Abstract
The influence of trichloroethylene (TCE) on a mixed culture of four different toluene-degrading bacterial strains (Pseudomonas putida mt-2, P. putida F1, P. putida GJ31, and Burkholderia cepacia G4) was studied with a fed-batch culture. The strains were competing for toluene, which was added at a very low rate (31 nmol mg of cells [dry weight]−1 h−1). All four strains were maintained in the mixed culture at comparable numbers when TCE was absent. After the start of the addition of TCE, the viabilities of B. cepacia G4 and P. putida F1 and GJ31 decreased 50- to 1,000-fold in 1 month. These bacteria can degrade TCE, although at considerably different rates. P. putida mt-2, which did not degrade TCE, became the dominant organism. Kinetic analysis showed that the presence of TCE caused up to a ninefold reduction in the affinity for toluene of the three disappearing strains, indicating that inhibition of toluene degradation by TCE occurred. While P. putida mt-2 took over the culture, mutants of this strain which could no longer grow on p-xylene arose. Most of them had less or no meta-cleavage activity and were able to grow on toluene with a higher growth rate. The results indicate that cometabolic degradation of TCE has a negative effect on the maintenance and competitive behavior of toluene-utilizing organisms that transform TCE.
A limited number of chlorinated aliphatic hydrocarbons can support bacterial growth by serving as a source of carbon and energy (17). In these cases, treatment of polluted sites or waste streams can be performed by using systems in which the number of desirable microorganisms increases because they proliferate at the expense of the contaminants. However, many chlorinated compounds are biodegradable only by cometabolic conversion. In this case, the xenobiotic compound does not cause a selective increase in the population of the active microorganisms. A primary substrate must be present for growth and maintenance, which, however, does not necessarily select for the desired degradative activity. Besides, conversion of the cosubstrate could lead to toxic products which inhibit the most active organisms.
The cometabolic conversion of trichloroethylene (TCE) by nonspecific oxygenases of aerobic bacteria is an example of a process which may harm the bacteria that execute the degradation reaction. The reaction uses reducing equivalents (9), and TCE can be a competitive inhibitor of the conversion of the primary growth substrate, which is usually also required to induce the oxidative enzyme. Furthermore, TCE conversion can be accompanied by toxic effects; i.e., cell damage may occur due to nonspecific reactions of TCE conversion products with cell components, as was shown with cultures of Pseudomonas putida F1 and Methylosinus trichosporium OB3b (27, 35). These toxic effects can lead to a large increase in maintenance energy demand, as was observed in a nongrowing, toluene-limited, fed-batch culture of Burkholderia cepacia G4, which converted TCE during cultivation on toluene as the primary substrate (22). However, despite these negative effects, cometabolic conversion is the only possibility for aerobic biodegradation of TCE and deserves significant attention, since no organisms which can grow on this compound are known.
When organisms are applied for cometabolic TCE degradation during in situ bioremediation processes, they might frequently face situations with very low substrate concentrations. Also, in reactors employing biofilms, the amount of growth substrate that is added is usually kept as low as possible to prevent the formation of excess biomass. However, under energy-limiting conditions, negative effects of TCE conversion, such as an increased maintenance coefficient, could give TCE degraders a large disadvantage compared to organisms that also use the primary substrate but do not degrade TCE. This could limit the long-term stability of a treatment process and lead to reduced conversion rates.
The purpose of the work described in this paper was to determine the competitiveness of TCE-degrading bacteria at very low primary substrate concentrations in the absence and presence of TCE. Toluene was taken as a model primary substrate.
Toluene degradation can start with oxidation of the methyl group or by direct oxidation of the aromatic ring. The first route is used by P. putida mt-2, which converts toluene via benzylalcohol, benzaldehyde, and benzoate to catechol (1). Direct oxidation of the aromatic ring is performed by, for example, B. cepacia G4 (25, 26) and P. putida F1 (34). B. cepacia G4 converts toluene via o-cresol to 3-methylcatechol (3MC) by two subsequent monooxygenase steps (32). P. putida F1 uses a dioxygenase to convert toluene to toluene dihydrodiol, which is subsequently oxidized to 3MC (34).
To study the effect of TCE on the competition for small amounts of toluene between microorganisms, four different toluene degraders were cultivated together in a fed-batch culture, which allows the use of very low toluene concentrations (5). Two of the strains (B. cepacia G4 and P. putida F1) are well known for their ability to cometabolically convert TCE when toluene is the primary substrate. P. putida mt-2 is unable to degrade TCE, while P. putida GJ31 (23) converts TCE slightly. The results show that when TCE was added to the culture, P. putida mt-2 and mutants thereof became the dominant organisms, which caused a loss of the TCE degradation capacity.
MATERIALS AND METHODS
Nomenclature.
The following parameters are used in this paper: ad, specific decay rate (minute−1); C, substrate concentration (micromolar); H, dimensionless Henry’s coefficient; kLa, mass transfer rate coefficient (minute−1); Ki, inhibition constant (micromolar); Km, Michaelis-Menten constant (micromolar); Ks, Monod constant (micromolar); μmax, maximal specific growth rate (minute−1); rmax, maximal specific substrate conversion rate (micromoles milligram of cells [dry weight]−1 minute−1); t, time (minutes); V, volume (liters); X, concentration of biomass (milligrams of cells [dry weight] liter−1); and Y, growth yield (milligrams of cells [dry weight] micromole−1). Subscripts denote the following parameters: g, gas phase; l, liquid phase; and 0, time zero.
Bacterial strains.
B. cepacia G4 (26) was a gift from M. S. Shields (U.S. Environmental Protection Agency, Gulf Breeze, Fla.). P. putida F1 (34) was provided by L. P. Newman (University of Minnesota, St. Paul). P. putida GJ31 was previously characterized (23), and P. putida mt-2 (1) was a gift from W. Duetz (RIVM, Bilthoven, The Netherlands).
Culture conditions.
The organisms were grown in a fed-batch fermentor with a working volume of 2.5 liters. The mineral medium (MMV) contained (per liter) 5.3 g of Na2HPO4 · 12H2O, 1.4 g of KH2PO4, 0.2 g of MgSO4 · 7H2O, 1.0 g of (NH4)2SO4, 1 ml of a vitamin solution (16), and 5 ml of a trace element solution. The trace element solution contained (per liter) 780 mg of Ca(NO3)2 · 4H2O, 200 mg of FeSO4 · 7H2O, 10 mg of ZnSO4 · 7H2O, 10 mg of H3BO3, 10 mg of CoCl2 · 6H2O, 10 mg of CuSO4 · 5H2O, 4 mg of MnSO4 · H2O, 3 mg of Na2MoO4 · 2H2O, 2 mg of NiCl2 · 6H2O, and 2 mg of Na2WO4 · 2H2O. Each component of the medium was autoclaved separately, except the phosphates and the trace elements, which were autoclaved together, and the vitamin solution, which was filter sterilized prior to addition. The pH was adjusted to 7.2 with autoclaved 0.5 M NaOH and 0.25 M H2SO4. The temperature was set at 28°C, and the impeller speed was set at 1,500 rpm.
Toluene was supplied to the culture via the gas phase. This was done by leading an airflow via a glass filter (P3; Elgebe, Leek, The Netherlands) through ice-cold toluene prior to addition to the culture. TCE was added via the gas phase from a gas cylinder containing 475 ppm of TCE in air (AGA Gas BV, Amsterdam, The Netherlands). Extra water-saturated air was added to the culture to supply sufficient oxygen. The flow rates of the gases are given in Table 1. All gas flows were filter sterilized before addition to the culture. Flows were controlled with mass flow controllers (type F201C-FA-11-V; 0 to 5 ml min−1, 0 to 20 ml min−1, and 0 to 500 ml min−1; Bronkhorst High-Tec B.V., Veenendaal, The Netherlands). The outgoing gas stream was led through a water column under slight overpressure, which facilitated the detection of possible leakage.
TABLE 1.
Substrate supply parameters
| Parameter (unit) | Value in fed-batch culture grown on:
|
|
|---|---|---|
| Toluene (0–40 days) | Toluene + TCE (40–83 days) | |
| Airflow (ml min−1) | 53.2 | 31.8 |
| Toluene flow (ml min−1) | 2.0 | 2.0 |
| TCE flow (ml min−1) | 22.5 | |
| [Toluene] in ingoing gas stream (μM)a | 13.3 | 13.0 |
| [TCE] in ingoing gas stream (μM)a | 7.1 | |
Average taken from gas chromatography measurements.
Analysis of the culture.
The culture density of each sample was estimated by measuring the optical density at 450 nm (OD450) on a Hitachi 100-60 spectrophotometer and by determining the protein concentration as described by Lowry et al. (21) with bovine serum albumin as a standard. The dry weight of the culture was determined after 40 days by centrifuging duplicate 100-ml samples of culture (15 min, 6,000 × g, 4°C), washing the pellets with the same volume of cold demineralized water, and drying the pellets to a constant weight in a preweighed aluminum cup for 2 to 3 days at 80°C.
The viability of the culture was determined by diluting a culture sample and counting the number of colonies (CFU) after the sample was plated on 1.5% agar plates containing either rich medium (nutrient broth [NB] plates) or mineral medium to which 10 mg of yeast extract liter−1 was added (MMY) instead of the vitamin solution. One day after plating, 5 to 10 μl of pure toluene (Tol plates) was added on a paper filter disk in the cover of the petri dish. This was done to prevent the immediate exposure of the cells to high toluene concentrations. The plates were incubated at 30°C. Colonies on rich plates appeared after 1 to 2 days, and those on Tol plates were visible after 3 to 4 days.
The different strains in the mixed fed-batch culture were distinguished on NB plates by colony morphology and growth rate and by plating diluted culture samples on selective plates followed by counting the CFU on each plate. The selective plates were made by adding a particular volatile carbon source on a paper filter disk which was placed in the cover of MMY plates 1 day after plating. The plates were incubated at 30°C. Colonies appeared after 3 to 4 days. Of the four strains studied, only P. putida GJ31 is able to grow on chlorobenzene plates. P. putida mt-2 could be identified by growth on p-xylene plates, and B. cepacia G4 could be identified by growth on o-cresol plates. The number of P. putida F1 cells was determined by the difference between the viability on m-cresol plates (on which both P. putida F1 and B. cepacia G4 grow) and the viability on o-cresol plates (on which P. putida F1 does not grow). Also, P. putida F1 and B. cepacia G4 could be distinguished on m-cresol plates because the colonies of B. cepacia G4 were larger than the colonies of P. putida F1.
The percentage of cells of each strain that formed colonies on NB plates and were also able to grow on toluene and on a selective substrate was measured by replica plating colonies obtained from NB plates onto Tol plates and onto selective plates. Replica-plated colonies on Tol plates were screened for the presence of catechol 2,3-dioxygenase (C23O) by spraying the plates with a 100 mM catechol solution. Positive colonies of P. putida mt-2 turn yellow due to conversion of catechol to 2-hydroxymuconic semialdehyde (HMS) (36). Mutants unable to grow on any of the selective substrates were further analyzed for the ability to grow on other substrates in batch cultures containing 1 or 5 mM substrate in MMV.
Plasmid extractions were done by using a modified method of Kado and Liu (18), as described by Duetz et al. (7). Southern hybridizations and chemiluminescent detection of plasmid DNA digested with EcoRI, SalI, or XhoI were performed with a digoxigenin-labeled probe containing the promoter region of the meta operon (Pm) and xylXYZ′ or containing xylTE′ (GenBank accession number M64747) as described by the manufacturer (Boehringer, Mannheim, Germany). The probes were obtained from the plasmids pGSH3537 (2.8-kb SacI-KpnI fragment) and pAW31 (1.2-kb SalI fragment) (5a).
The decay rate of the strains in the fed-batch culture exposed to TCE was calculated by using the formula xt/x0 = e−adt, in which xt is the viability on a selective plate at time t and x0 is the viability on a selective plate at time zero, when TCE addition to the fed-batch culture started. ad is the specific decay rate (2).
Protein profiles of the strains were made by centrifuging 60 μl of an overnight NB culture, resuspending the pellet in 10 μl of 1× loading buffer (31), and applying the mixture after boiling (5 min) on a 12.5% polyacrylamide gel containing sodium dodecyl sulfate. The gels were stained with Coomassie brilliant blue.
Enzyme assays.
Mutants of P. putida mt-2 were analyzed for the presence of enzymes of the meta- and ortho-cleavage pathways. For this, cells were grown overnight on 1 mmol of toluene h−1 in a 2.5-liter fed-batch culture. Cells were harvested by centrifugation and washed with either ice-cold mineral medium (MM) or ice-cold 0.1 M Tris-HCl (pH 7.5) containing 0.1 mM 1,4-dithiothreitol (TD). After resuspension of the cells in a small volume of MM or TD, they were used for the enzyme assays, which were performed at 30°C.
The cells which were washed and resuspended in MM were used immediately for oxygen uptake experiments. For this, cells were added to a small, magnetically stirred incubation vessel to which an oxygen electrode (O2 sensor type 12/220; Ingold, Urdorf, Switzerland) was connected. After determination of the endogenous oxygen consumption rate of the resting cell suspension, a concentrated solution of benzoate, m-toluate, or p-toluate was added to give a final concentration of 5 mM. The difference between the oxygen consumption rates before and after addition of the substrate was used to calculate the specific rate of oxidation of the substrate in micromoles gram of cells (dry weight)−1 minute−1.
The cells which were washed and resuspended in TD were disrupted by sonification and centrifuged for 30 min in an Eppendorf centrifuge (10,000 × g, 4°C). The clear supernatant solution was used as a source of crude cell extract. The protein content of the extract was determined with Coomassie brilliant blue, with bovine serum albumin as a standard.
Catechol 1,2-dioxygenase (C12O), C23O, 2-hydroxymuconic semialdehyde hydrolase (HMSH), and 2-hydroxymuconic semialdehyde dehydrogenase (HMSD) activities in the crude cell extract were measured as described previously (23). The conversion of catechol, 3MC, and 4-methylcatechol (4MC) to their corresponding meta-cleavage products was monitored at 375, 388, and 382 nm respectively. The extinction coefficients of HMS, 2-hydroxy-6-oxohepta-2,4-dienoic acid (HODA), and 2-hydroxy-5-methylmuconic semialdehyde (HMMS) are 36,000, 16,800, and 31,500 liters mol−1 cm−1, respectively (30). HMS, HODA, and HMMS were prepared in situ by incubation of solutions containing catechol, 3MC, or 4MC, respectively, in 45 mM phosphate buffer (pH 7.4) with purified C23O of P. putida mt-2 and were used to determine HMSH and HMSD activities. C23O was purified from a crude extract of P. putida mt-2 by using an acetone precipitation step followed by ion-exchange and hydrophobic interaction chromatographies.
Analytical methods.
TCE and toluene were measured in the gas phase on a CP 9001 gas chromatograph equipped with a CP Sil 5 CB column (length, 25 m; diameter, 0.53 mm) (Chrompack, Middelburg, The Netherlands) and a flame ionization detector. The carrier gas (helium) pressure was 150 kPa, and the oven temperature was 100°C.
Chloride was measured with a colorimetric assay (3).
Estimation of kinetic parameters.
The kinetic parameters (Ks and μmax) of the strains used in the mixed fed-batch culture were estimated from substrate depletion curves obtained with growing batch cultures (29). For this, the strains were grown on toluene in a 3-liter bioreactor on 0.75 liter of mineral medium at 30°C at a stirring speed of 1,050 rpm. The depletion of toluene was measured by on-line analysis of the toluene concentration in the headspace of the batch culture by gas chromatography. The headspace was continuously circulated at a rate of 100 to 120 ml min−1 with a micromembrane pump (NMP 02LU; KNF Neuberger GmbH, Freiburg-Munzingen, Germany). After passage through a Valco six-port sampling injector (Vici AG, Schenkon, Switzerland) to which a 35-μl sample loop was connected, the gas was injected back into the culture. Samples of 35 μl were automatically injected into the gas chromatograph every 5 min.
The obtained substrate depletion curves were described with a model in which the Monod equation and gas-liquid mass transfer of the substrate are incorporated, with biomass (X) and gas and liquid phase concentrations (Cg and Cl) as variables. The volumes of the gas and liquid phases (Vg and Vl) were 0.75 and 2.25 liters, respectively. Values between 0.048 and 0.057 mg of cells (dry weight) μmol−1 were taken for the yield (Y) of the different strains and were determined from batch cultures of each strain growing on 1 mM toluene. The dimensionless Henry’s coefficient (H) was determined as described by Diks (6) (Htoluene, 30°C = 0.27; HTCE, 30°C = 0.5). The mass transfer coefficient (kLa) for toluene was determined to be 0.14 min−1 by a procedure described by van Hylckama Vlieg et al. (33). The model consists of three equations: X = X0 + [(Cg,0 − Cg)Vg + (Cl,0 − Cl)Vl]Y/Vl, dCg/dt = −kLa(Cg/H − Cl)Vl/Vg, and dCl/dt = kLa(Cg/H − Cl) − μmaxCl/(Cl + Ks)X/Y.
The parameters Ks, μmax, and the initial concentrations of toluene in the gas and liquid phases (Cg,0 and Cl,0) were fitted to the numerically integrated equations by using the episode routine in Scientist for Windows 2.0 (Micromath Scientific Software, Salt Lake City, Utah). The square of the difference between the measured and fitted values was multiplied by 1/(Cg + 0.1) at each time point. The sum of these relative squares was minimized. This way, the data points at lower toluene concentrations have the same weight as the data points at higher toluene concentrations, while the more inaccurate values close to the detection limit (35 nM) are less important. The data points from the first hour were usually omitted to ensure that the system was in equilibrium.
The TCE-degrading capacities of the strains were tested with toluene-grown cells obtained from a 2-liter overnight fed-batch culture grown at a toluene addition rate of about 500 μmol h−1 to a density of 0.15 to 0.4 mg (dry weight) ml−1. Cells were harvested by centrifugation and resuspended in MM to a final volume of 25 ml. The depletion of TCE was measured by on-line analysis of the TCE concentration in the headspace of the resting cell-suspension, which was placed in a 120-ml stainless steel incubation vessel at 30°C. TCE concentrations were determined every minute in the same way as for the toluene depletion experiments described above. The culture was stirred at 1,000 rpm. The first-order rate constant of TCE degradation, which is equal to the maximum rate of substrate conversion (rmax) divided by the Michaelis-Menten constant (Km), was determined from the first-order region of the TCE depletion curve. This was done by plotting the natural logarithm of the TCE concentration in the gas phase against time and multiplying the slope of this line by −(VgH + Vl)/XVl.
RESULTS
Competition for toluene.
The competitive capacities of the toluene-degrading strains P. putida GJ31, P. putida mt-2, P. putida F1, and B. cepacia G4 were studied with a mixed fed-batch culture at a very low toluene concentration. For this, cells from batch cultures of each strain were added to the fed-batch bioreactor, and the mixed culture was grown at a toluene addition rate of about 600 μmol h−1 until the culture reached an OD450 of 3. The toluene load was then reduced to 44 μmol h−1. Previous experiments with B. cepacia G4 had shown that at this combination of toluene load and culture density, the specific growth rate on toluene is very low (22).
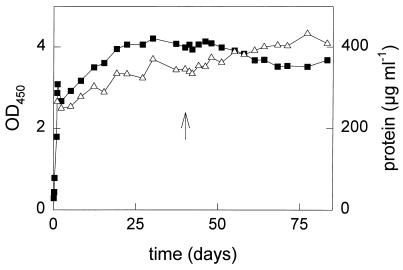
During 2 weeks, the culture density of the mixed fed-batch culture slowly increased to an OD450 of 4, after which it became constant (Fig. 1), meaning that hardly any net growth occurred and that all toluene added was used for maintenance of the culture. After approximately 1 week of slow growth, no toluene could be detected in the outgoing gas stream (detection limit, 35 nM), which means that more than 99.7% of the added toluene was converted (Fig. 2A) and that growth was not limited due to exhaustion of other nutrients. The dry weight of the culture was determined after 40 days, and the toluene conversion of the hardly growing, mixed fed-batch culture was calculated to be 31 nmol mg of cells (dry weight)−1 h−1.
FIG. 1.
Culture density of the fed-batch culture cultivated on toluene in the absence and presence of TCE. The OD and protein concentration were determined as a measure of the culture density. Symbols: ▪, OD450; ▵, protein concentration; ↑, time at which TCE addition was started.
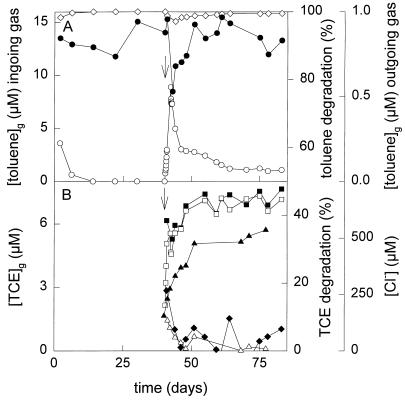
FIG. 2.
Conversion of toluene (A) and TCE (B) by the fed-batch culture. The concentrations of toluene and TCE in the ingoing and outgoing gas streams were determined by gas chromatography. The chloride concentration in the culture medium was determined with a colorimetric assay. Symbols: •, toluene in the ingoing gas stream; ○, toluene in the outgoing gas stream; ◊, percentage of toluene degraded by the culture; ▪, TCE in the ingoing gas stream; □, TCE in the outgoing gas stream; ⧫, percentage of TCE degraded by the culture (calculated from the concentrations of TCE in the in- and outgoing gas streams); ▴, chloride concentration; ▵, percentage of TCE degraded by the fed-batch culture (calculated from chloride measurements); ↓, time at which TCE addition was started.
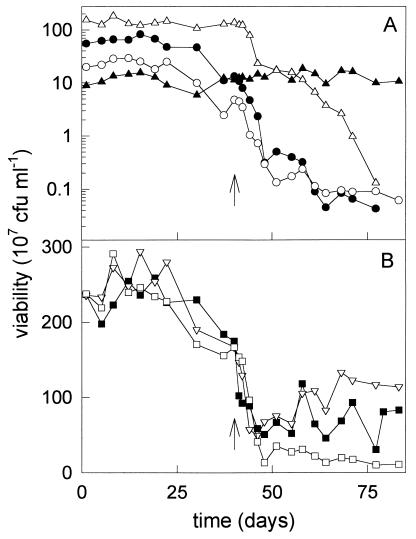
The population composition of the culture was determined by counting the colonies on selective plates (Fig. 3A). This corresponded to the viability of each strain on NB plates, which could be determined based on colony morphology. Just after the reduction of the toluene load from about 600 to 44 μmol h−1, the contribution of each strain to the total viability of the mixed culture varied between 4% (for P. putida mt-2) and 64% (for P. putida F1). During the cultivation at a very low toluene concentration, the population composition hardly changed, and all four strains were maintained for at least 40 days in the culture. The sum of the viabilities of the strains on selective plates was similar to the total viability of the mixed culture, as measured on NB and Tol plates (Fig. 3B). This means that all four strains were able to grow on their selective substrate. This was confirmed by replica plating colonies derived from NB plates to selective plates.
FIG. 3.
Population composition (A) and viability (B) of the fed-batch culture over time, determined as CFU on different agar plates. Symbols: •, P. putida GJ31 on chlorobenzene plates; ▴, P. putida mt-2 on p-xylene plates; ○, B. cepacia G4 on o-cresol plates; ▵, P. putida F1, determined as the difference between the viability on m-cresol and o-cresol plates; ▪, viability on NB plates, ▿, viability on Tol plates; □, sum of viabilities of the four strains on selective plates; ↑, time at which TCE addition was started.
Competition for toluene in the presence of TCE.
To study the influence of TCE on the competitive behavior of the four strains at very low toluene concentrations, TCE was added to the culture at a rate of 24 μmol h−1, starting after 40 days of fed-batch cultivation on toluene. During TCE addition, the culture density slowly decreased to an OD450 of 3.6 at 83 days. Immediately after the start of TCE addition, the toluene concentration in the outgoing gas stream increased to a maximum of around 500 nM after 2 days of TCE addition. It then slowly decreased to approximately twice the detection limit (Fig. 2A). Approximately 17% of the incoming TCE was degraded after 1 day, but this became less than 3% after 4 days (Fig. 2B). The chloride concentration slowly increased in the fed-batch culture (Fig. 2B).
Within a few days after the start of the addition of TCE, the viability of B. cepacia G4, measured as CFU on o-cresol plates, decreased with a specific decay rate of 0.0037 h−1 (r2 = 0.70). P. putida GJ31 and P. putida F1 showed similar behavior. The viability on chlorobenzene plates and the difference in viability on m-cresol plates and o-cresol plates decreased with specific decay rates of 0.0064 (r2 = 0.84) and 0.0068 (r2 = 0.94) h−1, respectively. The viability of P. putida mt-2 on p-xylene plates remained at a constant level (Fig. 3A). These results show that only a small amount of TCE could be converted by the mixed culture and that the TCE degradation capacity decreased over time, since the TCE-converting organisms were outcompeted by P. putida mt-2.
Accumulation of mutants.
After 1 week of TCE addition, a difference arose between the sum of the viabilities of the strains on selective plates and the viabilities of the mixed culture on NB plates and on Tol plates (Fig. 3B). To check if all the cells were still able to grow on one of the selective substrates, colonies derived from NB plates were replica plated on selective plates. An increasing amount of colonies was no longer able to grow on p-xylene, o-cresol, m-cresol, or chlorobenzene, indicating that these colonies are mutants.
The colony morphologies of these mutants were similar to that of P. putida mt-2. The protein profiles of the mutants were also found to be similar to the profile of the wild-type P. putida mt-2 by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown), indicating that they were mutants of P. putida mt-2. The mutants were further examined by replica plating on Tol plates and on the selective plates. Cells grown on Tol plates were checked for C23O (XylE) activity by spraying with catechol. If XylE is active, the colonies rapidly become yellow due to the conversion of catechol to the yellow HMS.
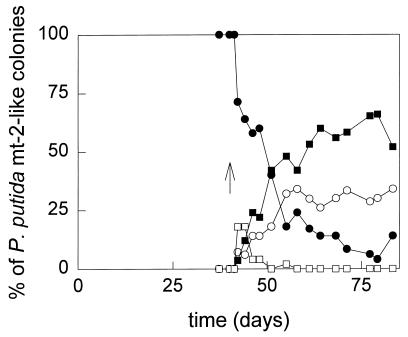
Three classes of mutants were distinguished. One class of mutants was no longer able to grow on toluene or on any of the other selective substrates. The mutants possessed a plasmid that was much smaller than the wild-type TOL plasmid. It did not hybridize with the Pm-xylXYZ′ and xylTE′ probes, indicating that it lacks the 39-kb fragment encoding the catabolic genes for toluene and xylene growth. Loss of this fragment also occurs during growth on benzoate (see, e.g., reference 37). Immediately after the addition of TCE, this phenotype was observed for approximately 20% of the P. putida mt-2-like colonies. However, this class of mutants disappeared after 1 week (Fig. 4).
FIG. 4.
Appearance of mutants of P. putida mt-2 during fed-batch cultivation. Colonies with a morphology similar to that of P. putida mt-2 on NB plates were replica plated on Tol plates and selective plates to determine their phenotype. Activity of XylE was determined by spraying Tol plates with replica-plated colonies with a catechol solution and checking for the formation of the yellow product HMS. Symbols: •, colonies able to grow on p-xylene and toluene and forming HMS after being sprayed with catechol (Tol+ p-Xyl+ XylE+ [wild-type]); □, colonies unable to grow on p-xylene and toluene (Tol− p-Xyl−); ▪, colonies able to grow on toluene, unable to grow on p-xylene, and not forming HMS when sprayed with catechol (Tol+ p-Xyl− XylE−); ○, colonies able to grow on toluene, unable to grow on p-xylene, and forming of HMS after being sprayed with catechol (Tol+ p-Xyl− XylE+); ↑, time at which TCE addition was started.
A second class of mutants was XylE+ and could still grow on toluene, m-xylene, and m-toluate but not on p-xylene or p-toluate. Approximately one-third of the mutants which could still grow on toluene but not on the selective plates belonged to this class (Tol+ p-Xyl− XylE+) (Fig. 4). The specific rate at which these mutants appeared was estimated from the increase in their number over time by multiplying the number of P. putida mt-2-like colonies on NB plates by the percentage of Tol+ p-Xyl− XylE+ mutants, which was determined by replica plating at each time point, and was found to be 0.032 h−1 (r2 = 0.65). No differences in plasmid size could be seen on agarose gels, and the plasmid still hybridized with the xylTE′ and Pm-xylXYZ′ probes. Southern hybridizations of EcoRI-, XhoI-, or SalI-digested plasmid DNA with the Pm-xylXYZ′ and xylTE′ probes also revealed no differences. However, in a crude extract of toluene-grown cells of one of these mutants (P. putida mt-2M11), the activities of three enzymes of the meta-cleavage pathway with different substrates were approximately one-fifth of the activities found with wild-type cells (Table 2). Also, cells of P. putida mt-2M11 which were grown on toluene no longer oxidized p-toluate (Table 3). In P. putida mt-2, p-toluate is oxidized by a toluate dioxygenase (XylXYZ) which can also convert benzoate and m-toluate. The xylXYZ genes are located in the meta operon of the TOL plasmid of P. putida mt-2, directly behind the promoter of the operon (1). The chromosomally encoded equivalent of XylXYZ is a benzoate dioxygenase, which can convert only benzoate and m-toluate (15, 28). The data suggest that the mutants of class 2 have a small mutation in the promoter region of the meta operon, which could not be detected by Southern analysis but which influences the expression of the meta pathway and leads to the absence of activity of XylXYZ.
TABLE 2.
Specific activities of enzymes of the ortho and meta pathways in crude extracts of P. putida mt-2 and mutants that originated from this strain
| Enzyme | Substrate | Sp act (U g of protein−1) of enzyme in crude extracts of P. putida:
|
|||
|---|---|---|---|---|---|
| mt-2 | mt-2M11 | mt-2M10 | mt-2M4 | ||
| C12O | Catechol | 22 | 29 | 77 | 55 |
| C23O | Catechol | 4,466 | 878 | 46 | 0 |
| 3-MC | 1,316 | 299 | 20 | 0 | |
| 4-MC | 2,403 | 421 | 30 | NDa | |
| HMSH | HMS | 184 | 26.6 | 2.5 | 0 |
| HODA | 529 | 85.6 | 4.3 | ND | |
| HMMS | 165 | 24.4 | 1.4 | ND | |
| HMSD | HMS | 110 | 33.2 | 0.4 | 0 |
| HODA | 46.5 | 4.2 | 0.4 | ND | |
| HMMS | 150 | 35.0 | 1.1 | ND | |
ND, not determined.
TABLE 3.
Oxygen uptake by washed resting cell suspensions of P. putida mt-2 and of mutants which originated from this strain
| Substrate | Oxygen consumption (μmol g of cells [dry weight]−1 min−1) by cells of P. putida:
|
|||
|---|---|---|---|---|
| mt-2 | mt-2M11 | mt-2M10 | mt-2M4 | |
| Benzoate | 197 | 84 | 269 | 100 |
| m-Toluate | 112 | 37 | 22 | 5.7 |
| p-Toluate | 101 | —a | — | — |
—, no activity detectable.
The third class of mutants was still able to grow on toluene but not on p-xylene or p-toluate. When colonies of this class were sprayed with catechol, little or no yellow coloration occurred. The specific rate at which these Tol+ p-Xyl− XylE− mutants appeared was 0.064 h−1 (r2 = 0.90). Most of these mutants grew very poorly on m-xylene and m-toluate, while the medium became brown, which indicates the accumulation of a (substituted) catechol. One of the mutants (P. putida mt-2M4) was unable to grow on m-xylene at all. It had a plasmid which was slightly smaller than the TOL plasmid of P. putida mt-2 and did not hybridize with a Pm-xylXYZ′ or a xylTE′ probe. No activity of enzymes of the meta pathway could be detected (Table 2). This suggested that P. putida mt-2M4 lacks all or a large part of the genes of the meta-cleavage pathway. A similar mutation was found by Brinkmann et al. (4) during unlimited growth of P. putida on toluene.
The other mutants of the third phenotypic class contained a plasmid which still hybridized with both probes and seemed to have the same size as the wild-type plasmid. However, Southern analysis showed that both the SalI and EcoRI fragments of the plasmid encoding part of xylX and the promoter region were about 0.4 kb smaller than the corresponding wild-type fragments. The activities of enzymes of the meta pathway were largely reduced in a crude extract of a toluene-grown mutant of this class (P. putida mt-2M10) (Table 2), indicating that the deletion caused a large reduction of the expression of the meta pathway. None of the mutants of class 3 could oxidize p-toluate (Table 3), indicating that XylXYZ is not active in these cells.
Kinetic analysis of toluene-degrading strains.
To determine the kinetic basis for the observed population changes and appearance of mutants, the kinetic parameters (μmax and Ks) of each strain were determined by on-line gas chromatographic analysis of toluene depletion from the headspaces of batch cultures. An example of a depletion curve is presented in Fig. 5A. The data show that B. cepacia G4 has a much lower affinity (μmax/Ks) for toluene than the other three strains, while P. putida GJ31 has the best kinetics for growth on toluene (Table 4).
FIG. 5.
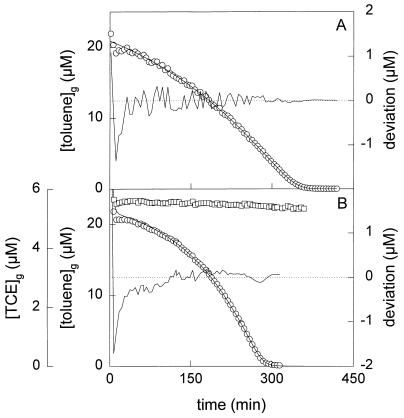
Toluene depletion curves obtained with batch cultures of P. putida mt-2 growing on toluene in the absence of TCE (A) and P. putida GJ31 growing on toluene in the presence of TCE (B). The concentrations of toluene (○) and TCE (□) in the headspace of the culture were determined. The lines through the open circles are the fitted toluene depletion curves. The other lines show the deviation of the data points from the fitted curve.
TABLE 4.
Effect of TCE on the Monod parameters for growth on toluenea
| Strain | Substrate(s) | Ksobs (μM) | μmax (h−1) | Affinity (μmax/Ksobs) |
|---|---|---|---|---|
| B. cepacia G4 | Toluene | 26.4b | 0.30b | 0.011 |
| Toluene + TCEc | 74.7 | 0.39 | 0.005 | |
| P. putida F1 | Toluene | 1.5b | 0.65b | 0.433 |
| Toluene + TCE | 11.2 | 0.60 | 0.054 | |
| P. putida mt-2 | Toluene | 3.3 | 0.25 | 0.075 |
| Toluene + TCE | 7.3 | 0.28 | 0.038 | |
| P. putida GJ31 | Toluene | 0.4 | 0.53 | 1.325 |
| Toluene + TCE | 3.7 | 0.53 | 0.143 |
The parameters were calculated from toluene depletion curves as described in Materials and Methods.
Average from two curves.
TCE was added at concentrations similar to that in the fed-batch culture.
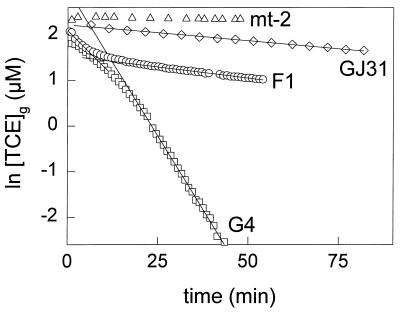
The TCE-degrading capacities of the strains were determined from TCE depletion curves measured with toluene-grown cells (Fig. 6). The first-order rate constants of TCE degradation were 52 and 0.6 ml mg of cells (dry weight)−1 h−1 for B. cepacia G4 and P. putida GJ31, respectively. For P. putida F1 the rate constant of TCE degradation decreased rapidly from 14 to 1.7 ml mg of cells (dry weight)−1 h−1. Wackett and coworkers also observed a rapid decrease of the rate over time, which was probably caused by the formation of toxic intermediates (34, 35). P. putida mt-2 showed no detectable TCE degradation (Fig. 6). The data show that P. putida mt-2 is not able to degrade TCE, while P. putida GJ31 degrades TCE much slower than P. putida F1 and B. cepacia G4, which are well known for their TCE-degrading capacities.
FIG. 6.
Logarithmic plots of TCE depletion by resting cell suspensions of B. cepacia G4 and P. putida F1, GJ31, and mt-2. The cells were grown on toluene. The concentrations of TCE in the headspace of the cultures were measured. TCE depletion by 0.43 mg (dry weight) of B. cepacia G4 ml−1 (□), 1.0 mg (dry weight) of P. putida F1 ml−1 (○), 1.9 mg (dry weight) of P. putida GJ31 ml−1 (◊), and 3.9 mg (dry weight) of P. putida mt-2 ml−1 (▵) is shown. The lines show the TCE depletion curves that were fitted through the data located in linear regions of the curves for B. cepacia G4, P. putida F1, and P. putida GJ31.
To study the effect of TCE on the kinetics of toluene utilization, toluene depletion curves were made in the presence of TCE, which was added to similar concentrations as with the fed-batch culture. B. cepacia G4 significantly converted TCE during growth on toluene, whereas the other three strains hardly converted TCE (Fig. 5B). The apparent affinities for toluene decreased in the presence of TCE (Table 4). For P. putida mt-2, this decrease was twofold. However, the affinities for toluene of P. putida F1 and GJ31 became almost 1 order of magnitude lower in the presence of TCE. Together with B. cepacia G4, these strains disappeared from the fed-batch culture after the start of TCE addition and are able to (slightly) degrade TCE.
The TCE depletion curve of P. putida GJ31 showed first-order behavior. This means that the half-saturation constant (KmTCE) of TCE conversion is much higher than the initial TCE concentration in the experiment, which was 18 μM in the liquid phase. TCE was present at 11 μM in the liquid phase during the preparation of the toluene depletion curve for strain GJ31. With these numbers, a model for competitive inhibition {Ksobs = Ks(1 + [TCE]/Ki)} predicts an increase in the observed Monod constants (Ksobs) with a factor of less than 1.6 if Ki were identical to KmTCE. However, the increase in Ksobs which was determined from toluene depletion curves in the presence of TCE was much higher (Table 4), indicating that the inhibition of toluene utilization by TCE cannot be described by a simple model for competitive inhibition. Landa et al. (19) also found that this model could not describe cometabolic TCE degradation by B. cepacia G4 in continuous culture. Instead, they observed that the inhibition constant of TCE for the conversion of toluene was higher than the Km of TCE conversion.
The kinetic parameters for growth on toluene of some mutants of P. putida mt-2 are given in Table 5. Mutants belonging to both class 2 and class 3 were analyzed. All five mutants had a higher μmax and a higher affinity than wild-type P. putida mt-2. The presence of TCE still caused a small decrease in the affinity for toluene (data not shown).
TABLE 5.
Monod parameters for growth on toluene of some mutants of P. putida mt-2a
| Mutant phenotype (strain) | Ks (μM) | μmax (h−1) | Affinity (μmax/Ks) |
|---|---|---|---|
| Tol+p-Xyl− XylE+ (mt-2M11) | 2.4 | 0.43 | 0.179 |
| Tol+p-Xyl− XylE+ (mt-2M20) | 2.9 | 0.42 | 0.145 |
| Tol+p-Xyl− XylE− (mt-2M10) | 3.3 | 0.46 | 0.139 |
| Tol+p-Xyl− XylE− (mt-2M12) | 3.7 | 0.47 | 0.127 |
| Tol+p-Xyl− XylE− (mt-2M4) | 5.3 | 0.50 | 0.094 |
The parameters were calculated from toluene depletion curves as described in Materials and Methods.
DISCUSSION
The four toluene-degrading strains used in this study (B. cepacia G4, P. putida mt-2, P. putida GJ31, and P. putida F1) had to compete for very low concentrations of toluene when they were cultivated together in a fed-batch culture. In situations like this, hardly any growth of the culture is allowed. However, shifts in the population composition are still possible. Zambrano et al. (38), for example, described the takeover of a stationary culture by a mutant of Escherichia coli. Likewise, we observed that mutants which had lost pTOM took over a culture of B. cepacia G4 which was exposed to TCE while being starved for carbon and energy (22). Under the conditions that were used here, all four strains were able to maintain themselves in the fed-batch culture at a rather constant viability for at least 40 days.
The strains that were present in the fed-batch culture showed large differences in the individual kinetic parameters μmax and Ks. If the capacities of the strains to compete during severe toluene limitation were determined by their kinetic properties, one would expect the strain with the highest affinity for toluene to take over the culture. Since no important changes in the population composition were observed, these kinetic parameters did not predict the outcome of the competition. This means that other factors also determined the competitive capacities of the strains. Bacteria could, for example, increase their affinity for toluene by a stronger induction of the enzymes involved in toluene degradation (20) or could produce compounds which inhibit other species. Factors like the amount of energy needed for maintenance purposes and the growth yield on products of lysed cells can also influence the survival of the strains. Also in continuous culture, the outcome of competition experiments cannot always be predicted by the affinity of each strain. Despite the lower affinity of B. cepacia G4 for toluene, it could win over P. putida mt-2 (8).
After TCE addition, only P. putida mt-2, which does not degrade TCE, remained in the culture. The numbers of cells of B. cepacia G4, P. putida F1, and P. putida GJ31 decreased, and TCE degradation diminished rapidly. We previously found that a nongrowing culture of B. cepacia G4 could cometabolically degrade TCE in a fed-batch culture as long as toluene was added (22). However, this resulted in a large increase in the maintenance energy demand of B. cepacia G4, most likely due to toxic effects of TCE conversion. Cytotoxicity by TCE conversion products has also been observed for P. putida F1 (35). Such toxic effects could give TCE-converting organisms a selective disadvantage, especially at the low toluene concentrations used.
Compared to that of B. cepacia G4, the first-order rate constant for TCE degradation of P. putida GJ31 suggests that at least 85 times less TCE will be converted by this strain, which is likely to be even less because of its superior kinetics for toluene degradation. In spite of that, the observed decay rates for P. putida GJ31 and F1 after TCE addition were even higher than the decay rate for B. cepacia G4. Analysis of toluene depletion curves made in the presence of concentrations of TCE similar to that in the fed-batch culture showed that the affinity (μmax/Ksobs) of P. putida GJ31 for toluene strongly decreased. Such a decrease was also observed for P. putida F1. For B. cepacia G4 the affinity decreased only 2.2-fold. However, during the kinetic measurements, B. cepacia G4 degraded a considerable amount of TCE, which leads to a large underestimation of Ksobs.
Although the affinity for toluene is not the sole parameter determining the outcome of the competition, reduction of affinity could still lower the competitiveness of an individual strain. Quantitatively, the effect of TCE on the Ksobs for toluene as determined in depletion experiments could not be described by competitive inhibition. For P. putida GJ31 and F1, the effect of TCE was larger than expected, which may be due to enzyme inactivation or product inhibition in addition to competitive inhibition. Our results indicate that the actual amount of TCE that can be converted by the organisms is not the only factor that determines their fate in the population, because the kinetics for the degradation of TCE did not correspond to the effect of this compound on the kinetics for growth on toluene and on the survival of the different strains.
After TCE addition was started, the competitive capacity of P. putida mt-2 was further improved by mutations which allowed this organism to grow on toluene at a higher rate, but this also resulted in the loss of the capacity to grow on p-xylene. The mutants did not convert p-toluate, indicating that they do not have any active XylXYZ. Also, the expression levels of enzymes further down in the meta pathway were strongly reduced. When the mutants grow on toluene, the enzymes of the upper pathway (1) will convert toluene to benzoate, after which the absence of XylXYZ and the reduction of the expression of the meta pathway probably allow benzoate to be degraded mainly via the ortho pathway, which is known to result in a higher growth rate (1, 4). The p-Xyl− mutants were detected after 2 days of TCE addition (Fig. 4). By that time, the toluene concentration in the outgoing gas stream had increased to ∼500 nM, probably due to the decay of B. cepacia G4 and P. putida F1 and GJ31. Since the mutants of P. putida mt-2 have an elevated growth rate on toluene, they could probably take over the culture more rapidly than wild-type P. putida mt-2.
During the 40 days of toluene addition in the absence of TCE, no p-Xyl− mutants were detected, although they could have had an improved fitness compared to P. putida mt-2. Since the mutants were observed soon after the start of TCE addition, the appearance of the mutants seemed to be a direct effect of this, for example, because TCE has some mutagenic effect which increases the overall rate of mutations or because TCE specifically inhibits a component of wild-type P. putida mt-2. XylXYZ might be such a component, since none of the mutants of the different classes had activity of this dioxygenase, while the expression levels of other enzymes of the meta operon differed considerably.
Although several studies describe the potential of toluene and phenol degraders for successful remediation of TCE (10, 12–14), our results indicate that the application of microorganisms that cometabolically degrade TCE carries a high risk of takeover of the desired population by organisms that are less sensitive to inhibitory effects of TCE. Fries et al. (11) showed that there is a large variety in the capacity to degrade TCE among toluene- and phenol-degrading microorganisms isolated from the Moffett field, and they also expected that organisms which do not degrade TCE will eventually take over the population. Indeed, Munakata-Marr et al. (24) recently observed a gradual decline in the breakdown of TCE in phenol-fed microcosms containing aquifer material from the Moffett field, while degradation of phenol remained complete. This was probably caused by a shift in the population towards phenol degraders that did not degrade TCE.
Stable degradation may require the stimulation of a specific group of organisms. This might be achieved by using a less common primary substrate which can be degraded only by enzymes that also convert TCE and for which no alternatives exist. o-Cresol might be such a primary substrate. It is degraded by the same TCE-degrading toluene monooxygenase (TOM) as toluene in B. cepacia G4 (32). TOM-containing organisms were found to dominate in the TCE-contaminated Moffett field (11), which indicates that the endogenous population can degrade TCE with o-cresol. In case of groundwater treatment with continuously operated bioreactors, separation of degradation and growth is another alternative to overcome instability problems and is currently under study in our lab.
ACKNOWLEDGMENTS
This work was financed by grants from the Dutch IOP Environmental Biotechnology program and the EC environment program.
We acknowledge Uwe Dehmel for providing the plasmids with the xylTE′ and Pm-xylXYZ′ genes. Wouter Duetz assisted with the P. putida mt-2 plasmid isolations.
REFERENCES
- 1.Assinder S J, Williams P A. The TOL plasmids: determinants of the catabolism of toluene and the xylenes. Adv Microb Physiol. 1990;31:1–69. doi: 10.1016/s0065-2911(08)60119-8. [DOI] [PubMed] [Google Scholar]
- 2.Bailey J E, Ollis D F. Biochemical engineering fundamentals. 2nd ed. New York, N.Y: McGraw-Hill Inc.; 1986. [Google Scholar]
- 3.Bergmann J G, Sanik J. Determination of trace amounts of chlorine in naphtha. Anal Chem. 1957;29:241–243. [Google Scholar]
- 4.Brinkmann U, Ramos J L, Reineke W. Loss of the TOL meta-cleavage pathway functions of Pseudomonas putida strain PaW1 (pWWO) during growth on toluene. J Basic Microbiol. 1994;5:303–309. doi: 10.1002/jobm.3620340503. [DOI] [PubMed] [Google Scholar]
- 5.Chesbro W. The domains of slow bacterial growth. Can J Microbiol. 1988;34:427–435. doi: 10.1139/m88-075. [DOI] [PubMed] [Google Scholar]
- 5a.Dehmel, U. Personal communication.
- 6.Diks R M M. The removal of dichloromethane from waste gases in a biological trickling filter. Ph.D. thesis. Eindhoven, The Netherlands: Eindhoven University of Technology; 1992. [Google Scholar]
- 7.Duetz W A, Winson M K, van Andel J G, Williams P A. Mathematical analysis of catabolic function loss in a population of Pseudomonas putida mt-2 during non-limited growth on benzoate. J Gen Microbiol. 1991;137:1363–1368. doi: 10.1099/00221287-137-6-1363. [DOI] [PubMed] [Google Scholar]
- 8.Duetz W A, de Jong C, Williams P A, van Andel J G. Competition in chemostat culture between Pseudomonas strains that use different pathways for the degradation of toluene. Appl Environ Microbiol. 1994;60:2858–2863. doi: 10.1128/aem.60.8.2858-2863.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ensley B D. Biochemical diversity of trichloroethylene metabolism. Annu Rev Microbiol. 1991;45:283–299. doi: 10.1146/annurev.mi.45.100191.001435. [DOI] [PubMed] [Google Scholar]
- 10.Fan S, Scow K M. Biodegradation of trichloroethylene and toluene by indigenous microbial populations in soil. Appl Environ Microbiol. 1993;59:1911–1918. doi: 10.1128/aem.59.6.1911-1918.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fries M R, Forney L J, Tiedje J M. Phenol- and toluene-degrading microbial populations from an aquifer in which successful trichloroethene cometabolism occurred. Appl Environ Microbiol. 1997;63:1523–1530. doi: 10.1128/aem.63.4.1523-1530.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopkins G D, Munakata J, Semprini L, McCarty P L. Trichloroethylene concentration effects on pilot-scale in-situ groundwater bioremediation by phenol-oxidizing microorganisms. Environ Sci Technol. 1993;27:2542–2547. [Google Scholar]
- 13.Hopkins G D, Semprini L, McCarty P L. Microcosm and in situ field studies of enhanced biotransformations of trichloroethylene by phenol-utilizing microorganisms. Appl Environ Microbiol. 1993;59:2277–2285. doi: 10.1128/aem.59.7.2277-2285.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hopkins G D, McCarty P L. Field evaluation of in situ aerobic cometabolism of trichloroethylene and three dichloroethylene isomers using phenol and toluene as the primary substrates. Environ Sci Technol. 1995;29:1628–1637. doi: 10.1021/es00006a029. [DOI] [PubMed] [Google Scholar]
- 15.Ichihara A, Adachi K, Hosokawa K, Takeda Y. The enzymatic hydroxylation of aromatic carboxylic acids; substrate specificities of anthranilate and benzoate oxidases. J Biol Chem. 1962;237:2297–2302. [PubMed] [Google Scholar]
- 16.Janssen D B, Scheper A, Witholt B. Biodegradation of 2-chloroethanol by pure bacterial cultures. Prog Ind Microbiol. 1984;20:169–178. [Google Scholar]
- 17.Janssen D B, Witholt B. Aerobic and anaerobic degradation of halogenated aliphatics. In: Sigel H, Sigel A, editors. Metal ions in biological systems. 28. Degradation of environmental pollutants by microorganisms and their metalloenzymes. New York, N.Y: Marcel Dekker, Inc.; 1992. pp. 299–327. [Google Scholar]
- 18.Kado C I, Liu S-T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landa A S, Sipkema E M, Weijma J, Beenackers A A C M, Dolfing J, Janssen D B. Cometabolic degradation of trichloroethylene by Pseudomonas cepacia G4 in a chemostat with toluene as the primary substrate. Appl Environ Microbiol. 1994;60:3368–3374. doi: 10.1128/aem.60.9.3368-3374.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Law A T, Button D K. Modulation of affinity of a marine pseudomonad for toluene and benzene by hydrocarbon exposure. Appl Environ Microbiol. 1986;51:469–476. doi: 10.1128/aem.51.3.469-476.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 22.Mars A E, Houwing J, Dolfing J, Janssen D B. Degradation of toluene and trichloroethylene by Burkholderia cepacia G4 in growth-limited fed-batch culture. Appl Environ Microbiol. 1996;62:886–891. doi: 10.1128/aem.62.3.886-891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mars A E, Kasberg T, Kaschabek S R, van Agteren M H, Janssen D B, Reineke W. Microbial degradation of chloroaromatics: use of the meta-cleavage pathway for mineralization of chlorobenzene. J Bacteriol. 1997;179:4530–4537. doi: 10.1128/jb.179.14.4530-4537.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munakata-Marr J, Matheson V G, Forney L J, Tiedje J M, McCarty P L. Long-term biodegradation of trichloroethylene influenced by bioaugmentation and dissolved oxygen in aquifer microcosms. Environ Sci Technol. 1997;31:786–791. [Google Scholar]
- 25.Nelson M J K, Montgomery S O, Mahaffey W R, Pritchard P H. Biodegradation of trichloroethylene and involvement of an aromatic pathway. Appl Environ Microbiol. 1987;53:949–954. doi: 10.1128/aem.53.5.949-954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson M J K, Montgomery S O, O’Neill O J, Pritchard P H. Aerobic metabolism of trichloroethylene by a bacterial isolate. Appl Environ Microbiol. 1986;52:383–384. doi: 10.1128/aem.52.2.383-384.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oldenhuis R, Oedzes J Y, van der Waarde J J, Janssen D B. Kinetics of chlorinated hydrocarbon degradation by Methylosinus trichosporium OB3b and toxicity of trichloroethylene. Appl Environ Microbiol. 1991;57:7–14. doi: 10.1128/aem.57.1.7-14.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reineke W, Knackmuss H-J. Chemical structure and biodegradability of halogenated aromatic compounds. Substituent effects on 1,2-dioxygenation of benzoic acid. Biochim Biophys Acta. 1978;542:412–423. doi: 10.1016/0304-4165(78)90372-0. [DOI] [PubMed] [Google Scholar]
- 29.Robinson J A, Tiedje J M. Nonlinear estimation of Monod growth kinetic parameters from a single substrate depletion curve. Appl Environ Microbiol. 1983;45:1453–1458. doi: 10.1128/aem.45.5.1453-1458.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sala-Trepat J M, Murray K, Williams P A. The metabolic divergence in the meta cleavage of catechols by Pseudomonas putida NCIB 10015. Physiological significance and evolutionary implications. Eur J Biochem. 1972;28:347–356. doi: 10.1111/j.1432-1033.1972.tb01920.x. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Shields M S, Montgomery S O, Cuskey S M, Chapman P J, Pritchard P H. Mutants of Pseudomonas cepacia G4 defective in catabolism of aromatic compounds and trichloroethylene. Appl Environ Microbiol. 1991;57:1935–1941. doi: 10.1128/aem.57.7.1935-1941.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Hylckama Vlieg J E T, de Koning W, Janssen D B. Transformation kinetics of chlorinated ethenes by Methylosinus trichosporium OB3b and detection of unstable epoxides by on-line gas chromatography. Appl Environ Microbiol. 1996;62:3304–3312. doi: 10.1128/aem.62.9.3304-3312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wackett L P, Gibson D T. Degradation of trichloroethylene by toluene dioxygenase in whole-cell studies with Pseudomonas putida F1. Appl Environ Microbiol. 1988;54:1703–1708. doi: 10.1128/aem.54.7.1703-1708.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wackett L P, Householder S R. Toxicity of trichloroethylene to Pseudomonas putida F1 is mediated by toluene dioxygenase. Appl Environ Microbiol. 1989;55:2723–2725. doi: 10.1128/aem.55.10.2723-2725.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wigmore G J, Bayly R C, Di Berardino D. Pseudomonas putida mutants defective in the metabolism of the products of meta fission of catechol and its methyl analogues. J Bacteriol. 1974;120:416–423. doi: 10.1128/jb.120.1.31-37.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams P A, Taylor S, Gibb L E. Loss of the toluene-xylene catabolic genes of TOL plasmid pWWO during growth of Pseudomonas putida on benzoate is due to a selective growth advantage of ‘cured’ segregants. J Gen Microbiol. 1988;134:2039–2048. doi: 10.1099/00221287-134-7-2039. [DOI] [PubMed] [Google Scholar]
- 38.Zambrano M M, Siegele D A, Almiron M, Tormo A, Kolter R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 1993;259:1757–1759. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]