Abstract
Pseudomonas sp. strain ADP contains the genes, atzA, -B, and -C, that encode three enzymes which metabolize atrazine to cyanuric acid. Atrazine-catabolizing pure cultures isolated from around the world contain genes homologous to atzA, -B, and -C. The present study was conducted to determine whether the same genes are present in an atrazine-catabolizing bacterial consortium and how the genes and metabolism are subdivided among member species. The consortium contained four or more bacterial species, but two members, Clavibacter michiganese ATZ1 and Pseudomonas sp. strain CN1, collectively mineralized atrazine. C. michiganese ATZ1 released chloride from atrazine, produced hydroxyatrazine, and contained a homolog to the atzA gene that encoded atrazine chlorohydrolase. C. michiganese ATZ1 stoichiometrically metabolized hydroxyatrazine to N-ethylammelide and contained genes homologous to atzB and atzC, suggesting that either a functional AtzB or -C catalyzed N-isopropylamine release from hydroxyatrazine. C. michiganese ATZ1 grew on isopropylamine as its sole carbon and nitrogen source, explaining the ability of the consortium to use atrazine as the sole carbon and nitrogen source. A second consortium member, Pseudomonas sp. strain CN1, metabolized the N-ethylammelide produced by C. michiganese ATZ1 to transiently form cyanuric acid, a reaction catalyzed by AtzC. A gene homologous to the atzC gene of Pseudomonas sp. strain ADP was present, as demonstrated by Southern hybridization and PCR. Pseudomonas sp. strain CN1, but not C. michiganese, metabolized cyanuric acid. The consortium metabolized atrazine faster than did C. michiganese individually. Additionally, the consortium metabolized a much broader set of triazine ring compounds than did previously described pure cultures in which the atzABC genes had been identified. These data begin to elucidate the genetic and metabolic bases of catabolism by multimember consortia.
Bacteria of different genera, existing in close proximity, are thought to aid each other in growth and survival via gene transfer and metabolic cross-feeding. The latter case has been relatively well studied with bacteria that provide amino acids or vitamins to other strains with biosynthetic deficiencies (34). More recently, there has been interest in elucidating microbial metabolic cooperativity that is functional in the catabolism of organic compounds. Many reports of interdependent catabolism involve anaerobic consortia (22, 36). Anaerobic catabolism often provides less energy to bacteria than does corresponding aerobic metabolism and thus requires higher metabolic efficiency, brought about by metabolic cooperativity. In contrast, aerobic catabolism has been elucidated principally with pure cultures studied individually. Aerobic bacterial pure cultures have provided most of the genes and enzymes that comprise the present knowledge base of bacterial catabolism. Studies of microbial catabolism have increasingly focused on the biodegradation of industrial and agricultural chemicals. The latter compounds are largely insecticides and herbicides.
Atrazine is the most widely used s-triazine herbicide; it is utilized globally to control broadleaf weeds. Atrazine has been deployed only over the last 40 years and was previously considered to be nonmetabolizable by the majority of soil bacteria. During the first 35 years of its use, bacterial atrazine catabolism was proposed to occur largely via N-dealkylation reactions, resulting in the accumulation of aminotriazine compounds in both soils and laboratory media (3–5, 11, 20, 21). More recently, pure cultures of bacteria that catabolize atrazine to CO2 have been described (8, 26, 27, 30, 37).
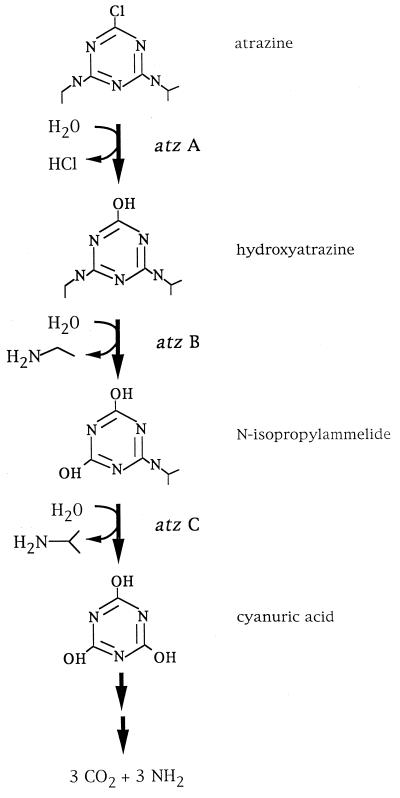
The nearly simultaneous reports of atrazine-mineralizing pure cultures by five research groups (8, 26, 27, 30, 37) after years of unsuccessful efforts suggested a recent evolutionary origin and distribution of atrazine degradation genes. Consistent with this, all of the recently identified atrazine-degrading bacteria, isolated from around the world, have been shown to contain similar genes that encode enzymes which catabolize atrazine to cyanuric acid (16) (see Fig. 1). Cyanuric acid can be used by many soil bacteria as the sole nitrogen source (10–12, 19, 23). The enzymes for atrazine catabolism to cyanuric acid are encoded by the atzABC genes, which are found on a self-transmissible plasmid in Pseudomonas sp. strain ADP, the best characterized atrazine-metabolizing bacterium studied at the molecular level (7, 16, 17, 26, 32). Moreover, multiple insertion sequence-like elements have been identified in DNA flanking the atz genes. These studies are beginning to yield insights into atrazine gene evolution and dispersion.
FIG. 1.
Atrazine catabolic pathway identified in Pseudomonas sp. strain ADP, showing the first three enzymatic reactions encoded by atzABC (7, 17, 32).
These data also provide the tools for investigating bacterial atrazine genes in situ or in microbial consortia cultured in the laboratory on atrazine. For example, an atrazine-catabolizing consortium was reported in 1994 (3), but that predated the identification of catabolic genes and pure cultures which metabolize atrazine to carbon dioxide. More recently, a stable aerobic consortium was obtained from an agricultural soil and characterized with respect to its ability to catabolize atrazine (1, 2).
The present study was conducted to determine whether the genes and metabolism of the consortium (1, 2) resembled those found in recently described atrazine-metabolizing pure cultures. Our results show that different consortium members separately contained the atzA, -B, and -C genes. Coupled with biochemical studies, this revealed the interspecies metabolic interactions relevant to atrazine catabolism by the consortium. Our findings begin to provide a framework for understanding how catabolic pathways may evolve and the different conditions under which pure-culture or consortial metabolism may be selected for during the global recycling of organic matter.
MATERIALS AND METHODS
Chemicals.
From the following sources, we obtained the chemicals indicated (the purity is given parenthetically): ChemServices (West Chester, Pa.), atrazine (98%); and Novartis (Greensboro, N.C.), simazine (97.3%), prometone (99.1%), ametryn (98.2%), simetryn (98.5%), prometryn (99.5%), atratone (98.9%) propazine (97.5%), hydroxyatrazine (97.8%), N-ethylammelide (95%), and N-isopropylammelide (97%). Cyanuric acid was obtained from Aldrich Chemical Co. (Milwaukee, Wis.). Uniformly ring-labeled [14C]atrazine (7.8 mCi/mmol) was purchased from Sigma Chemical Company (St. Louis, Mo.) and had a purity of >98%.
Bacteria and growth media.
The consortium studied in this work was previously isolated by enrichment from an agricultural soil which had a 15-year history of atrazine application (1, 2). The consortium consisted of a Clavibacter strain, a Pseudomonas strain, and at least two other unidentified bacterial strains (2, 15). It was maintained and grown on mineral salts (MS) medium containing glucose (1,000 mg/liter) as a carbon source and atrazine (100 mg/liter) as the sole source of nitrogen. An atrazine concentration of 100 mg/liter was used in all experiments unless indicated otherwise. MS medium consisted of 10 mM K2HPO4, 3 mM NaH2PO4, 1 mM MgSO4, and 10 ml of chloride-free trace element stock solution, which contained the following (in milligrams per liter); CaSO4, 200; FeSO4 · 7H2O, 200; MnSO4 · H2O, 20; NaMoO4 · 2H2O, 10; CuSO4, 20; CoSO4 · 7H2O, 10; and H3BO3, 2. In experiments with other s-triazine compounds and their metabolites, compounds were substituted for atrazine at the same concentration. The degradation of some s-triazines was also screened on MS medium containing Noble agar by a plate clearing-zone assay as previously described (18, 26), with atrazine and other s-triazines at concentrations of 500 mg/liter. All the s-triazines used in this study were stable during autoclaving, based on high-pressure liquid chromatography (HPLC) analysis, except for cyanuric acid, which was added as a filter-sterilized solution after autoclaving.
Growth and metabolism experiments.
Cells were grown in 50 ml of MS-atrazine medium containing uniformly ring-labeled [14C]atrazine at a concentration of 10,000 dpm ml−1 in 250-ml Erlenmeyer flasks equipped with NaOH traps for 14CO2 collection (2). Flasks were incubated at room temperature on a rotary shaker at 200 rpm. Precultures were grown in nonradiolabeled MS-atrazine medium for 4 days and inoculated into radiolabeled MS-atrazine medium by transfers of 10% culture volumes to new flasks. Cell growth was monitored by measuring absorbance at 600 nm with a Lambda 2 UV-visible light spectrometer (Perkin-Elmer Corp., Norwalk, Conn.). Atrazine and metabolites were determined by HPLC as described below. The mineralization of atrazine was determined by quantification of 14CO2 trapped in 2 N NaOH. The radioactivities in samples were measured with an LS 5000 TD liquid scintillation counter (Beckman Instruments, Irvine, Calif.) after the addition of 15 ml of Liquiscint liquid scintillation cocktail (National Diagnostics, Atlanta, Ga.). Measurements of [14C]atrazine mineralization and metabolite formation were done at 0, 6, 12, 24, 36, and 48 h after inoculation.
Resting-cell assays.
Cells were pregrown in MS-atrazine medium with atrazine at 250 mg liter−1 or in a control medium with atrazine at the same concentration and 10 mM NH4NO3. Cells were incubated for 2 days or until all the atrazine was removed from the solution, as determined by HPLC analysis. Cells were washed three times in phosphate-buffered saline (8.5 g of NaCl per liter, 0.3 g of KH2PO4 per liter, and 0.6 g of Na2HPO4 per liter [pH 7]) and centrifuged at 14,000 × g for 10 min. Washed cells were resuspended in phosphate-buffered saline containing atrazine at 33 mg liter−1 with 30,000 dpm of [14C]atrazine ml−1. The cell suspension had an A600 of 2.4, which corresponded to a protein concentration of 1.2 mg ml−1. One-milliliter aliquots were collected at 5, 20, 40, 60, 90, 120, 180, and 240 min and at 12 h after the addition of radiolabeled atrazine. Resting cells were removed from the solution by centrifugation, and the supernatant was analyzed for total radioactivity by liquid scintillation counting and for the remaining atrazine content and metabolite formation by HPLC. Identical suspensions were used to measure 14CO2 evolution in separate flasks equipped with NaOH traps. Samples were taken at 30, 60, 90, 120, 180, and 240 min and at 12 h after the addition of radiolabeled atrazine. Resting-cell cultures were incubated under stationary conditions at room temperature.
DNA manipulations.
Total genomic and plasmid DNAs were isolated as previously described (33). The nucleotide sequence was determined from both strands by using a PRISM ready reaction dyedeoxy terminator cycle sequencing kit (Perkin-Elmer Corp.) and a DNA sequencer (model 373A; Applied Biosystems, Foster City, Calif.). The Genetics Computer Group (Madison, Wis.) sequence analysis software package was used for all DNA and protein sequence comparisons and alignments. Southern blotting and hybridizations were performed as previously described (33). A 0.6-kb ApaI/PstI fragment from pMD4 (18), a 1.5-kb BglII fragment from pATZB-2 (7), and a 2.0-kb EcoRI/AvaI fragment from pTD2.5 (32) were used as probes for atzA, -B, and -C sequences as previously described (16). DNA probes were labeled with [α-32P]dCTP by using a Rediprime random primer labeling kit (Amersham Life Science, Arlington Heights, Ill.) according to the manufacturer’s instructions. The PRISM ready reaction dyedeoxy terminator cycle sequencing kit protocol (no. 401388, revision B; Perkin-Elmer Corp.) was used to prepare DNA samples for sequence analysis on a DNA sequencer (model 373A; Applied Biosystems).
PCR analysis and primer sequences.
Total genomic or plasmid DNA (500 ng) was used as the template for PCR. PCR-generated fragments were amplified with Taq DNA polymerase (Gibco BRL, Gaithersburg, Md.) (33), separated from primers on a 1.0% agarose gel, and purified from the gel slice by using the Wizard DNA cleanup system (Promega, Madison, Wis.). Custom primers were designed specifically for atzB (5′GTTGAGGTGAACTG3′ and (5′AGACTCGACGAAGGTT3′) and for atzA and atzC (16) by using the Primer Designer package (version 2.01; Scientific and Educational Software, State Line, Pa.). Primers were synthesized by Gibco BRL. To sequence the atzA and atzC PCR products, the 0.5- and 0.6-kb PCR products, respectively, were purified from gels by using the Geneclean II system (Bio 101 Inc., Vista, Calif.). The nucleotide sequences of PCR products were obtained as described above.
Preparation of whole cells and crude cell extracts.
Overnight cultures (1.5 liters) of Clavibacter michiganese, Pseudomonas sp. strain CN1, and the consortium were centrifuged at 12,000 × g for 10 min at 4°C. Cell pellets were washed twice with 25 mM MOPS (morpholinepropanesulfonic acid) buffer (pH 6.9) and resuspended in the same buffer on ice. Cold cell suspensions, prepared as described above, were subjected to three consecutive freeze-thaw cycles, followed by sonication with a Biosonik sonicator (Bronwill Scientific, Rochester, N.Y.). Sonication was carried out three times for 30 s each at 80% probe intensity, with intermittent cooling on ice. Sonicated cell suspensions were centrifuged at 12,000 × g for 10 min at 4°C to obtain protein extracts.
Analytical methods.
The HPLC method used for simultaneous analysis of atrazine and its metabolites was adapted from that of Rustum et al. (31) and employed UV detection at 215 and 230 nm after separation by reverse-phase chromatography on a C8 column. A 1050 series high-pressure liquid chromatograph (Hewlett-Packard Co., Fullerton, Calif.) was used for all separations. The solvents used were acetonitrile and 0.01 M KH2PO4 (pH 2.0), with a mobile-phase flow rate of 1 ml min−1. The time course of elution (ratio of acetonitrile to KH2PO4 buffer) was as follows: 0 to 10 min, 5:95; 10 to 21 min, 15:85; 21 to 31 min, 70:30; 31 to 32 min, 40:60; and 32 to 35 min, 5:95. The retention times determined for purified standards were as follows: atrazine, 28.4 min; hydroxyatrazine, 26.5 min; deethylatrazine, 24.8 min; deisopropylatrazine, 18.4 min; and 2-chloro-4,6-diamino-s-triazine, 3.17 min. Fractions containing metabolites were collected on a fraction collector (model Z110; Bio-Rad Laboratories, Hercules, Calif.). The total amount of atrazine or metabolite was calculated on the basis of the specific activity of [14C]atrazine used in each experiment.
The chloride released during atrazine metabolism was measured with a chloride electrode (model 94-17b; Orion Research Inc., Boston, Mass.).
The atrazine metabolite produced by C. michiganese ATZ1 was identified by mass spectometry. A 100-ml culture of C. michiganese ATZ1 was grown in MS medium containing 100 ppm of atrazine as the sole carbon and nitrogen source. Cells were removed by centrifugation, and the supernatant was collected for analysis. The metabolite was purified from the supernatant by HPLC as described above and analyzed by mass spectrometry. Electrospray ionization mass spectrometry was accomplished with a MAT 900 mass spectrometer (Finnigan, San Jose, Calif.). The orifice voltage was maintained at 10 V, and the needle voltage was maintained at 3.6 kV. The capillary temperature was 250°C.
RESULTS
Analysis of the consortium members for atz genes.
The atrazine-degrading consortium resembled Pseudomonas sp. strain ADP in that it catabolized uniformly ring-labeled [14C]atrazine to 14CO2 at an approximately 75% yield (data not shown). These data, coupled with our previous observations that all recently isolated atrazine-degrading bacteria contain genes homologous to atzA, -B, and -C from Pseudomonas sp. strain ADP (Fig. 1), prompted us to do the genetic studies described here. The first gene in the atrazine degradation pathway in Pseudomonas sp. strain ADP, atzA, encodes atrazine chlorohydrolase (AtzA), which converts atrazine to hydroxyatrazine (17). The second gene, atzB, encodes a hydrolytic deamination reaction that results in the formation of N-isopropylammelide (7), and this product is used by a hydrolytic deaminating enzyme that is encoded by the atzC gene, resulting in the formation of cyanuric acid (32).
The PCR technique was used to amplify sequences internal to the atzA, -B, and -C genes that were potentially present in total genomic DNA derived from C. michiganese ATZ1 or Pseudomonas sp. strain CN1. The size of the PCR products expected with atzA and -B primers was 0.5 kb, whereas the atzC primers produced a 0.6-kb product (16). Southern hybridization was further used to confirm the PCR results. PCR analysis suggested that C. michiganese was genetically the most versatile pure-culture isolate from the consortium, with homologs to the atzA, -B, and -C genes (Table 1). The sizes of the PCR products from atzA and atzB were the same as that of the product generated with DNA from Pseudomonas sp. strain ADP. Moreover, the 500-bp atzA PCR fragment had 100% DNA sequence identity to the corresponding gene in Pseudomonas sp. strain ADP, strongly suggesting that C. michiganese has the ability to catalyze atrazine dechlorination to hydroxyatrazine (see below). In contrast, the PCR product obtained with atzC gene primers was only 0.3 kb (half the expected size), and the sequenced PCR product showed only 44% identity with the comparable region from the Pseudomonas sp. strain ADP atzC gene. These data suggest that C. michiganese most likely catabolizes atrazine via reactions similar to the first two metabolic steps of Pseudomonas sp. strain ADP (Fig. 1), but the ability of a gene with only 44% sequence identity to encode the AtzC reaction is equivocal.
TABLE 1.
Identification of genes in pure-culture isolates from the atrazine-degrading consortium that are homologous to atzABC from Pseudomonas sp. strain ADP
| Strain | PCR product (kb)
|
Sequence identity (%)
|
||||
|---|---|---|---|---|---|---|
| atzA | atzB | atzC | atzA | atzB | atzC | |
| C. michiganese ATZ1 | 0.5 | 0.5 | 0.3 | 100 | NDa | 44 |
| Pseudomonas sp. strain CN1 | 0.5 | —b | 0.6 | 99 | ND | 99 |
ND, not determined.
—, no product was detected in PCR and Southern hybridization analyses.
Pseudomonas sp. strain CN1 contained sequences homologous to the atzA and -C genes, as determined by PCR and Southern hybridization analyses (Table 1). The DNA sequences of both PCR products revealed identities of 99% with the atzA and -C genes from Pseudomonas sp. strain ADP. These data, coupled with the absence of a gene homologous to atzB, indicated that Pseudomonas sp. strain CN1 may be able to continue the metabolism initiated by C. michiganese and that it may also carry out atrazine dechlorination in parallel with the latter strain.
Atrazine metabolism by C. michiganese.
The genetic data suggested that C. michiganese initiates consortial atrazine catabolism by carrying out reactions similar to the first two metabolic steps observed with Pseudomonas sp. strain ADP (Fig. 1). In pure-culture growth experiments with C. michiganese in MS-glucose medium containing uniformly ring-labeled [14C]atrazine, all the atrazine was removed after 4 days. However, no detectable 14CO2 was released. Since the consortium released 75% of the label as CO2, these data suggested that atrazine catabolism by C. michiganese was incomplete, a result consistent with the suggestion from PCR and sequence data that the relatively low-homology atzC gene may not function in catalyzing the third reaction in the pathway. Atrazine degradation in a growing culture, however, was accompanied by stoichiometric release of the chloride ion (data not shown), indicating a conversion of atrazine to hydroxyatrazine similar to that found in Pseudomonas sp. strain ADP.
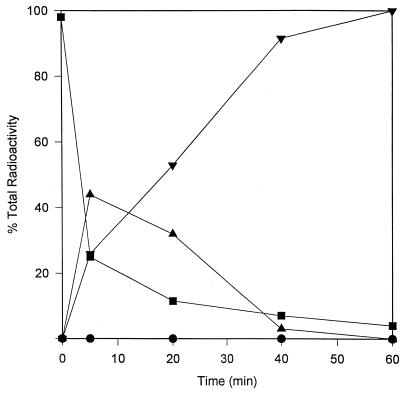
Resting-cell assays were conducted to determine the possible metabolites produced by these reactions (Fig. 2). Transient formation of hydroxyatrazine was observed, with a maximum of 45% of the total radioactivity accumulating as this metabolite at 5 min. Hydroxyatrazine decreased to below detectable limits within 60 min, with concomitant accumulation of an unknown metabolite comprising 98% of the total radioactive compounds. These same metabolites were observed by HPLC analysis with resting cells of the consortium (data not shown). However, with the consortium, the unknown metabolite did not accumulate.
FIG. 2.
Resting-cell suspensions of C. michiganese ATZ1 with uniformly ring-labeled [14C]atrazine and analysis of culture filtrates by HPLC, as described in Materials and Methods. The materials analyzed were atrazine (▪), hydroxyatrazine (▴), 14CO2 (•), and an unknown metabolite (▾).
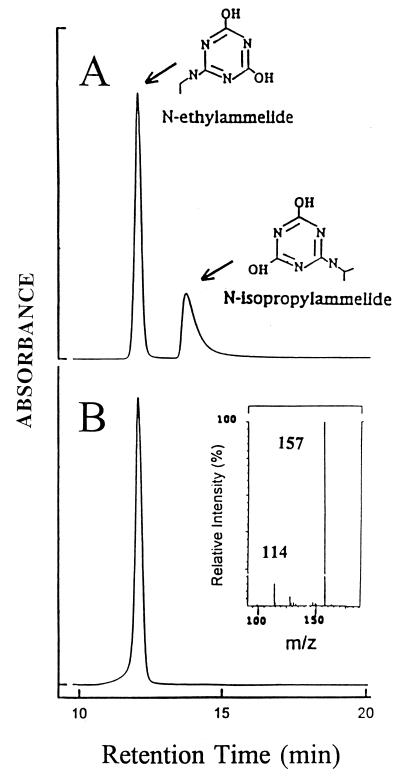
The unknown metabolite accumulated by C. michiganese ATZ1 was identified by HPLC retention time and mass spectrometry (Fig. 3). The mass spectrum showed a diagnostic ion at m/z 157 (m + H), which is identical to that of the standard N-ethylammelide. Another prominent ion at m/z 114 could be accounted for by the loss of an ethylamino fragment (NC2H5).
FIG. 3.
Identification of the accumulating unknown metabolite produced by C. michiganese ATZ1 as N-ethylammelide by HPLC and mass spectrometry. (A) Authentic standards N-isopropylammelide and N-ethylammelide. (B) Unknown metabolite by HPLC, with its mass spectrum shown in the inset.
C. michiganese ATZ1 utilized both ethylamine and isopropylamine as the sole nitrogen source and was also able to metabolize isopropylamine as the sole carbon and nitrogen source. Moreover, C. michiganese was able to grow on atrazine as the sole carbon and nitrogen source, albeit slowly and to only a limited optical density. Strain ATZ1 was unable to grow on cyanuric acid as a nitrogen source.
Catabolism of atrazine metabolites by Pseudomonas sp. strain CN1.
Pseudomonas sp. strain CN1 did not grow on atrazine as either a carbon or nitrogen source. Moreover, no detectable chloride release was measured in resting-cell assays of Pseudomonas sp. strain CN1 incubated with 100 ppm of atrazine for 72 h. These data suggested that strain CN1 lacked a functional AtzA and thus could not initiate the catabolism of atrazine. Western blotting experiments with antibody raised against purified AtzA protein from Pseudomonas sp. strain ADP revealed a cross-reacting protein with a molecular weight of 33,000 in strain CN1 (data not shown). The functional AtzA has a molecular weight of 52,421 (17), indicating that a truncated AtzA-like protein may be present in strain CN1.
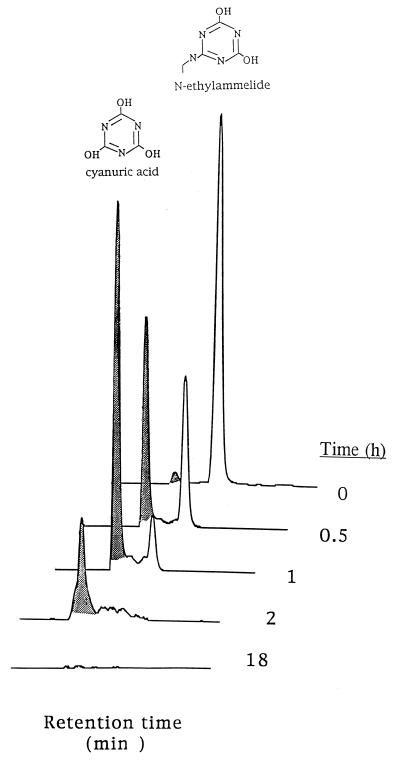
Pseudomonas sp. strain CN1 complemented the atrazine metabolism of C. michiganese by carrying out catabolic steps subsequent to the AtzA and -B reactions shown in Fig. 1. Here, strain CN1 was observed to express functional AtzC activity. Experiments with protein extracts obtained from strain CN1 showed that N-ethylammelide was hydrolyzed to cyanuric acid, which was then further degraded to levels below the detection limit after 18 h (Fig. 4). In separate experiments, Pseudomonas sp. strain CN1 was shown to hydrolyze N-isopropylammelide. With cell extracts from Pseudomonas sp. strain CN1, N-ethylammelide was preferred over N-isopropylammelide as the substrate. After incubation with 160 ppm of N-ethylammelide and N-isopropylammelide for 10 min at 25°C, 4 and 93% of substrates, respectively, were detectable at the end of the experiment. The ability to transform N-isopropylammelide to cyanuric acid was previously shown for Escherichia coli clones expressing atzC from Pseudomonas sp. strain ADP (32). Furthermore, cell extract from recombinant E. coli expressing atzC hydrolyzed both N-ethylammelide and N-isopropylammelide (14).
FIG. 4.
Transformation of N-ethylammelide and cyanuric acid by cell extracts from Pseudomonas sp. strain CN1, as demonstrated by HPLC.
Unlike C. michiganese, Pseudomonas sp. strain CN1 grew on cyanuric acid as the sole nitrogen source, suggesting that it was at least partially responsible for the atrazine-mineralizing capability of the consortium. It was also capable of utilizing ethylamine or isopropylamine as the sole nitrogen source when it was grown in MS liquid medium with glucose as a carbon source. However, no growth was detected when 100 mM ethylamine or isopropylamine was provided as the sole carbon and nitrogen source.
Success of the consortium compared to individual bacteria.
Pseudomonas sp. strain CN1 is not able to initiate atrazine metabolism and thus would not be expected to survive with atrazine as the sole carbon or nitrogen source. In contrast, C. michiganese can grow on atrazine as the sole carbon or nitrogen source and shares this property with the consortium as a whole. In this context, it is important to consider how the consortium may improve the overall metabolism of atrazine.
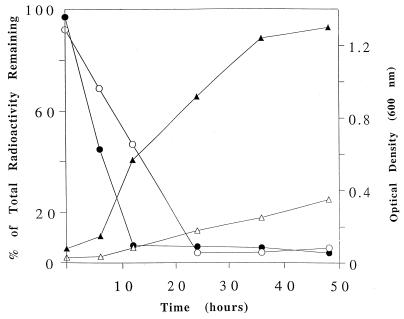
In comparative growth experiments, the consortium performed significantly better, both in attaining maximal cell density and in removing atrazine from growing cultures (Fig. 5). This difference was particularly striking with respect to growth, which was likely due to greater metabolism of atrazine providing increased opportunities to harvest nitrogen and energy for growth.
FIG. 5.
Comparison of C. michiganese growth (▵) and atrazine catabolism (○) with consortium growth (▴) and atrazine catabolism (•) on MS medium with glucose and atrazine.
Growth comparisons among C. michiganese ATZ1, Pseudomonas sp. strain CN1, a mixture of the two strains, and the consortium provided additional insights (Table 2). The consortium, but neither strain individually or C. michiganese and Pseudomonas sp. strain CN1 in coculture, could grow on ethylamine as the sole carbon and nitrogen source. The consortium grew more rapidly than did C. michiganese alone on isopropylamine. These data suggested that the consortium metabolizes alkylamines more efficiently and that this underlies the better growth of the consortium on atrazine.
TABLE 2.
Catabolism of atrazine and its metabolites by pure cultures of strains ATZ1 and CN1 and the consortium
| Strain(s) | Growth ona:
|
|||||
|---|---|---|---|---|---|---|
| Atrazine | Hydroxyatrazine | N-Ethylammelide | Cyanuric acid | Ethylamine | Isopropylamine | |
| C. michiganese ATZ1 | +* | + | − | − | + | +* |
| Pseudomonas sp. strain CN1 | − | NDb | + | + | + | + |
| C. michiganese ATZ1 + Pseudomonas sp. strain CN1 | +* | + | + | + | + | +* |
| Consortium | +* | + | + | + | +* | +* |
+, growth on substrate; −, no growth on substrate; ∗, growth on substrate as a carbon and nitrogen source.
ND, not determined.
Consortium catabolism of s-triazines other than atrazine.
The consortium was grown on MS medium with various s-triazine compounds at concentrations of 500 μg/ml in agar medium and 100 μg/ml in liquid medium. The following s-triazines were catabolized by >95%: ametryn, prometryn, simazine, propazine, simetryn, and atratone.
DISCUSSION
It is increasingly important to understand the genetic basis of how microbial consortia function to collectively catabolize organic compounds. Moreover, the study of catabolic-gene distribution in different bacteria can contribute significantly to our understanding of how bacteria evolve new metabolic functions.
In a classic study (34), Senior et al. determined the individual metabolic roles of five members in a microbial consortium that metabolized the herbicide Dalapon. In their study, a single enzymatic dechlorination reaction generated the intermediary metabolite pyruvate and therefore the metabolism that sustained the consortium was not based on shared catabolic reactions but on cross-feeding with other metabolites.
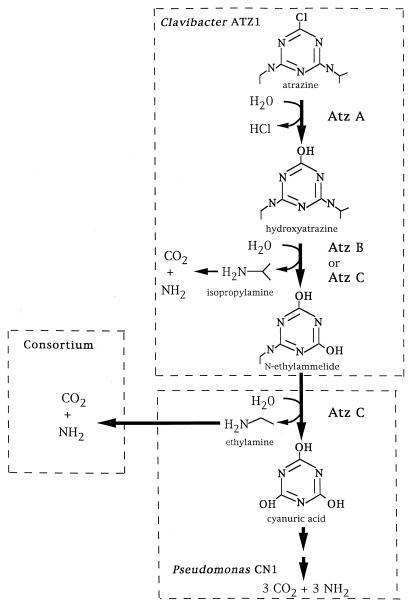
The present study demonstrates that sequential steps in atrazine catabolism can be carried out consecutively by two members of a microbial consortium and that overlapping sets of genes in each underlie the observed physiology. Figure 6 illustrates the individual contributions of consortium members, with major roles being played by the Clavibacter and Pseudomonas strains, which sequentially catalyze atrazine side-chain removal and ring cleavage, respectively. The data suggest that the Clavibacter and Pseudomonas strains and all the mixed-consortium members have the metabolic capability to metabolize alkylamines nominally as sole nitrogen sources. The consortium is most effective with both alkylamines and use either substrate as a carbon source. The capability to metabolize alkylamines efficiently may be important in environments where the buffering capacity is limited and the pH would rise markedly if amines accumulated. This has previously been observed with Pseudomonas sp. strain ADP, which metabolizes alkylamines very slowly and is toxified by atrazine metabolites in the absence of a medium with a very high buffering capacity (14). The observation here that the consortium collectively metabolized atrazine faster than did individual strains is consistent with the idea that maintaining low steady-state concentrations of alkylamines may be a selective pressure for maintaining the stable consortium. Further studies will help to determine the validity of this hypothesis. However, this type of metabolism has precedence in fatty acid-fermenting consortia, where hydrogen partial pressure must be kept very low by interspecies hydrogen transfer in order to make anaerobic fatty acid oxidation thermodynamically favorable (36). In these instances, the organisms form very stable cocultures and cannot be grown separately in the laboratory.
FIG. 6.
Atrazine catabolism by the atrazine-degrading consortium, with the individual roles of consortium members indicated.
It is interesting that the second and third reactions of atrazine catabolism catalyzed by pure-culture Pseudomonas sp. strain ADP (Fig. 1) and the consortium (Fig. 6) are inverted, but the overall metabolic logic of ultimately producing cyanuric acid is maintained. This requires a difference in the specificity of only one enzyme, AtzB, since we have demonstrated here that AtzC can hydrolyze N-ethylammelide and N-isopropylammelide. The atzB genes detected by PCR are suggested to have homologies high enough to yield a strong band in a high-stringency PCR, followed by gel electrophoresis, and the DNA fragment obtained had the same molecular weight. However, small sequence differences may reasonably be expected to change the specificity of AtzB to preferentially remove the N-isopropylamine side chain rather than the N-ethylamine side chain of atrazine. C. michiganese is able to use N-isopropylamine, but not N-ethylamine, as its sole nitrogen and carbon source; this may provide the selective pressure for generating or maintaining an altered AtzB reaction specificity.
There are known examples of consortia in which metabolites generated by one strain are further transformed by others, allowing nitrogen or carbon assimilation by different consortium members. A laboratory coculture consisting of Rhodococcus corallinus and Pseudomonas sp. strain NRRL B12228 was established to metabolize the s-triazine compound deethylsimazine as the sole nitrogen source (12). This was the prelude to construction of a genetically engineered strain that was capable of metabolizing atrazine and contained genes encoding a Rhodococcus cytochrome P-450 mono-oxygenase active with atrazine and a Pseudomonas sp. s-triazine hydrolase active with the metabolites generated by mono-oxygenase-catalyzed reactions (35). In another example, a two-species coculture metabolizing 4-aminobenzenesulfonate, in which catechol-4-sulfonate generated by Hydrogenophaga paleronii S1 was metabolized as a carbon source by Agrobacterium radiobacter S2, was recently described (13). In addition, a coculture catabolizing carbaryl was generated by mixing bacteria that released 1-naphthol and metabolized it to CO2 (9).
The present study extends previous work by demonstrating the individual metabolic and genetic contributions of consortium members that use a proposed recently evolved catabolic pathway (16). Atrazine and related s-triazine herbicides have been in commercial use for approximately 40 years. The wide use of s-triazine herbicides has led to their detection as contaminants in groundwater (6, 28, 29) and to point source soil contamination problems where these herbicides have been spilled. Previously, many isolates and mixed cultures that partially degrade atrazine have been found (3, 10); more recently, several bacterial pure cultures which can completely mineralize atrazine and other s-triazines have been isolated (8, 26, 27, 30, 37). In 1995, Mandelbaum et al. (26) isolated a single atrazine-mineralizing bacterium from a mixture of bacteria originally reported to be a consortium (24, 25), which suggested that the isolate arose from gene transfer which occurred in the mixed culture. The possibility of this has been heightened by our observation that the atzABC genes are located on a 96-kb plasmid, with at least two genes having flanking regions with high homologies to known insertion sequence elements (16). Thus, the present study may offer a window to the evolution of a catabolic pathway by beginning to reveal how genes move from a consortium to individual strains and how mixed cultures containing metabolically cooperating genes may be stably maintained.
ACKNOWLEDGMENT
This work was supported by a grant from Novartis Crop Protection (to L.P.W. and M.J.S.).
REFERENCES
- 1.Alvey S, Crowley D E. Influence of organic amendments on biodegradation of atrazine as a nitrogen source. J Environ Qual. 1995;24:1156–1162. [Google Scholar]
- 2.Alvey S, Crowley D E. Survival and activity of an atrazine-mineralizing bacterial consortium in rhizosphere soil. Environ Sci Technol. 1996;30:1596–1603. [Google Scholar]
- 3.Assaf N A, Turco R F. Accelerated biodegradation of atrazine by a microbial consortium is possible in culture and soil. Biodegradation. 1994;5:29–35. doi: 10.1007/BF00695211. [DOI] [PubMed] [Google Scholar]
- 4.Behki R, Topp E, Dick W, Germon P. Metabolism of the herbicide atrazine by Rhodococcus strains. Appl Environ Microbiol. 1993;59:1955–1959. doi: 10.1128/aem.59.6.1955-1959.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behki R M, Khan S U. Degradation of atrazine by Pseudomonas: n-dealkylation and dehalogenation of atrazine and its metabolites. J Agric Food Chem. 1986;34:746–749. [Google Scholar]
- 6.Belluck D A, Benamin S L, Dawson T. Groundwater contamination by atrazine and its metabolites: risk assessment, policy, and legal implications. In: Somasundaram L, Coats J R, editors. Pesticide transformation products: fate and significance in the environment. Washington, D.C: American Chemical Society; 1991. pp. 254–273. [Google Scholar]
- 7.Boundy-Mills K, de Souza M L, Mandelbaum R M, Wackett L P, Sadowsky M J. The atzB gene of Pseudomonas sp. strain ADP encodes the second enzyme of a novel atrazine degradation pathway. Appl Environ Microbiol. 1997;63:916–923. doi: 10.1128/aem.63.3.916-923.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouquard C, Ouazzani J, Prome J-C, Michel-Briand Y, Plesiat P. Dechlorination of atrazine by a Rhizobium sp. isolate. Appl Environ Microbiol. 1997;63:862–866. doi: 10.1128/aem.63.3.862-866.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapalamadugu S, Chaudry G R. Hydrolysis of carbaryl by a Pseudomonas sp. and construction of a microbial consortium that completely metabolizes carbaryl. Appl Environ Microbiol. 1991;57:744–750. doi: 10.1128/aem.57.3.744-750.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook A M. Biodegradation of s-triazine xenobiotics. FEMS Microbiol Rev. 1987;46:93–116. [Google Scholar]
- 11.Cook A M, Beilstein P, Grossenbacher H, Hutter R. Ring cleavage and degradative pathway of cyanuric acid in bacteria. Biochem J. 1985;231:25–30. doi: 10.1042/bj2310025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook A M, Hutter R. Deethylsimizine: bacterial dechlorination, deamination, and complete degradation. J Agric Food Chem. 1984;32:581–587. [Google Scholar]
- 13.Dangmann E, Stolz A, Kuhm A E, Hammer A, Feigel B, Noisommit-Rizzi N, Rizzi M, Reuss M, Knackmuss H J. Degradation of 4-aminobenzenesulfonate by a two-species bacterial coculture. Physiological interactions between Hydrogenophaga palleronii S1 and Agrobacterium radiobacter S2. Biodegradation. 1996;7:223–229. doi: 10.1007/BF00058181. [DOI] [PubMed] [Google Scholar]
- 14.de Souza, M. L. Unpublished data.
- 15.de Souza, M. L., and D. Newcombe. Unpublished data.
- 16.de Souza, M. L., M. J. Sadowsky, J. Seffernick, B. Martinez, and L. P. Wackett. Unpublished data.
- 17.de Souza M L, Sadowsky M J, Wackett L P. Atrazine chlorohydrolase from Pseudomonas sp. strain ADP: gene sequence, enzyme purification, and protein characterization. J Bacteriol. 1996;178:4894–4900. doi: 10.1128/jb.178.16.4894-4900.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Souza M L, Wackett L P, Boundy-Mills K L, Mandelbaum R T, Sadowsky M J. Cloning, characterization, and expression of a gene region from Pseudomonas sp. strain ADP involved in the dechlorination of atrazine. Appl Environ Microbiol. 1995;61:3373–3378. doi: 10.1128/aem.61.9.3373-3378.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eaton R W, Karns J S. Cloning and analysis of s-triazine catabolic genes from Pseudomonas sp. strain NRRLB-12227. J Bacteriol. 1991;173:1215–1222. doi: 10.1128/jb.173.3.1215-1222.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erickson E L, Lee K H. Degradation of atrazine and related s-triazines. Crit Rev Environ Contam. 1989;19:1–13. [Google Scholar]
- 21.Giardina M C, Giardi M T, Filacchioni G. Atrazine metabolism by Nocardia: elucidation of initial pathway and synthesis of potential metabolites. Agric Biol Chem. 1982;46:1439–1445. [Google Scholar]
- 22.Ianotti E L, Kafkewitz D, Wolin M J, Bryant M P. Glucose fermentation products in Ruminococcus albus grown in continuous culture with Vibrio succinogenes: changes caused by interspecies transfer of H2. J Bacteriol. 1973;114:1231–1240. doi: 10.1128/jb.114.3.1231-1240.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jutzi K, Cook A M, Hutter R. The degradative pathway of the s-triazine melamine. Biochem J. 1982;208:679–684. doi: 10.1042/bj2080679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mandelbaum R T, Wackett L P, Allan D L. Mineralization of the s-triazine ring of atrazine by stable bacterial mixed cultures. Appl Environ Microbiol. 1993;59:1695–1701. doi: 10.1128/aem.59.6.1695-1701.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandelbaum R T, Wackett L P, Allan D L. Rapid hydrolysis of atrazine to hydroxyatrazine by soil bacteria. Environ Sci Technol. 1993;27:1943–1947. [Google Scholar]
- 26.Mandelbaum R T, Wackett L P, Allan D L. Isolation and characterization of a Pseudomonas sp. that mineralizes the s-triazine herbicide atrazine. Appl Environ Microbiol. 1995;61:1451–1457. doi: 10.1128/aem.61.4.1451-1457.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moscinski J K, Jayachandran K, Moorman T B. Abstracts of the 96th General Meeting of the American Society for Microbiology 1996. Washington, D.C: American Society for Microbiology; 1996. Mineralization of the herbicide atrazine by Agrobacterium radiobacter; p. 458. [Google Scholar]
- 28.Pick F E, van Dyk L P, Botha E. Atrazine in ground and surface water in maize production areas of the Transvaal, South Africa. Chemosphere. 1992;25:335–341. [Google Scholar]
- 29.Radosevich M, Crawford J J, Traina S J, Oh K H, Tuovinen O H. Sorption and degradation of pesticides and organic chemicals in soil. SSSA special publication no. 32. Madison, Wis: SSSA; 1993. Biodegradation of atrazine and alachlor in subsurface sediments; pp. 33–41. [Google Scholar]
- 30.Radosevich M, Traina S J, Hao Y, Tuovinen O H. Degradation and mineralization of atrazine by a soil bacterial isolate. Appl Environ Microbiol. 1995;61:297–301. doi: 10.1128/aem.61.1.297-302.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rustum A M, Ash S, Saxena A. Reversed-phase high-performance liquid chromatographic method for the determination of soil-bound [14C]-atrazine and its radiolabeled metabolites in a soil metabolism study. J Chromatogr. 1990;22:209–218. [Google Scholar]
- 32.Sadowsky M J, Tong Z, de Souza M L, Wackett L P. AtzC is a new member of the amidohydrolase protein superfamily strain and is homologous to other atrazine-metabolizing enzymes. J Bacteriol. 1998;180:152–158. doi: 10.1128/jb.180.1.152-158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 34.Senior E, Bull A T, Slater J H. Enzyme evolution in a microbial community growing on the herbicide Dalapon. Nature. 1976;263:476–479. doi: 10.1038/263476a0. [DOI] [PubMed] [Google Scholar]
- 35.Shao Z O, Seffens W, Mulbry W, Behki R M. Cloning and expression of the s-triazine hydrolase gene (trzA) from Rhodococcus corallinus and development of Rhodococcus recombinant strains capable of dealkylating and dechlorinating the herbicide atrazine. J Bacteriol. 1995;177:5748–5755. doi: 10.1128/jb.177.20.5748-5755.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thauer R K, Jungermann K, Decker K. Energy conservation in chemoautotrophic bacteria. Bacteriol Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yanze-Kontchou C, Gschwind N. Mineralization of the herbicide atrazine as a carbon source by a Pseudomonas strain. Appl Environ Microbiol. 1994;60:4297–4303. doi: 10.1128/aem.60.12.4297-4302.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]