Abstract
Several genes in the trichothecene biosynthetic pathway of Fusarium sporotrichioides have been shown to reside in a gene cluster. Sequence analysis of a cloned DNA fragment located 3.8 kb downstream from TRI5 has led to the identification of the TRI11 gene. The nucleotide sequence of TRI11 predicts a polypeptide of 492 residues (Mr = 55,579) with significant similarity to members of the cytochrome P-450 superfamily. TRI11 is most similar to several fungal cytochromes P-450 (23 to 27% identity) but is sufficiently distinct to define a new cytochrome P-450 gene family, designated CYP65A1. Disruption of TRI11 results in an altered trichothecene production phenotype characterized by the accumulation of isotrichodermin, a trichothecene pathway intermediate. The evidence suggests that TRI11 encodes a C-15 hydroxylase involved in trichothecene biosynthesis.
Trichothecenes are sesquiterpenoid toxins that are produced by several genera of filamentous fungi, including Fusarium (22). Trichothecene toxins are thought to act by inhibiting protein synthesis (9, 13) and are highly cytotoxic to many eukaryotes. The contamination of feedstuffs with trichothecenes has been associated with instances of mycotoxicoses and is recognized as an important agricultural problem. Fusarium species are the major source of the trichothecenes found in grains. Recently, it has been shown that the production of trichothecenes enhances the virulence of Fusarium species on wheat (5).
The trichothecene biosynthetic pathway begins with the cyclization of farnesyl pyrophosphate by the enzyme trichodiene synthase to form trichodiene (Fig. 1). Trichodiene undergoes multiple oxygenations involving molecular oxygen and as many as four different esterifications (3) to form trichothecene products, such as T-2 toxin. Like other fungal terpenoids, the trichothecenes are produced as a large family of structurally related compounds, with individual Fusarium species typically producing distinctive trichothecene profiles. Oxygenation steps in trichothecene biosynthesis are of particular interest since the degree of oxygenation greatly alters trichothecene toxicity. At least one trichothecene oxygen, the 12,13-epoxide, is required for toxicity (2). In Fusarium sporotrichioides, the initial oxygenation of trichodiene in the biosynthesis of T-2 toxin (Fig. 1) is catalyzed by a cytochrome P-450 monooxygenase encoded by TRI4 (6). Hydroxylation of C-15 is the first oxygenation step employing an intermediate containing the core trichothecene structure (3). The product of the C-15 hydroxylase, 15-decalonectrin, is most likely the last common intermediate of the various Fusarium trichothecene pathways. Biosynthesis of deoxynivalenol, the trichothecene most commonly detected in grains, is thought to branch off of the T-2 toxin pathway at this intermediate.
FIG. 1.
Diagram of the trichothecene pathway in F. sporotrichioides. OAc, acetate.
Trichothecene pathway genes appear to be closely linked in F. sporotrichioides (12). At least four genes in the pathway have been identified on a single cosmid clone, and evidence for additional genes has been reported (7). Recently, the clustering of genes involved in the macrocyclic trichothecene pathway of Myrothecium roridum has also been demonstrated (20), indicating that gene clustering is widespread for fungal trichothecene pathways. Here, we report the characterization of the TRI11 gene from the trichothecene gene cluster in F. sporotrichioides.
MATERIALS AND METHODS
Strains, media, and culture conditions.
F. sporotrichioides NRRL 3299 was obtained from the USDA/ARS Culture Collection at the National Center for Agricultural Utilization Research, Peoria, Ill. Cultures were grown in YPG medium (0.3% yeast extract, 1% peptone, 2% glucose) for DNA isolation and in GYEP medium (5% glucose, 0.1% yeast extract, 0.1% peptone) for trichothecene and RNA analyses (23). All cultures were inoculated to a final concentration of 106 conidia/ml and incubated at 28°C on a gyratory shaker (200 rpm).
Cosmid mapping, plasmid constructions, and fungal transformation.
Cosmid 9-1 was mapped with respect to SacI restriction sites by cloning individual SacI fragments larger than 1.0 kb into pGEM-7Zf+ (Promega, Inc.). The ends of each cloned SacI fragment were then sequenced with plasmid-specific primers. On the basis of these sequences, primers were designed in the reverse orientation approximately 200 to 300 bp downstream from the ends of each fragment. Subsequently, these primers were used in PCRs with cosmid 9-1 as the template in all pairwise combinations to determine the physical relationships of the SacI fragments. The amplification of a PCR product with a particular primer pair indicated that two SacI fragments were contiguous. All PCR products were less than 600 bp, suggesting that any SacI fragments that were excluded from the initial cloning step must be less than 200 bp. The TRI11 gene was originally cloned as a 9.5-kb SacI fragment (pFSC3-3) and then further localized on a 2.9-kb SacI-HindIII fragment that was subcloned from pFSC3-3 into pBluescript II KS(−) (Stratagene). The resulting plasmid, pFSC3-6, contained the entire TRI11 gene. Gene disruption of TRI11 was accomplished by cloning the BamHI-XbaI fragment of pFSC3-6, consisting of a doubly truncated portion of the TRI11 coding region, into the fungal transformation vector pUCH2-8 to yield plasmid pTRI11D1. The transformation vector pUCH2-8 was constructed by inserting the SalI-HindIII fragment of pUCH1, which contains promoter 1 from Cochliobolus heterostrophus (21) fused to the hygromycin phosphotransferase open reading frame, into the SalI-HindIII sites of pBluescript II KS(−) (Stratagene). Transformation of F. sporotrichioides and the subsequent selection and isolation of transformants were performed according to a procedure described previously (18).
Northern blots.
To isolate RNA, cultures were grown in GYEP medium for 23 h and harvested by filtration. These growth conditions support trichothecene biosynthesis and result in high-level expression of several pathway gene mRNAs (6). The mycelial mats (approximately 0.5 to 1.0 g) were immediately ground in liquid N2, and RNA was isolated with an RNaid kit (Bio 101) by the acid-phenol procedure described in the manufacturer’s product literature. Northern blotting was carried out as described elsewhere (17) by using a 32P-labeled probe made from the SacI-HindIII fragment cloned in pFSC3-6.
cDNA isolation.
The TRI11 cDNA coding sequence was amplified by PCR using Pfu polymerase (Stratagene) and a cDNA library as the template. The cDNA library was constructed in the yeast expression vector pYES2 (Invitrogen) with F. sporotrichioides RNA harvested from a 23-h GYEP medium-grown culture (12). The primers for PCR were 677 (5′-GCGAAGCTTCATGTTCCAATACTCCCTGTGG-3′) and 678 (5′-CCCGAATTCTCCTAACAAGGGTAACAGCC-3′), which correspond to the 5′ and 3′ ends of the TRI11 coding region, respectively. The resulting PCR product was extracted from an agarose gel band with Gene Clean (Bio 101) and cloned with a Prime PCR Cloner kit (5′→3′, Inc.). The 3′ end of the TRI11 cDNA was amplified by anchored PCR using the cDNA library as a template and primers specific for the TRI11 coding sequence and the cDNA cloning vector pYES2. The primers for PCR were 614 (5′-GAGAATCCTTCAGTTGTC-3′) and 513 (5′-GCGTGAATGTAAGCGTGAC-3′). The resulting PCR product was sequenced directly.
Analysis of fungal transformants.
Transformants were analyzed by PCR using two different primer pairs. One primer pair consisted of primer 453 (5′-GTAAATGTCGAGCTTCCG-3′), which corresponds to a portion of the TRI11 sequence not present in pTRI11D1, and primer 248 (5′-CTATGCCTACAGCATCCAGG-3′), which corresponds to the 3′ end of the HygB gene in the transformation vector pUCH2-8. Only transformants carrying a copy of the disrupter vector that had integrated at the TRI11 locus would be expected to produce a product with these primers. All six transformants analyzed produced the expected fragment of 2,345 bp following PCR. No products from reactions using NRRL 3299 and pTRI11D1 DNA as the template were seen.
PCR and DNA sequencing.
The procedures employed for amplification of DNA fragments by PCR have been described previously (17). The amplified fragments were purified with Gene Clean (Bio 101) and then used directly for cloning or as templates for sequencing with the Taq DYEdeoxy sequencing kit (Applied Biosystems). Sequencing reaction products were analyzed with an Applied Biosystems model 377 automated DNA sequencer.
Computer analyses.
Sequence similarity searches of the PIR 49.0 and SWISS-PROT 33.0 databases were performed by using the FASTA (16) and BLAST (1) programs. Alignments between individual sequences were performed with CLUSTAL W, version 1.6 (19).
Nucleotide sequence accession number.
The nucleotide sequence of TRI11 has been submitted to GenBank under accession no. AF011355.
RESULTS
Mapping of cosmid 9-1.
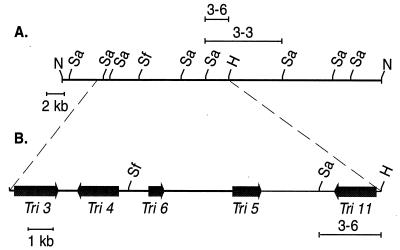
A single cosmid, cosmid 9-1 (Fig. 2), contains at least four trichothecene pathway genes (8, 12). We prepared a detailed restriction map of this cosmid by cloning all of the SacI fragments and the two NotI-SacI fragments present at the ends of this cosmid. No NotI sites were observed in cosmid 9-1, other than the two NotI sites that flank the BamHI insertion site of the cosmid cloning vector. The ends of each cloned fragment were sequenced to permit the design of PCR primers in the opposite direction. Pairwise combinations of these primers in PCRs employing cosmid 9-1 as a template revealed the order and orientation of all nine SacI fragments. The ends of contiguous fragments were identified by the production of PCR products of the expected size.
FIG. 2.
(A) Restriction map of cosmid 9-1. All of the SacI and SfiI sites in cosmid 9-1 are shown; the only HindIII site shown is the site used to define the fragment cloned into pFSC3-6. SacI fragment 3-3 is also indicated. (B) Map of the trichothecene pathway gene cluster showing the location of pathway gene coding regions. The SacI and HindIII sites of the fragment in pFSC3-6 are shown in addition to the SfiI site. Sa, SacI; Sf, SfiI; H, HindIII.
Isolation and characterization of TRI11.
A 9.5-kb SacI fragment designated 3-3 (Fig. 2A) and located downstream of TRI5 was the focus of our study. The 2.9-kb SacI-HindIII fragment nearest TRI5 (fragment 3-6) was subcloned from 3-3 to yield plasmid pFSC3-6. This plasmid was used to probe a Northern blot of RNA from a 23-h GYEP medium-grown culture (data not shown), and a 2-kb band was observed. Subsequent analysis of fragment 3-6 revealed a probable coding region of approximately 1.6 kb interrupted by several introns. The putative coding region was named TRI11, and its location relative to other pathway genes is shown in Fig. 2B.
Sequences at the predicted ends of the coding region were used to design primers (677 and 678) for the amplification of a TRI11 cDNA from an F. sporotrichioides cDNA library. The resulting cDNA sequence predicted a protein of 492 amino acids (Mr = 55,579) and a genomic sequence containing four introns ranging in size from 49 to 76 bp.
The 3′ end of the TRI11 cDNA was amplified by anchored PCR using primer 677 and a primer corresponding to sequences on the cloning vector for the cDNA library. Analysis of the sequence showed that the TRI11 transcript extends 190 bases beyond the translation stop codon.
Disruption of TRI11.
To investigate the role of TRI11 in trichothecene biosynthesis, TRI11− mutants were generated by gene disruption using a plasmid, pTRI11D1, carrying a doubly truncated portion of the TRI11 coding region. Homologous integration of pTRI11D1 at the TRI11 gene should result in the generation of two nonfunctional copies of TRI11, each carrying either a 5′ or a 3′ truncation.
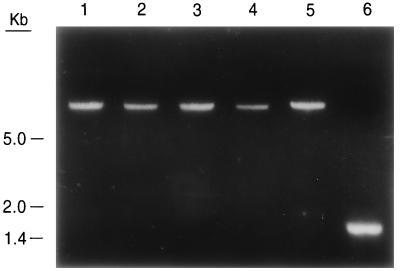
Following transformation, six hygromycin-resistant transformants were isolated. Integration of pTRI11D1 within TRI11 was confirmed by PCR using primers located outside the TRI11 fragment (677 and 678) on pTRI11D1 (Fig. 3). PCR of DNA from five transformants yielded products of a size consistent with the presence of pTRI11D1 within TRI11 (approximately 7.8 kb) and considerably larger than the PCR product from NRRL 3299 DNA (1.8 kb). All five transformants had an altered trichothecene production phenotype. Cultures of F. sporotrichioides NRRL 3299 grown on GYEP medium for 3 days accumulate primarily T-2 toxin (Fig. 4B). When the TRI11 disruptants were grown under the same conditions and analyzed as described elsewhere (11), the major trichothecene found was isotrichodermin (Fig. 4A), an intermediate in trichothecene biosynthesis (3).
FIG. 3.
Analysis of TRI11 gene disruptants in F. sporotrichioides by PCR. Transformant and NRRL 3299 DNA preparations were used as templates for PCRs primed with oligonucleotides 677 and 678. PCR products were separated on a 1% agarose gel (40 mM Tris-acetate, 1 mM EDTA) and stained with ethidium bromide. Lanes 1 to 5, PCR products from five different transformant DNAs; lane 6, PCR product from F. sporotrichioides NRRL 3299 DNA.
FIG. 4.
Chromatogram of F. sporotrichioides culture extracts from a TRI11− transformant (A) and parent strain NRRL 3299 (B). Cultures were grown in GYEP medium for 3 days and analyzed under identical conditions by gas-liquid chromatography as described previously (11). The locations of isotrichodermin and T-2 toxin are indicated.
Comparisons between TRI11 and other cytochromes P-450.
The TRI11 gene product (TRI11) was compared to proteins in the PIR database by using BLAST and FASTA (1, 16). Both comparisons indicated that TRI11 is most similar to fungal cytochrome P-450-type enzymes. An alignment between the protein sequences for TRI11 and ord1 (25), a cytochrome P-450 present in the aflatoxin pathway of Aspergillus parasiticus, showed the highest overall similarity, with 27% identity. In addition, alignments with ord1 revealed 36% identity over a region of 250 amino acids. A high level of similarity was also observed with benzoate para-hydroxylase (24) from Aspergillus niger (CYP53, 26% identity) and the pisatin demethylase (10) from Nectria haematococca (CYP57, 23% identity). TRI11 and TRI4 (6), another trichothecene pathway cytochrome P-450, showed only 23% identity.
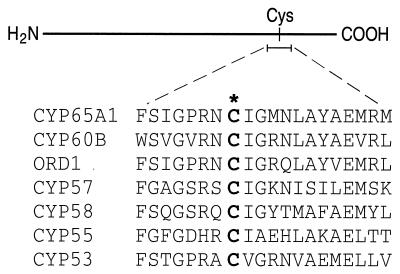
TRI11 contains a number of residues that appear to be conserved in all cytochromes P-450. Most significantly, it contains the highly conserved sequence motif that constitutes the heme-binding domain of cytochromes P-450. A universally conserved cysteine residue within this motif serves as the fifth ligand for the coordination of heme iron at the monooxygenase active site. TRI11 contains a 10-amino-acid sequence starting at Phe431 (Fig. 5) which is in perfect agreement with the heme-binding domain consensus sequence. This sequence is also found to be aligned with the corresponding sequence in other cytochromes P-450, suggesting that Cys438 functions as the fifth ligand in the heme-binding domain of TRI11. Several other residues are also found to be highly conserved in cytochromes P-450, although their functions are unknown (14, 15); the corresponding residues in TRI11 are Gly298, Glu354, and Arg357.
FIG. 5.
Alignments between the conserved heme-binding motifs of TRI11 and other fungal cytochromes P-450. The conserved cysteinyl residues are indicated (boldface). The fungal cytochromes P-450, species, and gene names (and GenBank accession numbers) are as follows: CYP65A1 from F. sporotrichioides, TRI11 (AF011355); CYP60B from Aspergillus nidulans, stcL (U34740); ord1 from A. parasiticus (L40839); CYP57 from N. haematococca, pdm2 (P38364); CYP58 from F. sporotrichioides, TRI4 (U22462); CYP55 from Fusarium oxysporum (M63340); and CYP53 from A. niger, bphA (A22974).
DISCUSSION
TRI11 is the fifth gene to be identified within the trichothecene pathway gene cluster of F. sporotrichioides. On the basis of a restriction map we constructed for cosmid 9-1 (Fig. 2), TRI11 is located approximately 3.8 kb from TRI5 and is transcribed in the opposite direction. Disruption of TRI11 results in an altered trichothecene profile that is consistent with the participation of the TRI11 gene product in trichothecene biosynthesis. Transformants lacking a functional copy of TRI11 do not accumulate T-2 toxin and other late-pathway trichothecenes characteristic of F. sporotrichioides NRRL 3299 but instead accumulate the earlier-pathway intermediate isotrichodermin. Finally, expression of TRI11 is coincident with the expression of other pathway genes, as shown by the detection of a TRI11 transcript in RNA isolated from cultures grown under conditions known to support the expression of other pathway genes (6).
Whole-cell feeding experiments involving a TRI11− gene disruption mutant were earlier shown to result in the efficient conversion of 15-decalonectrin to T-2 toxin (11). 15-Decalonectrin is the likely enzymatic product of TRI11 and differs from isotrichodermin only in the presence of the C-15 hydroxyl group (Fig. 1). These experiments suggest that the accumulation of isotrichodermin in TRI11− mutants is due to their inability to convert isotrichodermin to 15-decalonectrin and that TRI11 encodes an oxygenase responsible for the hydroxylation of C-15 in trichothecene biosynthesis. While isotrichodermin is the only trichothecene observed in 3-day cultures of TRI11− mutants (Fig. 4A), metabolites of isotrichodermin accumulate in cultures grown for longer periods (11). Oxygenated products of isotrichodermin, such as 8-hydroxytrichodermin and 3,4,8-trihydroxytrichothecene, that are not intermediates in trichothecene biosynthesis have been detected in 7-day cultures of a TRI11− mutant.
Sequence similarity analyses suggest that TRI11 encodes a cytochrome P-450 monooxygenase. Several highly conserved residues characteristic of cytochromes P-450 are present and properly positioned in TRI11, including the cysteinyl peptide involved in heme binding. Although the similarity of TRI11 to known cytochromes P-450 is sufficient to justify its inclusion in the cytochrome P-450 superfamily, it is different enough from members of other cytochrome P-450 families (<40% identical) to warrant its designation as the first member of a new cytochrome P-450 family, CYP65A1.
TRI11 is the second cytochrome P-450 in the trichothecene pathway to be identified. The other cytochrome P-450, TRI4, catalyzes an unspecified oxygenation reaction involving trichodiene (6). Interestingly, TRI11 and TRI4 do not show a high degree of similarity, indicating that they are not the result of a recent gene duplication event.
The presence of a C-15 hydroxyl group is characteristic of most Fusarium trichothecenes and all of the macrocyclic trichothecenes. Recently, it was reported that microsomal fractions from Fusarium culmorum can convert isotrichodermin to 15-decalonectrin, 7α-hydroxyisotrichodermin, and 8α-hydroxyisotrichodermin (26). The reaction conditions used in these experiments were consistent with the established requirements for cytochrome P-450 monooxygenases; however, the oxygenation of isotrichodermin did not appear to be inhibited by known P-450 inhibitors such as carbon monoxide and cyanide. On the basis of these results, it was concluded that cytochromes P-450 were not likely to be involved in the later oxygenation steps of trichothecene biosynthesis. Our identification of TRI11 as the C-15 hydroxylase in F. sporotrichioides indicates that cytochromes P-450 do catalyze some late pathway steps. Other evidence supporting the participation of cytochromes P-450 after the isotrichodermin step in the pathway include the observation that all of the oxygens linked directly to the trichodiene carbon skeleton are derived from molecular oxygen (4). The apparent failure of known P-450 inhibitors to affect the cell-free oxygenation of isotrichodermin may reflect differences in the sensitivity of these enzymes to the inhibitors or the need for different experimental conditions to demonstrate their effectiveness. Identification of TRI11 as a cytochrome P-450 involved in trichothecene biosynthesis further emphasizes the importance of this group of enzymes in mycotoxin biosynthetic pathways.
ACKNOWLEDGMENTS
We thank Marcie Moore and Kim MacDonald for their excellent technical assistance.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Colvin E W, Cameron S. Synthesis and biological evaluation of a trichothecene epi-epoxide, 3a,4b,15-triacetoxy-12,13-epi-epoxytrichothec-9-ene. J Chem Soc Chem Commun. 1986;1986:1642–1643. [Google Scholar]
- 3.Desjardins A E, Hohn T M, McCormick S P. Trichothecene biosynthesis in Fusarium species: chemistry, genetics, and significance. Microbiol Rev. 1993;57:595–604. doi: 10.1128/mr.57.3.595-604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desjardins A E, Plattner R D, Vanmiddlesworth F. Trichothecene biosynthesis in Fusarium sporotrichioides: origin of the oxygen atoms of T-2 toxin. Appl Environ Microbiol. 1986;51:493–497. doi: 10.1128/aem.51.3.493-497.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desjardins A E, Proctor R H, Bai G, McCormick S P, Shaner G, Buechley G, Hohn T M. Reduced virulence of trichothecene-nonproducing mutants of Gibberella zeae in wheat field tests. Mol Plant-Microbe Interact. 1996;9:775–781. [Google Scholar]
- 6.Hohn T M, Desjardins A E, McCormick S P. The Tri4 gene of Fusarium sporotrichioides encodes a cytochrome P450 monooxygenase involved in trichothecene biosynthesis. Mol Gen Genet. 1995;248:95–102. doi: 10.1007/BF02456618. [DOI] [PubMed] [Google Scholar]
- 7.Hohn T M, Desjardins A E, McCormick S P, Proctor R H. Biosynthesis of trichothecenes, genetic and molecular aspects: issues involving toxic microorganisms. In: Eklund M, Richard J L, Katsutoshi M, editors. Molecular approaches to food safety. Fort Collins, Colo: Alaken, Inc.; 1995. pp. 239–247. [Google Scholar]
- 8.Hohn T M, McCormick S P, Desjardins A E. Evidence for a gene cluster involving trichothecene pathway biosynthetic genes in Fusarium sporotrichioides. Curr Genet. 1993;24:291–295. doi: 10.1007/BF00336778. [DOI] [PubMed] [Google Scholar]
- 9.Iglesias M, Ballesta J P G. Mechanism of resistance to the antibiotic trichothecin in the producing fungi. Eur J Biochem. 1994;223:447–453. doi: 10.1111/j.1432-1033.1994.tb19012.x. [DOI] [PubMed] [Google Scholar]
- 10.Maloney A P, VanEtten H D. A gene from the plant pathogen Nectria haematococca that encodes the phytoalexin-detoxifying enzyme pisatin demethylase defines a new cytochrome P450 family. Mol Gen Genet. 1994;243:506–514. doi: 10.1007/BF00284198. [DOI] [PubMed] [Google Scholar]
- 11.McCormick S P, Hohn T M. Accumulation of trichothecenes in liquid cultures of a Fusarium sporotrichioides mutant lacking a functional trichothecene C-15 hydroxylase. Appl Environ Microbiol. 1997;63:1685–1688. doi: 10.1128/aem.63.5.1685-1688.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCormick S P, Hohn T M, Desjardins A E. Isolation and characterization of Tri3, a gene encoding 15-O-acetyltransferase from Fusarium sporotrichioides. Appl Environ Microbiol. 1996;62:353–359. doi: 10.1128/aem.62.2.353-359.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLaughlin C S, Vaughn M H, Campbell J M, Wei C M, Stafford M E. Inhibition of protein synthesis by trichothecenes. In: Rodricks J V, Hesseltine C W, Mehlman M A, editors. Mycotoxins in human and animal health. Park Forest, Ill: Park Forest. Pathotoxin Publishers; 1977. pp. 263–273. [Google Scholar]
- 14.Nelson D R, Strobel H W. On the membrane topology of vertebrate cytochrome P450 proteins. J Biol Chem. 1988;263:6038–6050. [PubMed] [Google Scholar]
- 15.Nelson D R, Strobel H W. Secondary structure prediction of 52 membrane-bound cytochromes P450 shows a strong structural similarity to P450cam. Biochemistry. 1989;28:656–660. doi: 10.1021/bi00428a036. [DOI] [PubMed] [Google Scholar]
- 16.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Proctor R H, Hohn T M. Aristolochene synthase. Isolation, characterization, and bacterial expression of a sesquiterpenoid biosynthetic gene (Ari1) from Penicillium roqueforti. J Biol Chem. 1993;268:4543–4548. [PubMed] [Google Scholar]
- 18.Salch Y P, Beremand M N. Gibberella pulicaris transformants: state of transforming DNA during asexual and sexual growth. Curr Genet. 1993;23:343–350. doi: 10.1007/BF00310897. [DOI] [PubMed] [Google Scholar]
- 19.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trapp, S. C., T. M. Hohn, S. P. McCormick, and B. B. Jarvis. Characterization of the macrocyclic trichothecene gene cluster in Myrothecium roridum. Mol. Gen. Genet., in press. [DOI] [PubMed]
- 21.Turgeon B G, Garber R C, Yoder O C. Development of a fungal transformation system based on selection of sequences with promoter activity. Mol Cell Biol. 1987;7:3297–3305. doi: 10.1128/mcb.7.9.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ueno Y. Trichothecene mycotoxins: mycology, chemistry, and toxicology. Adv Nutr Sci. 1980;3:301–351. [Google Scholar]
- 23.Ueno Y, Sawano M, Ishii K. Production of trichothecene mycotoxins by Fusarium species in liquid shake culture. Appl Microbiol. 1975;30:4–9. doi: 10.1128/am.30.1.4-9.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Gorcom R F M, Boschloo J G, Kuijvenhoven A, Lange J, van Vark A J, Bos C J, van Balken J A M, Pouwels P H, van den Hondel C A M J J. Isolation and molecular characterization of the benzoate-para-hydroxylase gene (bphA) of Aspergillus niger: a member of a new gene family of the cytochrome P450 superfamily. Mol Gen Genet. 1990;223:192–197. doi: 10.1007/BF00265053. [DOI] [PubMed] [Google Scholar]
- 25.Yu J J, Chang P K, Cary J W, Wright M, Bhatnagar D, Cleveland T E, Payne G A, Linz J E. Comparative mapping of aflatoxin pathway gene clusters in Aspergillus parasiticus and Aspergillus flavus. Appl Environ Microbiol. 1995;61:2365–2371. doi: 10.1128/aem.61.6.2365-2371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zamir L O, Devor K A, Nikolakakis A, Sauriol F. Biosynthesis of the trichothecene 3-acetyldeoxynivalenol: cell-free hydroxylations of isotrichodermin. Can J Microbiol. 1996;42:828–834. doi: 10.1139/m96-104. [DOI] [PubMed] [Google Scholar]