.
Activities at ECORC were funded in part by a research grant, MII 95-01, from Research Branch, Agriculture and Agri-food Canada. Studies at Brock University and at HRIO were supported by funds from the CMGA, URIF grant BR24-001 from the Ontario Ministry of Education and Training, and a Food Systems 2002 grant from the Ontario Ministry of Agriculture, Food and Rural Affairs.
REFERENCES
- 1.Bissett J. A revision of the genus Trichoderma. I. Section Longibrachiatum sect. nov. Can J Bot. 1984;62:924–931. [Google Scholar]
- 2.Bissett J. A revision of the genus Trichoderma. II. Infrageneric classification. Can J Bot. 1991;69:2357–2372. [Google Scholar]
- 3.Bissett J. A revision of the genus Trichoderma. III. Section Pachybasium. Can J Bot. 1991;69:2373–2417. [Google Scholar]
- 4.Bissett J. A revision of the genus Trichoderma. IV. Additional notes on section Longibrachiatum. Can J Bot. 1991;69:2418–2420. [Google Scholar]
- 4a.Bissett, J. Unpublished data.
- 5.Goodin M M, Schlagnhaufer B, Romaine C P. Encapsidation of LaFrance disease-specific double-stranded RNAs in 36-nm isometric viruslike particles. Phytopathology. 1992;82:285–290. [Google Scholar]
- 6.Hudspeth M E S, Shumard D S, Tatti K M, Grossman L I. Rapid purification of yeast mitochondrial DNA in high yield. Biochim Biophys Acta. 1980;610:221–228. doi: 10.1016/0005-2787(80)90003-9. [DOI] [PubMed] [Google Scholar]
- 7.Meyer R J. Mitochondrial DNAs and plasmids as taxonomic characteristics in Trichoderma viride. Appl Environ Microbiol. 1991;57:2269–2276. doi: 10.1128/aem.57.8.2269-2276.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer W, Lieckfeldt E, Kuhls K, Freedman E Z, Börner T, Mitchell T G. DNA- and PCR-fingerprinting in fungi. In: Pena S D J, Chakraborty R, Epplen J T, Jeffreys A J, editors. DNA fingerprinting: state of the science. Basel, Switzerland: Birkhäuser Verlag; 1993. pp. 311–320. [DOI] [PubMed] [Google Scholar]
- 9.Morris E, Doyle O, Clancy K. A profile of Trichoderma species. I. Mushroom compost production. Mushroom Sci. 1995;14:611–618. [Google Scholar]
- 10.Morris E, Doyle O, Clancy K. A profile of Trichoderma species. II. Mushroom growing units. Mushroom Sci. 1995;14:619–625. [Google Scholar]
- 11.Murray M G, Thompson W F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muthumeenakshi S, Mills P R, Brown A E, Seaby D A. Intraspecific molecular variation among Trichoderma harzianum isolates colonising mushroom compost in the British Isles. Microbiology. 1994;140:769–777. doi: 10.1099/00221287-140-4-769. [DOI] [PubMed] [Google Scholar]
- 13.Muthumeenakshi S, Mills P R. Detection and differentiation of fungal pathogens of Agaricus bisporus. Mushroom Sci. 1995;14:603–610. [Google Scholar]
- 14.Reed K C, Mann D A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985;13:7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rifai M A. A revision of the genus Trichoderma. Mycol Pap. 1969;116:1–56. [Google Scholar]
- 16.Rinker D. Disease management strategies for Trichoderma mould: a summary of seminar by Don Betterley. Mushroom World. 1993;4:3–5. [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 18.Seaby D A. Infection of mushroom compost by Trichoderma species. Mushroom J. 1987;179:355–361. [Google Scholar]
- 19.White T J, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal DNA for phylogenetics. In: Innes M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 315–322. [Google Scholar]
- 20.Williams J G K, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Worrell J J. Media for the selective isolation of Hymenomycetes. Mycologia. 1991;83:296–302. [Google Scholar]
- 22.Wuest P, Bengtson G D. Penn State Handbook for commercial mushroom growers. University Park: The Pennsylvania State University; 1982. [Google Scholar]