Abstract
We describe a rapid oligonucleotide probe design strategy based on subtractive hybridization which yields probes for 16S rRNA or rRNA genes of individual members of microbial communities that are specific within the context of those communities. This strategy circumvents the need to sequence many similar or identical clones of dominant members of a community. Radioactively labeled subfragments of a cloned 16S rRNA gene sequence for which a probe is required (target) were hybridized with biotinylated total 16S ribosomal DNA (rDNA) amplified from the microbial community, and the hybrids formed were subsequently discarded. The remaining enriched fragments were used to screen a library consisting of cloned subfragments of the target sequence by colony hybridization in order to identify the variable regions of the 16S rRNA gene with the required specificity. The sequencing of random clones in one 16S rDNA library demonstrated that only those clones with 100% sequence identity with the probe fragment were detected by it. Moreover, sequencing of other, randomly selected, probe-positive clones revealed 100% sequence identity with the probe. Probes developed in this way tended to correspond to more variable regions of the 16S rRNA if the target sequences were similar to the sequences of other clones in the library and to less variable regions if the target sequences were phylogenetically isolated within the clone library. Although the absolute specificity of the latter probes, as assessed by comparison with available database sequences, was lower than the absolute specificity of the probes from the more variable regions, they were specific within the context of the environmental samples from which they were derived.
rRNA gene sequence analysis and probe development represent a powerful experimental approach for investigation of species structure, diversity, and population dynamics in natural microbial communities. The utility of a probe is directly related to its specificity for the target group of organisms. The principal difficulty in the application of this technology is the paucity of 16S rRNA sequence information on the microbial world, due to the fact that the majority of microorganisms have not yet been isolated and/or sequenced. The consequence of this is that probe design is based on incomplete information; the assumed specificity of a probe thus generally falls in time as more sequence information becomes available. The rational design of probes of a given specificity which are valid for the entire microbial world requires a much larger sequence data set than we presently have. The actual specificity of probes for any given habitat must therefore be assessed by sequencing many clones from that habitat.
An alternative approach might be the empirical design of probes with habitat-based specificity for given sequences within a particular environmental habitat. In this case, probe specificity would not be defined by comparison with known sequences in a database but rather would be defined by comparison with unknown sequences in the sample. This comparison would be done not by database interrogation but by experimental hybridization with total 16S ribosomal DNA (rDNA) amplified from DNA extracted from the sample. The probes generated by this approach would be specific only for the microbial community represented by the DNA of the sample used for the probe design or for repeat samples from an environmental habitat with a relatively constant microbial species composition.
In this report we describe a subtractive hybridization-based method generally applicable to empirical development of oligonucleotide probes with habitat-based specificities for the investigation of microbial communities.
MATERIALS AND METHODS
Bacterial isolates and samples.
The following three samples were used in this study: (i) sediment from the Spittelwasser, a highly polluted stream and tributary of the Elbe River in Germany; (ii) a biofilm which developed on glass plates immersed in the Elbe River; and (iii) a community enriched from Spittelwasser sediment and able to use 3-chlorobiphenyl (3CB) as a sole carbon source. The Spittelwasser sediment sample and the 3CB enriched culture were characterized by performing a sequence analysis of parts of the 16S rDNA clone libraries and were shown to contain mainly β-proteobacterial sequences. Analysis of cloned 16S rDNA amplification products from the 3CB enrichment culture revealed predominantly sequences of bacterial strains Y1 and 3.3 (see below) and some sequences of other members of the Proteobacteria (data not shown). The biofilm was investigated by hybridizing nucleic acid extracts and amplified rrn operons with group-specific probes; in this case, the sequences of members of the α subclass of the Proteobacteria were found to be the most abundant sequences (15). Bacterial strains Y1 and 3.3 cultured from the 3CB enrichment culture are members of the Alcaligenes faecalis and Burkholderia branches, respectively, of the β subclass of the Proteobacteria and provided the 16S rDNA target sequences for this study. Other target sequences used in this study were obtained from a 16S rDNA clone library of the Spittelwasser sediment sample in a pBluescript vector (Stratagene, Heidelberg, Germany) in Escherichia coli DH5α.
DNA isolation.
DNA was isolated from bacterial strains and enrichment cultures by the protocol of Wilson (20), modified as follows for application to environmental samples. After incubation with cetyltrimethylammonium bromide, lysates were repeatedly frozen in liquid nitrogen and thawed in a 65°C water bath three times, and then the DNA was precipitated with isopropanol after 2 M ammonium acetate was added, to reduce coprecipitation of humic substances. For PCRs DNA was dissolved in TE buffer (pH 8) and reprecipitated with sodium acetate and ethanol.
Preparation of target DNA.
The restriction fragments used in subtractive hybridization were designated the targets of subtraction. The 16S rDNAs of strains Y1 and 3.3 were amplified from 500 ng of genomic DNA with primers 16F27 and 16R1492, which are specific for the 5′ and 3′ ends of the 16S rRNA genes of members of the Bacteria (9). Insertions in pBluescript vectors were amplified from boiled lysates of the clones by using primers DX-F (5′-TACCGGGCCAAGAGTTTGATC-3′) and DX-R (5′-GGGGGGCCAACGGTTACCTT-3′), which are complementary to the junction sequences between the vector and the insert. The cycling program consisted of 60 s of denaturation (96°C), 30 s of primer annealing (55°C), and 180 s of extension (72°C) for 30 cycles and a final 10-min extension step. The PCR products from a 100-μl reaction mixture were separated from excess primers and nucleotides with a Microcon-100 concentrator (Amicon, Witten, Germany). The products were digested with restriction enzymes AciI, MspI, and TaqI at 37°C for several hours, followed by an additional 2 h at 65°C, the incubation temperature recommended for TaqI. The reaction mixture was extracted with phenol, and nucleic acids were precipitated by adding 10 volumes of butanol. The precipitate was suspended in 20 μl of 10 mM Tris (pH 8). To control for digestion of the target DNA and recovery of precipitated fragments, 2 μl of the solution was electrophoresed on a polyacrylamide gel (12% polyacrylamide in TBE), which was subsequently stained with ethidium bromide. Fragment concentrations were estimated by comparing bands on the stained gels.
A 5-μl portion of restriction fragments was labeled with [α-32P]dCTP and [α-32P]dGTP by filling in 5′ protruding termini with Klenow polymerase. Unincorporated nucleotides were removed by using a nucleotide removal kit (Qiagen, Hilden, Germany). Labeled fragments were precipitated with butanol and dried. Radioactivity was measured by Cerenkov counting of total reaction mixtures with a model LS1801 scintillation counter (Beckmann, Munich, Germany).
Preparation of fragment libraries.
Clone libraries were generated by cloning restriction fragments of the target DNA dephosphorylated by shrimp alkaline phosphatase (U.S. Biochemicals, Bad Homburg, Germany) into an AccI-linearized pBluescript SK(+) vector. Hybrid plasmids were introduced into E. coli DH5α by electroporation. Colony lifts were carried out by standard methods (14).
Synthesis and purification of subtracter DNA.
Subtracter DNA is biotinylated DNA used for capture of unspecific target fragments. In the application described here, the subtracter DNA consisted of the total 16S rDNA amplified from DNA extracted from an environmental sample. To ensure the highest possible sequence diversity in the subtracter DNA, PCRs were carried out with relatively large amounts of total DNA extracted from each environmental sample, and the products of several replicate reactions were combined for use as the subtracter DNA. 16S rDNA was amplified by PCR from DNA (ca. 500 ng) extracted from the microbial communities by using primers 16F27 and 16R1492 (9) biotinylated at the 5′ ends (MWGBiotech, Ebersberg, Germany). Each reaction mixture contained each deoxynucleoside triphosphate at a concentration of 800 μM, each primer at a concentration of 600 nM, and 2.5 U of Taq polymerase in a 100-μl volume. PCR products were separated from primers by using a QIAwell vacuum manifold (Qiagen) according to the manufacturer’s protocol for purification of nucleic acids, were precipitated with sodium acetate and ethanol, were washed in 70% ethanol, and were resuspended to 10% of the initial volume.
Solution hybridization.
Radioactively labeled target fragments were resuspended in 2 μl of 2.5× hybridization buffer (75% formamide, 25 mM Tris [pH 7.5], 2.5 mM EDTA), and 2 μl of a subtracter DNA solution was added. The mixture was heat denatured at 95°C for 2 min and chilled on ice, and 1 μl of 5 M NaCl was added. The reaction mixture was overlaid with mineral oil (U.S. Biochemicals) and incubated overnight at 56°C.
Removal of target-subtracter hybrids.
Streptavidin-coated magnetic beads (Dyna, Hamburg, Germany) were washed twice with phosphate-buffered saline (10 mM sodium phosphate [pH 7.4], 130 mM NaCl) containing 0.1% bovine serum albumin and once with TE buffer containing 1 M NaCl and then resuspended at a concentration of 2% (wt/vol) in TE buffer containing 1 M NaCl. After the oil layer was removed, the hybridization mixture was combined with 100 μl of binding buffer (TE buffer [pH 8] containing 1 M NaCl) and 50 μl of a magnetic bead suspension. After incubation of the mixture for 1 h at room temperature, the magnetic beads were removed with a magnet, and the supernatant fluid was transferred to a Microcon-10 concentrator. Then 400 μl of water was added, the column was centrifuged (7,500 × g, 20 min), and the nucleic acids in the supernatant fluid were precipitated with butanol and resuspended in 2 μl of 2.5× hybridization buffer for a subsequent cycle of hybridization (see above).
Membrane hybridization.
The 32P-labeled fragments remaining in the supernatant fluid after the final subtraction were used directly to screen colony lifts of the clone library of the target sequence fragments. Colony lifts were hybridized in a buffer containing 20% formamide, 1 M NaCl, 10 mM Tris (pH 7.5), 1 mM EDTA, and 0.1% sodium dodecyl sulfate at 45°C. The membranes were washed twice in 200 mM NaCl–20 mM Tris (pH 8)–0.1% sodium dodecyl sulfate at 45°C for 10 min. The hybridization patterns were visualized on X-ray film (Kodak, Rochester, N.Y.) exposed at −70°C by using an intensifier screen.
Sequence determination.
Plasmids were prepared from positive clones of the fragment library by using a Qia well kit (Qiagen) and were sequenced by using a model 373A DNA sequencer and a Taq cycle sequencing mixture according to the protocol of the manufacturer (Perkin-Elmer, Applied Biosystems, Weiterstadt, Germany).
Specificity of enriched probe fragments and oligonucleotide probes designed from the sequences of such fragments.
The specificity of probe fragments in a habitat was assessed by comparison with the 16S rDNA sequences determined from the clone library prepared from the habitat (17). The phylogenetic specificity of probe fragments was assessed by comparison with 16S rRNA and rRNA gene sequences from the Ribosomal Database Project (10) by using the Check_Probe function. This allowed us to map mismatches and facilitated the design of oligonucleotide probes for optimal regions of the target gene. Clones with full-length sequenced 16S rDNA inserts were used as controls for optimizing hybridization conditions for oligonucleotide probes, so that probes hybridized only to corresponding sequences exhibiting 100% identity. Oligonucleotide probes were used as probes for screening the clone library by using standard laboratory protocols (14). Some of the positive clones identified in this way were partially sequenced to assess probe specificity.
RESULTS
Subtractive hybridization strategy for habitat-based probes: empirical identification of 16S rDNA regions specific for individual members of microbial communities.
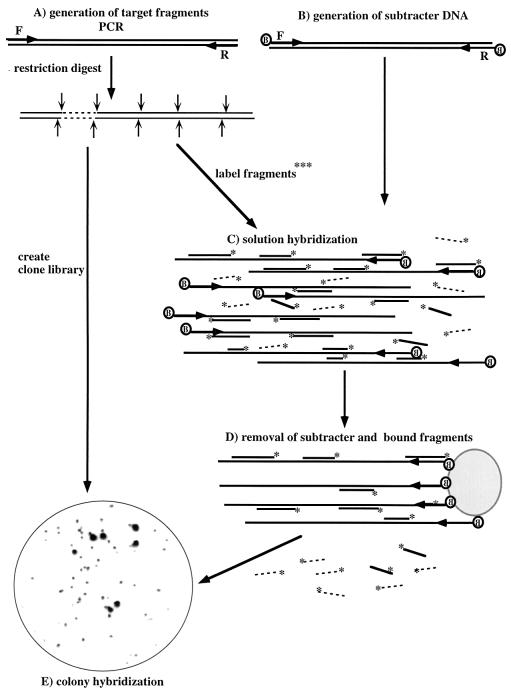
The procedure developed for enrichment of probe sequences specific for a cloned rDNA gene is depicted in Fig. 1. The target gene is amplified and cleaved by multiple four-base recognition restriction endonucleases. The fragments generated in this way on the one hand are used to generate a clone library and on the other hand are 5′ end labeled with 32P. Biotinylated subtracter DNA is generated by PCR amplification of 16S rDNA genes of the total DNA extracted from the environmental sample studied. Radioactively labeled fragments of the target sequence are hybridized in solution with the biotinylated full-length subtracter DNA, followed by removal of hybrids by using streptavidin-coated magnetic beads. The fragments remaining in the supernatant fluid are then subjected to a second round of subtractive hybridization. The twice-enriched target fragments remaining in the supernatant fluid are then used directly to screen the clone library consisting of the target fragments by colony hybridization. Strongly positive clones are sequenced. Comparison of the sequenced clones with data bank sequences by using the Check_Probe function of the Ribosomal Database Project (10) allows currently available phylogenetic information to be taken into account for design of optimal probes targeted to the enriched region of the gene. Note that the fragments remaining in the supernatant fluid after subtractive hybridization cannot themselves be used directly for probing as they react, albeit weakly, with all clones in the sample library, presumably because they are contaminated with a small percentage of nonspecific fragments.
FIG. 1.
Subtractive hybridization strategy used to obtain habitat-based probes. A, Generation of restriction endonuclease-generated subfragments from PCR-amplified target DNA from a 16S rDNA clone or bacterial strain, creation of a clone library of target subfragments, and radioactive labeling of subfragments for hybridization; B, generation of biotinylated subtracter DNA by PCR amplification of 16S rDNA from the total DNA extracted from the environmental sample by using two 5′-biotinylated primers; C, solution hybridization of radioactively labeled target fragments and biotinylated subtracter DNA; D, removal of subtracter-target hybrids by binding to streptavidin-coated beads; E, use of enriched target fragments for screening of the clone library of target fragments by colony hybridization. In the example shown a clone library of subfragments of Acidovorax-like sequence SW4 was screened with subfragments enriched by two hybridizations with subtracter DNA consisting of 16S rDNA amplified from DNA extracted from Spittelwasser sediment. F and R indicate forward and reverse primers, respectively; B indicates biotin; and ∗ indicates 32P end label. Radioactive target fragments removed by subtractive hybridization are shown as solid lines, whereas those which tend not to be removed by subtractive hybridization and which are thus specific for the target are shown as dashed lines.
Nonspecific losses of target sequences and subtraction optimization.
To estimate nonspecific losses of target fragments during hybridization to subtracter DNA, 100 ng (17 pmol) of the GAM 18-mer oligonucleotide (l-Sc-gProt-1027-a-A-18) (1, 11), a probe specific for the 23S rRNA of members of the γ subclass of the Proteobacteria, was radioactively labeled (14) and used as a target. Biotinylated 16S rDNA amplified from DNA of strain 3.3, which is a member of the β subclass of the Proteobacteria, was used as the subtracter DNA. The radioactivity remaining in the supernatant fluid after subtractive hybridization was reduced by about 50% in each round (Table 1). The degree of loss of specific fragments due to unspecific binding in this experiment may be considered to be a minimum since, in a normal subtraction experiment involving only 16S rRNA gene sequence partners, the sequence similarities between target and subtracter are greater.
TABLE 1.
Comparison of nonspecific and specific removal of target fragments as a function of the number of cycles of subtractive hybridization
| Round of hybridizationa | % of initial target label remaining in:
|
% Difference in fraction boundb | |
|---|---|---|---|
| Oligonucleotide GAMc | 16S rDNA fragments of isolate 3.3 | ||
| First | 55 | 41 | 14 |
| Second | 29 | 8 | 21 |
| Third | 14 | 3 | 11 |
The subtracter DNA used was 16S rDNA amplified from bacterial isolate 3.3. Note that hybridization of two differentially labeled but otherwise identical DNA molecules in equimolar concentrations statistically produces 50% homologous pairs and 50% hybrids, which results in some 50% of the target remaining in the supernatant after removal of hybrids. Therefore, significantly more than the observed 59% initial capture in the 16S rDNA fragments of isolate 3.3 might not have been expected. The additional capture in the subsequent rounds of subtraction resulted from the progressively increasing ratio of subtracter to target, since the same subtracter concentrations were used for all rounds.
Percentage of initial target label remaining in oligonucleotide GAM − 1 percentage of initial target label remaining in 16S rDNA fragments of isolate 3.3.
See Manz et al. (11).
For comparison, the 16S rDNA fragments of strain 3.3 were used as targets for subtraction with DNA having an identical sequence in order to estimate the efficiency of capture of target fragments by homologous subtracter sequences. There were equimolar amounts of subtracter and target DNAs (i.e., 2 pmol of subtracter DNA and 2 pmol of each restriction fragment DNA [24 fragments in all], resulting in 96 pmol of single-stranded target fragments) in the first round of hybridization, and the maximum difference between unspecific and specific binding was observed after the second cycle of hybridization (Table 1).
Specific losses resulting from target sequences in the subtracter.
The fact that 16S rRNA genes are highly conserved and exhibit a maximum of 35% sequence differences means that cross-reactivities are significantly higher in our system than in other applications of subtractive hybridization. Thus, in contrast to other applications (2, 5, 13, 16, 18, 19), the presence of full-length target sequences in subtracter DNA results in losses of the specific fragments enriched at each round of hybridization. To determine whether these losses significantly compromise the procedure, the 16S rDNA fragments of isolate Y1, a member of the A. faecalis branch in the β subclass of the Proteobacteria, were prepared as targets and used with subtracter DNA prepared from the 3CB enrichment culture from which strain Y1 was initially isolated. This enrichment culture was shown to contain mostly sequences of Y1 and 3.3, a member of the Burkholderia branch of the β subclass of the Proteobacteria, plus a few sequences of more distantly related members of the Proteobacteria. After two rounds of subtraction, enriched fragments were used to screen a fragment library of strain Y1 16S rDNA. Four positively reacting clones were sequenced and were found to have the same 55-nucleotide insert, spanning the region from positions 176 to 231 of the 16S rRNA (E. coli numbering [3]), which is one of the most variable regions in the 16S rRNA gene (18). Comparisons of the corresponding regions of the Y1 and 3.3 16S rDNA sequences revealed 15 nucleotide differences within this 55-nucleotide fragment. Thus, the presence of the target sequence in subtracter DNA, even as a major species, does not constitute a problem for the enrichment of specific target fragments. The proportions of individual sequences in subtracter DNA obtained from mixed populations with high sequence diversity, which is generally the case with environmental samples, are of course generally much lower than the proportions of individual sequences in the system which we analyzed.
Application of the procedure to obtain a specific probe for a clone derived from a sediment sample.
SW4 is a 16S rDNA sequence cloned from DNA extracted and amplified from the Spittelwasser sediment sample. This sequence was used as the target DNA for hybridization with subtracter DNA consisting of total 16S rDNA amplified from the same DNA sample. The sequence of the target DNA indicated that it was derived from a bacterium closely related to members of the Acidovorax branch of the β subclass of the Proteobacteria. An initial sequence analysis of 45 cloned 16S rDNAs obtained from this sediment sample showed that the majority of these 16S rDNAs belong to members of the β subclass of the Proteobacteria (data not shown). Subtractive hybridization by using the scheme shown in Fig. 1 led to the enrichment of a 68-nucleotide fragment representing a small hypervariable region corresponding to E. coli positions 964 to 1030 (3) within the 16S rRNA. This probe was used to screen the 45 sequenced clones of the 16S rDNA library obtained from the Spittelwasser sediment DNA extract. Positive hybridization signals were obtained exclusively with the six clones which exhibited sequence identity in the probe region and belonged to the SW4-Acidovorax sequence type (17). The probe gave no positive signals for any of the remaining 39 clones, whose sequences differed by 7 to 32 nucleotides in the probe region.
Comparison of the SW4 probe with sequences in the Ribosomal Database Project database led to the identification of two related sequences, both from Rhodoferax fermentans, a member of the β subclass of the Proteobacteria differing by four nucleotides in this region. Therefore, sequences in the currently known reference data set were not substantially more similar to the sequence of this enriched fragment than were the cloned and sequenced nontarget 16S rDNAs from the sediment sample used as the source for the subtracter DNA and hence were not useful for designing a probe specific for clone SW4.
Development of oligonucleotide probes based on sequences of enriched fragments.
The procedure described above yields probe fragments that are generated by restriction endonuclease cleavage and hence are variable in length. To standardize probe size and thus hybridization conditions, much shorter (18-nucleotide) oligonucleotide probes were designed from the enriched fragments with the aid of the Check-Probe program (9). One such oligonucleotide probe was synthesized and used to screen another 105 uncharacterized clones in the library made from the Spittelwasser sediment. This probe detected nine new clones, whose partial sequences revealed that they all belonged to the SW4-Acidovorax group. This information, in combination with the information on probe specificity obtained from the analysis of 45 randomly selected fully sequenced clones, demonstrated that the probe was highly specific for the Acidovorax clones within the context of the Spittelwasser sediment 16S rDNA clone library.
Influence of the composition of subtracter DNA on enriched probe sequences.
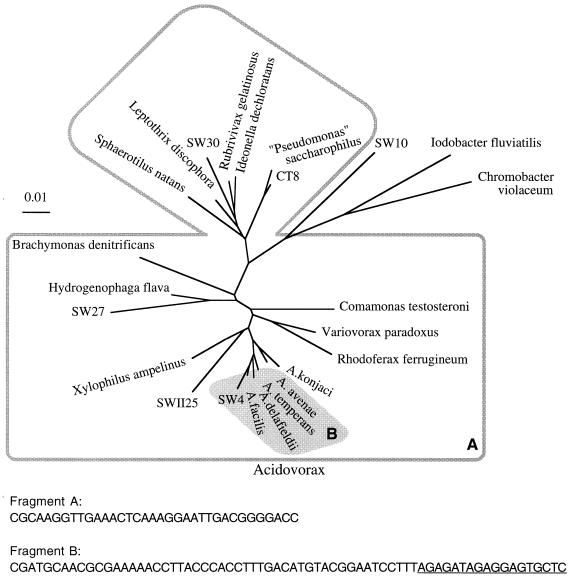
Two subtractive hybridization procedures were performed with target DNA from SW4. One was a repeat of the experiment described above with subtracter DNA amplified from Spittelwasser sediment DNA and resulted in enrichment of the same fragment. Another subtractive hybridization was performed with subtracter DNA amplified from DNA extracted from the biofilm grown in the Elbe River. Hybridization studies had previously shown that this subtracter DNA contains mainly sequences of members of the α subclass of the Proteobacteria (15). This subtractive hybridization led to the enrichment of two fragments, namely, the 68-nucleotide fragment described above and a 34-nucleotide fragment representing part of a more conserved region of the 16S rRNA (positions 897 to 933 [E. coli numbering]). The sequence of the more conserved fragment was identical to the sequences of the corresponding regions of all reference sequences from members of the Comamonadaceae branch of the β subclass of the Proteobacteria (Fig. 2) and to cloned sequences from Spittelwasser sediment which belonged to this group. Most of the 16S rRNA sequences of bacteria belonging to the β1 branch of the Proteobacteria show complete sequence conservation of this fragment. The fragment differed by only one or two nucleotides from the majority of the other cloned sequences from Spittelwasser sediment, which clustered in various subclasses of the Proteobacteria and the low-G+C-content gram-positive bacteria. The high proportion of Comamonadaceae-related sequences and the small sequence difference compared to many of the other sequences in the Spittelwasser 16S rDNA had obviously prevented the enrichment of this fragment when Spittelwasser 16S rDNA was the subtracter DNA. As there is no sequence information available for the Elbe River biofilm, we compared the sequence of the more conserved fragment to the database sequences of members of the most abundant group in this biofilm, namely, the α subclass of the Proteobacteria. This comparison revealed that most of these bacteria differed by two or three nucleotides in this region and that members of the genera Azospirillum and Acetobacter and the Rhodobacter-Rhodovulum group differed by only one nucleotide. Azospirillum spp. are nitrogen-fixing bacteria that mainly inhabit the rhizosphere, whereas Acetobacter spp. are typically isolated from ethanol-containing liquids and Rhodovulum spp. are typically of marine origin. Such bacteria would a priori not be expected in a river biofilm. Among this group of bacteria exhibiting sequence relatedness to the enriched fragment, only members of the Rhodobacter branch containing freshwater isolates might be expected to be present. The only fragment enriched by hybridization to subtracter consisting of predominantly β-proteobacterial sequences (Spittelwasser sediment) corresponded to a hypervariable region and therefore exhibited a specificity for a phylogenetically narrow group of bacteria. In contrast, the additional fragment enriched from the same target by hybridization to a subtracter consisting predominantly of sequences of members of the α subclass the Proteobacteria (biofilm community) corresponded to a more conserved region and was specific for a broader phylogenetic group (Fig. 2).
FIG. 2.
Phylogenetic specificity of the probes obtained from target sequence SW4. Two probe fragments were enriched from the SW4 target sequence by subtractive hybridization with subtracter DNA from Spittelwasser sediment and the Elbe River biofilm community. The dendrogram shows, within the framework of relevant reference strains, the phylogenetic relationships of the various 16S rDNAs cloned from the Spittelwasser sediment that group within the Comamonadaceae branch of the β subclass of the Proteobacteria and that react with the probe fragments. The clones in the larger group defined by the outline (area A) exhibit sequence identity with probe fragment A derived from the more conserved region of the 16S rDNA, whereas the clones in the shaded area (area B) react with probe fragment B derived from the hypervariable region of the Acidovorax 16S rDNA. The sequences (sense strands) of fragment A and fragment B are shown. The target sequence of the oligonucleotide designed from fragment B is underlined.
DISCUSSION
Investigations of microbial community structure and diversity in environmental samples generally include cloning and sequence determination of 16S rRNA genes obtained by PCR amplification of DNA extracted from the samples (7). Detection of minor members in a given environmental sample generally requires analysis of many isolates or cloned sequences, and the sequencing of clusters of identical or almost identical clones of dominant community members is time-consuming and labor-intensive. Two methods for sorting clones on the basis of similarity involve the determination of restriction endonuclease cleavage patterns and the analysis of sequence similarities by means of specific probes targeted to the 16S rDNA genes. The advantages of probes include (i) their convenience, (ii) their suitability for in situ studies, (iii) their ability to provide (semi)quantitative information, and (iv) the fact that they are phylogenetically based. However, probes are only as good as their specificities, and it is currently impossible to know the absolute specificity of any probe due to the paucity of reference information available. For example, several recently discovered bacterial lineages, such as the SAR11 cluster (7), whose members have been found in samples from different oceans (12), presently form isolated groups in current microbial phylogenetic schemes. Useful probes for such phylogenetically isolated sequences should include both probes that are highly specific for the known members and probes with a broader specificity to detect sequences that are related but not identical to the sequences of the known members. Neither type of probe can currently be reliably designed due to the paucity of sequence information on phylogenetically related organisms. Clearly, in the absence of adequate sequence information it is advantageous to take into account the unknown microbial background of the sampled environment by using the broadest possible sequence information in the sample. The sequence information to be analyzed is, within the bias restrictions of PCR amplifications, available in the 16S rDNA amplified from sample extracts. Subtractive hybridization between a cloned sequence from an environmental sample and the total 16S rDNA sequences in the sample can thereby lead to empirical selection of a region of the 16S rDNA for probe design on the basis of the sequence background of a particular habitat. We present here an assessment of the use of subtractive hybridization to develop probes specific for dominant organisms (e.g., strain Y1 in the 3CB enrichment culture) and minor organisms (e.g., the Elbe River biofilm organisms) in environmental samples.
In contrast to classical applications of subtractive hybridization (2, 5, 19) involving, for example, gene deletions (18), tissue-specific transcripts (13), and transcripts of genes transformed into cells (16), where there is little or no cross-hybridization between the sequence enriched and the subtracter sequences, the application described here involves two partners, both of which consist of well-conserved 16S rDNA which exhibits a maximal dissimilarity of about 35%. The hybridization thus identifies only sequence dissimilarities and does not indicate the presence or absence of a fragment. Moreover, the amplified 16S rDNA from the DNA extract of the sample in question which is used as subtracter contains some target DNA.
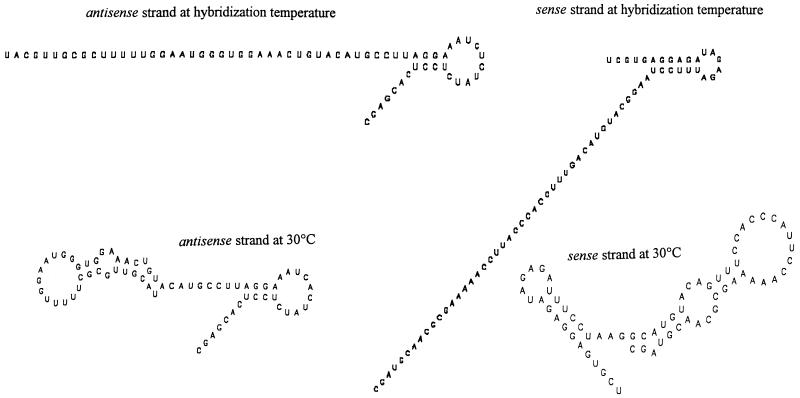
The differences between most known α-subclass proteobacterial 16S rRNA sequences and the fragment of the Acidovorax-like β-subgroup proteobacterial sequence enriched by subtractive hybridization with the biofilm 16S rDNA are small, only 2 or 3 nucleotides out of 34. Similar results were obtained in other experiments when targets with low similarities to the sequences of the dominant bacteria in the sample were used (data not shown). It may at first sight appear surprising that specific target fragments of this type can be enriched by nonoptimized, low-stringency hybridization. However, the displacement of short DNA fragments from hybrid duplexes by longer DNA fragments is a well-established procedure for detection of specific sequences in mixed samples of nucleic acids (4, 6) and is effective even for sequences differing by a single nucleotide when such sequences are very short. A similar strand displacement process also occurred in the solution hybridizations described here between short radioactively labeled target fragments and the 1.5-kb subtracter strands. In the case of the highly conserved 16S rDNA and when subtracter fragments are present at high concentrations, homologous regions displace short imperfectly complementary fragments. On the other hand, the highly ordered secondary structure of rDNA might reduce the accessibility of many potential hybridization sites, particularly for small fragments. Intrastrand duplexes within 16S rRNA are formed by both canonical and noncanonical base pairing, and similar but weaker structures can be formed in rDNA single strands. This is important since, in mixed-population PCR products, strands exactly complementary to each other meet only for the very abundant species and not for rare species. The more variable fragment of the Acidovorax-like sequence enriched by subtractive hybridization corresponds to a region in 16S rRNA which forms its own unit of secondary structure within the small-subunit rRNA. The enrichment of such fragments by subtractive hybridization might in principle therefore result from the formation of highly stable intrastrand secondary structures which prevent binding to subtracter DNA. To assess this possibility, the expected secondary structures of the sense and antisense strands of the enriched fragments for the temperature used for solution hybridization (56°C) and for a lower temperature (30°C) representing lower-stringency conditions were determined (8) (Fig. 3). At the hybridization temperature, little secondary structure was predicted, and only at temperatures of 30°C and lower was strong folding expected, but even under such conditions the helices of the secondary structures included noncanonical G-U pairs, which do not exist in DNA. It therefore seems unlikely that our procedure predominantly enriches fragments exhibiting a highly ordered secondary structure which restrict participation in hybridization with the subtracter DNA. Probably the worst-case situation in this regard is the generation of restriction endonuclease cleavage fragments containing entire regions known to form stable secondary structures. We have used the mfold program to analyze the predicted secondary structure under hybridization conditions (1 M NaCl, 56°C) of single-stranded DNA fragments representing the highly ordered 16S rRNA sequences at E. coli positions 240 to 268, 598 to 640, 1240 to 1299, and 1419 to 1481. Although short hairpins at positions 240 to 268 (8 bp; free energy = −0.2 kcal/mol), 598 to 640 (6 bp; −2.2 kcal/mol), 1240 to 1299 (4 bp; −3.3 kcal/mol), and 1419 to 1481 (11 bp; −1.7 kcal/mol) within longer sequences (60 bases) are predicted, all of the sequences except the sequence at positions 1419 to 1481 contain G-U pairs, the longest of which has 4 G-U pairs out of 11 bp. The short rather unstable secondary structures of these sequences therefore are not generally expected to be a problem for hybrid formation.
FIG. 3.
Predicted secondary structures of the narrow-specificity probe fragment obtained from an Acidovorax-like sequence by subtractive hybridization with 16S rDNA generated from Spittelwasser sediment. The probe specificity of the fragment (fragment B) is shown in Fig. 2. The predicted secondary structures of the sense and antisense strands at 30 and 56°C are shown; the latter temperature was used for subtractive hybridization. The NaCl concentrations included in the calculation were the same as those used in solution hybridization, although formamide was not included in the algorithm.
The results presented here demonstrate our ability to enrich for specific variable and conserved regions of cloned 16S rDNA. The use of bulk 16S rDNA containing many sequences similar to the target sequence as subtracter tends to lead to the enrichment of more hypervariable regions. When used as probes, these enriched fragments show strong overall specificity for the sequence originally used as the target. On the other hand, the use of target sequences for microbes belonging to an isolated taxon within the community led to the enrichment of additional, more conserved 16S rDNA fragments. These fragments are nevertheless distinct from the equivalent regions of the 16S rDNAs of the more dominant species present in the subtracter DNA. Thus, although such fragments may share sequence motifs with sequences of larger phylogenetic groups and thus exhibit low absolute specificity, they exhibit high specificity in the context of the relevant habitat. The β subclass of the Proteobacteria, members of which were used for the experiments presented here, is a bacterial group for which many 16S rRNA sequences are available. However, the main utility of the subtraction hybridization approach is the design of probes for minor and abundant microorganisms belonging to phylogenetically isolated and poorly characterized taxa which are present in the habitat under investigation.
ACKNOWLEDGMENTS
We thank Ed Moore and Heike Haas for stimulating discussions, Ingrid Schnellbacher for providing DNA extracts from biofilms, and the reviewer of an earlier version of the manuscript for many helpful suggestions.
This work was supported by grant 0319-433B from the German Ministry of Education and Research. K.N.T. expresses gratitude to the Fonds der Chemischen Industrie for generous support.
REFERENCES
- 1.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bjourson A J, Cooper J E. Isolation of Rhizobium loti strain-specific DNA sequences by subtraction hybridization. Appl Environ Microbiol. 1988;54:2852–2855. doi: 10.1128/aem.54.11.2852-2855.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brosius J, Palmer J L, Kennedy H P, Noller H F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins M, Fritsch E F, Ellwood M S, Diamond S E, Williams J I, Brewen J G. A novel diagnostic method based on DNA strand displacement. Mol Cell Probes. 1988;2:15–30. doi: 10.1016/0890-8508(88)90040-0. [DOI] [PubMed] [Google Scholar]
- 5.Darrasse A, Kotoujanansky A, Bertheau Y. Isolation by genomic subtraction of DNA probes specific for Erwinia carotovora subsp. atroseptica. Appl Environ Microbiol. 1994;60:298–306. doi: 10.1128/aem.60.1.298-306.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellwood M S, Collins M M, Fritsch E F, Williams J I, Diamond S E, Brewen J G. Strand displacement applied to assays with nucleic acid probes. Clin Chem. 1986;32:1631–1636. [PubMed] [Google Scholar]
- 7.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 8.Jaeger J A, Turner D H, Zuker M. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 1990;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- 9.Lane D. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 10.Maidak B J, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1996;24:82–85. doi: 10.1093/nar/24.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligonucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 12.Mullins T D, Britschgi T B, Krest R L, Giovannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplancton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 13.Rodriguez I R, Chader G J. A novel method for the isolation of tissue-specific genes. Nucleic Acids Res. 1992;20:3528. doi: 10.1093/nar/20.13.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 15.Schnellbacher I. Einsatz von phylogenetischen Sonden zur Untersuchung einer mikrobiellen Gemeinschaft in Elbe-Aufwuchs. Diploma thesis. Braunschweig, Germany: Technical University of Braunschweig; 1995. [Google Scholar]
- 16.Scott M R D, Westphal K-H, Rigby P W C. Activation of mouse genes in transformed cells. Cell. 1983;34:557–567. doi: 10.1016/0092-8674(83)90388-4. [DOI] [PubMed] [Google Scholar]
- 17.Stackebrandt E, Rainey F A. Partial and complete 16S rDNA sequences, their use in generation of 16S rEDNA phylogenetic trees and their implications in molecular ecological studies. In: Akkermans A D L, van Elsas J D, de Bruin F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. p. 3.1.1. [Google Scholar]
- 18.Straus D, Ausubel F M. Genomic subtraction for cloning DNA corresponding to deletion mutations. Proc Natl Acad Sci USA. 1990;87:1889–1893. doi: 10.1073/pnas.87.5.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welcher A A, Torres A R, Ward D C. Selective enrichment of specific DNA, cDNA and RNA sequences using biotinylated probes, avidin and copper-chelate agarose. Nucleic Acids Res. 1986;24:10027–10044. doi: 10.1093/nar/14.24.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. pp. 2.4.1–2.4.5. [DOI] [PubMed] [Google Scholar]