Abstract
Many strains of Pseudomonas syringae pv. syringae produce one of four classes of small cyclic lipodepsinonapeptides: syringomycins, syringostatins, syringotoxins, or pseudomycins. These metabolites are phytotoxic and growth inhibitory against a broad spectrum of fungi. Their production is dependent upon the expression of conserved biosynthesis and export genes syrB and syrD, respectively. PCR and oligonucleotide primers specific for a 752-bp fragment of syrB were used to identify cyclic lipodepsinonapeptide-producing strains of P. syringae pv. syringae. In contrast, PCR amplification with primers based on syrD did not always correlate with possession of the syrD gene, as indicated by Southern blot analysis, or with cyclic lipodepsinonapeptide production. Sequence comparisons of 400 nucleotides from the syrB PCR-amplified fragments showed 94% plot similarity among 27 strains. In a sequence phenogram, syringostatin and syringotoxin producers were grouped apart from syringomycin-producing strain B301D, with sequences that differed by eight and nine conserved base substitutions, respectively. PCR amplification of the 752-bp syrB fragment offers rapid and accurate detection of cyclic lipodepsinonapeptide-producing strains, and its sequence provides some predictive capabilities for identifying syringotoxin and syringostatin producers.
Many strains of Pseudomonas syringae pv. syringae are known to produce cyclic lipodepsipeptides as secondary metabolites. These metabolites consist of small forms (approximately 1,200 Da) (34) and large forms (approximately 2,500 Da) (3), with the latter collectively known as syringopeptins. The small forms are cyclic lipodepsinonapeptides (CLPs) that possess hydroxylated acyl chains and the conserved tetrapeptide sequence dehydroaminobutanoic acid-hydroxyaspartic acid-chlorothreonine-serine. They include the syringomycins (14, 31), syringostatins (13, 23), syringotoxins (5), and pseudomycins (6). Conceivably, other CLP forms remain undiscovered. A strain of Pseudomonas fuscovaginae was recently reported to produce syringotoxin (12), showing that CLPs are not produced exclusively by P. syringae pv. syringae.
The CLPs are considered to be plant virulence factors and antifungal agents (16). They affect plant membrane activities (11, 34) and induce necroses at relatively high concentrations (22), but the relationship of these effects to plant diseases has not been clearly established. Certain P. syringae pv. syringae mutants that do not produce syringomycin in vitro still cause varying degrees of disease symptoms (21, 26, 30, 35). On the other hand, a few P. syringae pv. syringae isolates that are nonpathogenic on pear and cherry fruit produce antifungal agents presumed to be syringomycin (17, 27), and wheat isolate P. syringae M1 produces syringomycin E but does not cause disease symptoms on wheat (1). Two P. syringae strains that produce syringomycin E (10) are employed as biocontrol agents against fungal diseases on postharvest citrus (9, 10). Purified CLPs have potent antifungal activities against a broad spectrum of fungi including human pathogens (20, 33).
D. C. Gross and colleagues have shown that the CLP production genes, syrB and syrD, are conserved among P. syringae pv. syringae strains (27, 28, 36). These genes are predicted to encode proteins that function in CLP synthesis and export, respectively. In view of the variabilities encountered in CLP production in vitro (27) and the difficulties in their chemical identification, the syrB and syrD genes offer a better means for identifying CLP producers. In addition, sequence differences in these genes offer the potential to identify strains that produce specific classes of CLPs, as suggested by restriction fragment length polymorphism (RFLP) analyses of Quigley and Gross (27).
In the present study, the earlier work of D. C. Gross and colleagues is extended by employing PCR protocols and oligonucleotide primers derived from the coding sequences of the syrB and syrD genes to quickly and reliably detect CLP-producing bacteria. In addition, PCR-amplified products from the syrB genes in 27 P. syringae strains were analyzed to determine relationships between base sequence and class of CLP produced.
MATERIALS AND METHODS
Bacterial strains.
The strains used in this study are listed in Table 1. Strains B301D, 475, SD19, 761-5, Ps268, B458, W4N5, W4N7, W4N47, and W4N103 were obtained from D. Gross (Washington State University, Pullman). Strains Lilac and SY12 were obtained from S. Thompson (Utah State University) and A. Isogai (Nara Institute of Science and Technology), respectively. Strains ESC-10 and ESC-11 were obtained from C. Bull (USDA Agricultural Research Service, Fresno, Calif.). P. syringae pv. tabaci and P. syringae pv. pisi were obtained from A. Anderson (Utah State University). Other strains were isolated locally (Table 1).
TABLE 1.
Strains used and their properties related to syrB and syrD and to CLP produced
| Strain | Plant source | syrB and syrD sequencesa (reference) | PCR signal for:
|
CLP produced, if known (reference) | |
|---|---|---|---|---|---|
| syrB | syrD | ||||
| P. syringae pv. syringae and related P. syringae strains | |||||
| B301D | Pear | Yes (27) | + | + | Syringomycin (4, 31) |
| 475 | Maize | Yes (27) | + | + | |
| SD19 | Sorghum | Yes (27) | + | + | |
| 761-5 | Foxtail | Yes (27) | + | + | |
| Ps268 | Lemon | Yes (27) | + | + | Syringotoxin (18) |
| SY12 | Lilac | Yes (27) | + | + | Syringostatin (24) |
| B458 | Orange | Yes | + | + | Syringotoxin (5) |
| W4N5 | Apple | Yes (27) | + | + | |
| W4N7 | Cherry | Yes (27) | + | + | |
| W4N47 | Cherry | No (27) | − | −c | |
| W4N103 | Cherry | Yes (27) | + | + | |
| Lilac | Lilac | Yes | + | + | Syringostatin (25) |
| M1 | Wheat | Yes (1) | + | + | Syringomycin (1) |
| M2 | Wheat | Yes | + | + | Syringomycin (32) |
| M3 | Wheat | Yes | + | + | |
| M4 | Wheat | Yes | + | + | |
| M5 | Wheat | Yes | + | + | |
| M6 | Wheat | Yes | + | + | |
| M7 | Wheat | Yes | + | + | |
| C1 | Cherry | No | − | − | None |
| C2 | Cherry | No | − | − | None |
| C3 | Cherry | Yes | + | + | C3A (25) |
| FP13 | Wheat | Yes | + | + | |
| FP14 | Wheat | Yes | + | + | |
| FP17 | Wheat | No | − | + | |
| FP18 | Wheat | Yes | + | + | |
| FP20 | Wheat | Yes | + | + | |
| FP21 | Wheat | Yes | + | + | |
| FP22 | Wheat | Yes | + | + | |
| ESC-10 | Apple | Yes (10) | + | + | Syringomycin (10) |
| ESC-11 | Pear | Yes (10) | + | + | Syringomycin (10) |
| pv. pisi | Unknown | No | − | −c | None |
| pv. tabaci | Unknown | No | − | −c | None |
| Other fluorescent pseudomonads from diseased plants | |||||
| SH-1 | Cherry | NDb | − | + | None |
| 95-39 | Cherry | ND | − | − | None |
| 95-54 | Lilac | ND | − | + | None |
| KS1 | Maize | No | − | − | None |
| KS2 | Maize | No | − | − | None |
As determined by Southern blotting.
ND, the test was not done.
A 446-bp syrD fragment was amplified at an annealing temperature of 60°C.
DNA isolation.
All strains were grown in potato dextrose broth (Difco Laboratories, Detroit, Mich.). Genomic DNA was isolated as described previously (2). After ethanol precipitation, nucleic acids were dissolved in sterile distilled water and stored at −20°C.
Southern blot analysis.
Detection of the syrB and syrD genes was performed by Southern blot analysis unless the presence of the genes was previously confirmed. Purified genomic DNAs were digested with the EcoRI restriction enzyme, electrophoresed on 0.8% agarose gels, and transferred to Hybond-N+ membranes (Amersham, Arlington Heights, Ill.). Plasmids p91 (pUC18 containing a 1-kb EcoRI-PstI fragment of syrB) and p9 (pUC19 containing a 510-bp SalI fragment of syrD) were obtained from D. Gross (Washington State University). The syrB and syrD fragments were purified by gel electrophoresis and hybridized with the enhanced chemiluminescence direct nucleic acid labeling and detection system (Amersham).
Oligonucleotide primers.
The complete sequences of the syrB (36) and syrD (28) genes from P. syringae pv. syringae B301D have been derived. Two 21-mer oligonucleotides for each sequence were selected for amplification by PCR with Oligo primer analysis software (National Biosciences, Inc., Plymouth, Minn.). For syrB, the oligonucleotide sequences were 5′-CTTTCCGTGGTCTTGATGAGG-3′ (primer B1) and 5′-TCGATTTTGCCGTGATGAGTC-3′ (primer B2). These primers were located 787 and 1,539 bp, respectively, into the open reading frame of the syrB gene and yielded a 752-bp product (34). For syrD, the oligonucleotide sequences were 5′-AAACCAAGCAAGAGAAGAAGG-3′ (primer D1) and 5′-GGCAATACCGAACAGGAACAC-3′ (primer D2). These primers were located 466 and 912 bp, respectively, into the open reading frame of the syrD gene and yielded a 446-bp product (28).
PCR amplification.
PCR amplification was performed in a total volume of 100 μl. Each reaction mixture contained 1× PCR reaction buffer (50 mM KCl, 10 mM Tris-HCl [pH 8.3]), a 0.5 μM concentration of each primer, a 200 μM concentration of each dNTP, 1.5 mM MgCl2, 0.025 U of Taq DNA polymerase per μl, 100 to 200 ng of genomic DNA, and 30 μl of mineral oil to prevent evaporation. The PCR was run for 35 cycles. Each cycle consisted of template denaturation at 94°C for 1.5 min, primer annealing at 60°C for 1.5 min, and DNA extension for 3.0 min at 72°C. After the cycling period was finished, an additional extension of 10 min at 72°C was included. Small aliquots (5 μl) of the PCR products were analyzed on 1% agarose gels. The remaining amplified DNAs were precipitated with three volumes of ethanol before digestion with the SalI restriction enzyme. The digested DNA fragments were also analyzed on agarose gels. Many samples were duplicated by using 10 μl of whole-cell suspensions in place of purified DNAs and/or increasing the annealing temperature to 62°C.
Sequencing of PCR products and analysis.
After amplification, the 752-bp syrB fragments from each strain were purified with a Wizard PCR Preps DNA purification system (Promega, Madison, Wis.). The sequence of each fragment was determined with primer B1 at the Utah State University Biotechnology Center (Logan). Sequences of 400 bp from 27 P. syringae strains were analyzed with the Wisconsin Sequence Analysis programs of the Genetics Computer Group (Madison, Wis.). The sequences were aligned with the Genetics Computer Group program PILEUP. Pairwise distances between the sequences were determined with the DISTANCES program by the Jukes-Cantor method. A phenogram was drawn based upon these distances with GROWTREE by the unweighted pair group method using arithmetic averages. Areas of similarity and divergence were determined by PLOTSIMILARITY. The sequence analysis started 810-bp into the open reading frame of syrB.
RESULTS
PCR analysis of bacterial strains.
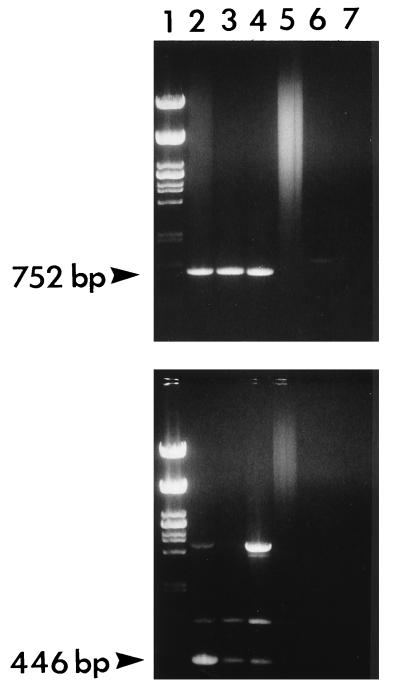
PCR amplification with the syrB primers yielded 752-bp fragments from all strains that were confirmed to have the syrB gene by Southern blot analyses (Table 1 and Fig. 1, upper panel). The 752-bp fragment was found to be absent when PCR was performed on DNAs isolated from strains that were confirmed syrB negative by Southern blot analyses. Occasionally, nonspecific fragments of various sizes would be amplified. However, no consistent pattern was seen for the occurrence of these fragments, as they were found in strains both positive and negative for syrB, and most were eliminated by increasing the annealing temperature to 62°C.
FIG. 1.
PCR amplification of the 752-bp fragment of syrB (upper panel) and the 446-bp fragment of syrD (lower panel). The primer annealing temperature was 62°C. Lane 1, λ-PstI-digested DNA; lane 2, P. syringae pv. syringae B301D; lane 3, related P. syringae W4N7; lane 4, P. syringae pv. syringae SY12; lane 5, W4N47; lane 6, P. syringae pv. pisi; lane 7, P. syringae pv. tabaci. Products were separated on 1.0% agarose gels.
As expected, the syrD primers amplified a 446-bp fragment (Fig. 1, lower panel). This amplification was seen with all strains that were confirmed to have syrD by Southern blot analyses, but it was not limited to these strains (Table 1). The syrD-negative strains for which the 446-bp fragment was amplified were P. syringae pv. tabaci, P. syringae pv.pisi, and P. syringae W4N47. This 446-bp fragment was not amplified in these strains when the annealing temperature was increased to 62°C (Fig. 1, lower panel). Another fragment of approximately 800 bp was often amplified.
The same results were obtained with whole-cell suspensions instead of purified genomic DNAs.
Restriction enzyme analysis of amplified products.
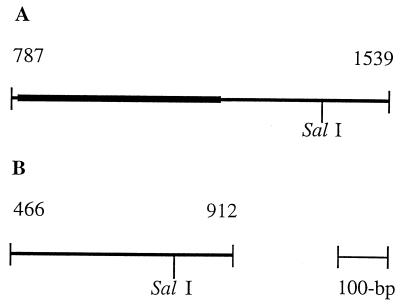
The SalI restriction enzyme site found in the B301D product (Fig. 2A) was not always present in the syrB-amplified products from other strains. SalI digestion resulted in three distinguishable RFLPs: (i) one 752-bp fragment (no SalI site), (ii) two fragments of 621 and 131 bp whose sizes were equivalent to those predicted by the B301D restriction map (Fig. 2A), and (iii) two fragments whose sizes were estimated to be 410 and 342 bp.
FIG. 2.
Restriction maps of the syrB (A) and syrD (B) regions of P. syringae pv. syringae B301D amplified by PCR. The boldface line indicates the 400-bp region of syrB that was sequenced.
The SalI restriction site in the syrD-amplified fragment of B301D (Fig. 2B) was conserved in all strains. The analysis yielded two fragments from all strains; the fragments were 321 and 125 bp in size.
Nucleotide sequence analysis.
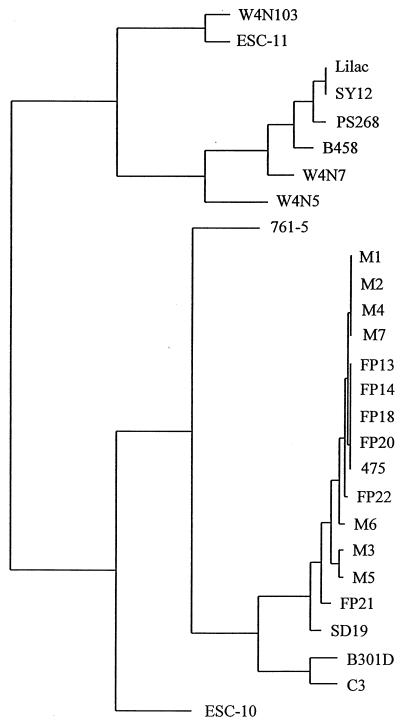
For the 27 strains with positive PCR signals for syrB, the sequence plot similarity between the 400-nucleotide fragments was 94%. Most of the differences were base substitutions. Percent differences in the sequences ranged from 12.5% between B301D and W4N5 to 0% among three different groups. Strains that were grouped together with identical sequences were Lilac and SY12 (syringostatin producers); M1, M2, M4, and M7; and FP13, FP14, FP18, and FP20 (Fig. 3). Strains within the latter two groups were isolated from the same plant hosts in similar locations, but strains Lilac and SY12 were isolated separately from lilac hosts in Kaysville, Utah, and Japan, respectively. The sequences of syringotoxin-producing strains Ps268 and B458 differed by 1.5%. Nucleotide analysis grouped the 27 strains into three major branches (Fig. 3). One branch contained the known syringostatin (Lilac and SY12)- and syringotoxin (Ps268 and B458)-producing strains. Compared to the sequence of syringomycin-producing strain B301D, the sequences of the syringostatin producers differed by 9.9% and those of the syringotoxin producers, Ps268 and B458, differed by 9 and 10.2%, respectively. Nine and eight base substitutions were unique to the syringostatin- and syringotoxin-producing strains, respectively, compared to B301D (Table 2). Other strains that were grouped with the syringostatin- and syringotoxin-producing strains were W4N7 and W4N5.
FIG. 3.
Phenogram of sequences of the 400-bp fragments of syrB from 27 different strains. The lengths of horizontal lines represent relative numbers of nucleotide substitutions assumed under the Jukes-Cantor model.
TABLE 2.
Comparison of syrB fragment base substitutions unique to syringostatin and syringotoxin producers with corresponding bases of syringomycin-producing strain B301D
| Open reading frame nucleotide position | Base of:
|
||||
|---|---|---|---|---|---|
| B301D | Syringostatin producer
|
Syringotoxin producer
|
|||
| Lilac | SY12 | PS268 | B458 | ||
| 887 | C | G | G | G | G |
| 936 | C | T | T | T | T |
| 987 | G | A | A | A | A |
| 1002 | A | G | G | G | G |
| 1011 | G | T | T | T | T |
| 1065 | G | C | C | C | C |
| 1083 | T | C | C | C | C |
| 1123 | G | A | A | G | G |
| 1129 | C | T | T | T | T |
Strains that have been identified as producing syringomycin were found throughout the other two branches. Strain C3, which produces a new CLP, C3A, was placed near syringomycin-producing B301D. The percent sequence difference between these strains was 2.8%.
DISCUSSION
A PCR method was developed and evaluated for its ability to identify P. syringae pv. syringae strains that produce CLPs. It is based on the known conservation of the peptide biosynthesis gene, syrB, in this group of organisms (27) and was shown to specifically detect strains that produce syringomycin, syringostatin, and syringotoxin. A similar PCR-based approach was developed and used to identify analogous coronatine-producing strains of P. syringae (8). In addition to their specificity and reliability, these methods offer the advantages of speed, sensitivity, and simplicity compared to approaches based on DNA hybridization. Thus, PCR amplification of the 752-bp syrB fragment documented in this report could be exploited for applications such as the detection and quantitation of CLP-producing bacteria in host plants. Examples of such applications are the use of PCR-based assays to detect Erwinia amylovora in fruit tree tissues for diagnosis of fire blight disease (7) and the detection and localization of phenazine and 2,4-diacetylphloroglucinol antibiotic-producing fluorescent pseudomonads in soils (29).
The sequences of the PCR-amplified syrB fragments from 27 syrB-positive strains were compared. As observed previously by Quigley and Gross (27) using genomic DNA hybridization analyses, syrB is polymorphic. This sequence polymorphism is likely related to variations in the CLPs produced by the different P. syringae strains. SyrB is postulated to be a peptide synthetase based on its shared homology with adenylate-forming enzymes that catalyze peptide antibiotic and siderophore biosyntheses (36). Specifically, six signature core sequences together resemble those of amino acid-activating domains of thiotemplate-employing peptide synthetases. One specific region (residues 310 to 333) falls within the PCR-amplified sequences analyzed in the present study and was suggested to be responsible for binding and activating serine for syringomycin biosynthesis (36). However, recent studies with enzymatically active preparations show that SyrB activates threonine (which is subsequently chlorinated) and not serine (15, 19). Analogous regions occur in peptide synthetases encoded by grsB and HTS-1 of Bacillus brevis and Cochliobolus carbonum, respectively, which activate proline, and EntF of Escherichia coli, which activates serine (36). Within this region of syrB, we observed two different base substitutions when comparing syringomycin-producing strain B301D with syringostatin- and syringotoxin-producing strains (Table 2). Syringostatin and syringotoxin differ from each other by a single amino acid (at corresponding positions, syringostatin has diaminobutanoic acid and syringotoxin has glycine) and differ significantly from syringomycin in hydroxyamino acid content (34). Syringostatin and syringotoxin, but not syringomycin, possess homoserine and threonine, and they contain one serine residue instead of syringomycin’s two. All possess chlorothreonine flanked by hydroxyaspartic acid (on the N-terminal side) and serine. If the syrB sequence variations observed in the present study do not contribute to these hydroxyamino acid differences, they may instead reflect evolutionary divergence of the conserved activation domain for chlorothreonine. A close genetic relatedness between syringostatin and syringotoxin producers was also noted by Quigley and Gross (27) based on genomic RFLP analyses.
Partial sequence analysis of the 752-bp syrB fragments offers some limited capability for predicting the kind of CLP produced. As mentioned above, four separately isolated syringostatin and syringotoxin producers showed high levels of sequence identity that led to close groupings in a sequence phenogram (Fig. 3). Two other P. syringae pv. syringae strains, W4N5 and W4N7, were also grouped closely with the syringostatin and syringotoxin producers. Although the CLPs from these two strains have not been chemically identified, the probability is high that they each synthesize either syringostatin or syringotoxin. In contrast, syringomycin-producing strains occur throughout the phenogram, thus precluding the use of this fragment sequence to predict producers of this CLP. A similar lack of sequence relatedness in syringomycin producers was observed by Quigley and Gross (27), who found that syringomycin-producing strains B301D and SC-1 occupied different branches of maximum-parsimony trees based on RFLP analyses with syrB and syrD probes.
Recently isolated P. syringae C3 produces a novel CLP with a unique amino acid composition (25) and, accordingly, is not grouped closely with producers of CLPs of known structure. In regard to plant source, P. syringae strains isolated from cereals formed a tight cluster in the sequence phenogram (Fig. 3). These included strains with the M and FP prefixes isolated from wheat and strains 475 and SD19 from maize and sorghum, respectively. However, the foxtail isolate, strain 761-5, differed significantly from the other monocot strains. A similar relationship among monocots was observed by Quigley and Gross (27) using RFLP maximum-parsimony analyses.
The syrD oligonucleotide primers were effective in detecting the presence of the syrD gene, but they were not always specific to known CLP-producing strains at an annealing temperature of 60°C. At the higher annealing temperature, 62°C, the syrD primers were more specific and may be useful for detecting new CLP-producing strains. The syrD gene is expected to code for an ATP-binding transporter (28), and the lack of specificity of the observed 446-bp fragment may reflect the ubiquity of this or a similar transporter. Also, the observation of additional amplified products derived from the syrD primers (Fig. 1) suggests the occurrence of other related transporter genes in CLP producers.
ACKNOWLEDGMENTS
We thank D. C. Gross (Washington State University) for providing the syrB sequence and strains and for insightful reviewing, N. Quigley (University of Tennessee) for DNA samples in early phases of this work, and Paul Wolf (Utah State University) for advice in sequence analyses.
This research was supported by the Utah Agriculture Experiment Station (Project 607) and the Utah State University Research Office.
Footnotes
Journal paper 6020 of the Utah Agricultural Experiment Station.
REFERENCES
- 1.Adetuyi F C, Isogai A, Di Giorgio D, Ballio A, Takemoto J Y. Saprophytic Pseudomonas syringae strain M1 of wheat produces cyclic lipodepsipeptides. FEMS Microbiol Lett. 1995;131:63–67. doi: 10.1016/0378-1097(95)00236-x. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1989. [Google Scholar]
- 3.Ballio A, Barra D, Bossa F, Collina A, Grgurina I, Marino G, Moneti G, Paci M, Pucci P, Segre A, Simmaco M. Syringopeptins, new phytotoxic lipodepsipeptides of Pseudomonas syringae pv. syringae. FEBS Lett. 1991;291:109–112. doi: 10.1016/0014-5793(91)81115-o. [DOI] [PubMed] [Google Scholar]
- 4.Ballio A, Barra D, Bossa F, DeVay J E, Grgurina I, Iacobellis N S, Marino G, Pucci P, Simmaco M, Surico G. Multiple forms of syringomycin. Physiol Mol Plant Pathol. 1988;33:493–496. [Google Scholar]
- 5.Ballio A, Bossa F, Collina A, Gallo M, Iacobellis N S, Paci M, Pucci P, Scaloni A, Segre A, Simmaco M. Structure of syringotoxin, a bioactive metabolite of Pseudomonas syringae pv. syringae. FEBS Lett. 1990;269:377–380. doi: 10.1016/0014-5793(90)81197-v. [DOI] [PubMed] [Google Scholar]
- 6.Ballio A, Bossa F, Di Giorgio D, Ferranti P, Paci M, Pucci P, Scaloni A, Segre A, Strobel G A. Novel bioactive lipodepsipeptides from Pseudomonas syringae: the pseudomycins. FEBS Lett. 1994;355:96–100. doi: 10.1016/0014-5793(94)01179-6. [DOI] [PubMed] [Google Scholar]
- 7.Bereswill S, Bugert P, Völksch B, Ullrich M, Bender C L, Geider K. Identification and relatedness of coronatine-producing Pseudomonas syringae pathovars by PCR analysis and sequence determination of the amplification products. Appl Environ Microbiol. 1994;60:2924–2930. doi: 10.1128/aem.60.8.2924-2930.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bereswill S, Pahl A, Bellemann P, Zeller W, Geider K. Sensitive and species-specific detection of Erwinia amylovora by polymerase chain reaction analysis. Appl Environ Microbiol. 1992;58:3522–3526. doi: 10.1128/aem.58.11.3522-3526.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bull C T, Stack J P, Smilanick J L. Pseudomonas syringae strains ESC-10 and ESC-11 survive on wounds on citrus and control green and blue molds of citrus. Biol Control. 1997;8:81–88. [Google Scholar]
- 10.Bull, C. T., M. L. Wadsworth, K. N. Sorensen, J. Y. Takemoto, R. K. Austin, and J. L. Smilanick. Syringomycin E produced by biological control agents controls green mold on lemons. Submitted for publication.
- 11.Di Giorgio D, Camoni L, Ballio A. Toxins of Pseudomonas syringae pv. syringae affect H+-transport across the plasma membrane of maize. Physiol Plant. 1994;91:741–746. [Google Scholar]
- 12.Flamand M-C, Pelsser S, Ewbank E, Maraite H. Production of syringotoxin and other bioactive peptides by Pseudomonas fuscovaginae. Physiol Mol Plant Pathol. 1996;48:217–231. [Google Scholar]
- 13.Fukuchi N, Isogai A, Nakayama J, Takayama S, Yamashita S. Isolation and structural elucidation of syringostatins, phytotoxins produced by Pseudomonas syringae pv. syringae lilac isolate. J Chem Soc Perkin Trans I. 1992;1:875–880. [Google Scholar]
- 14.Fukuchi N, Isogai A, Yamashita S, Suyama K, Takemoto J Y, Suzuki A. Structure of phytotoxin syringomycin produced by a sugar cane isolate of Pseudomonas syringae pv. syringae. Tetrahedron Lett. 1990;31:1589–1592. [Google Scholar]
- 15.Gross, D. C. 1997. Personal communication.
- 16.Gross D C. Molecular and genetic analysis of toxin production by pathovars of Pseudomonas syringae. Annu Rev Phytopathol. 1991;29:247–278. [Google Scholar]
- 17.Gross D C, Cody Y S, Proebsting E L, Jr, Radamaker G K, Spotts R A. Ecotypes and pathogenicity of ice-nucleation-active Pseudomonas syringae isolated from deciduous fruit tree orchards. Phytopathology. 1984;74:241–248. [Google Scholar]
- 18.Gross D C, DeVay J E. Production and purification of syringomycin, a phytotoxin produced by Pseudomonas syringae. Physiol Plant Pathol. 1977;11:13–28. [Google Scholar]
- 19.Guenzi E, Galli G, Grgurina I, Schloz-Schroeder B, Gross D, Grandi G. Abstracts of the 97th General Meeting of the American Society for Microbiology, 1997. Washington, D.C: American Society for Microbiology; 1997. Genetic and functional characterization of the syringomycin operon of Pseudomonas syringae pv. syringae. abstr. B-361; p. 90. [Google Scholar]
- 20.Harrison L, Teplow D B, Rinaldi M, Strobel G. Pseudomycins, a family of novel peptides from Pseudomonas syringae possessing broad-spectrum antifungal activity. J Gen Microbiol. 1991;137:2857–2865. doi: 10.1099/00221287-137-12-2857. [DOI] [PubMed] [Google Scholar]
- 21.Hrabak E M, Willis D K. Involvement of the lemA gene in production of syringomycin and protease by Pseudomonas syringae pv. syringae. Mol Plant-Microbe Interact. 1993;6:368–375. [Google Scholar]
- 22.Iacobellis N S, Lavermicocca P, Grgurina I, Simmaco M, Ballio A. Phytotoxic properties of Pseudomonas syringae pv. syringae toxins. Physiol Mol Plant Pathol. 1992;40:107–116. [Google Scholar]
- 23.Isogai A, Fukuchi N, Yamashita S, Suyama K, Suzuki A. Structures of syringostatins A and B, novel phytotoxins produced by Pseudomonas syringae pv. syringae isolated from lilac blights. Tetrahedron Lett. 1990;31:695–698. [Google Scholar]
- 24.Isogai A, Fukuchi N, Yamashita S, Suyama K, Suzuki A. Syringostatins, novel phytotoxins produced by Pseudomonas syringae pv. syringae. Agric Biol Chem. 1989;53:3117–3119. [Google Scholar]
- 25.Kim K-H. M. S. thesis. Logan: Utah State University; 1996. [Google Scholar]
- 26.Mazzola M, White F F. A mutation in the indole-3-acetic acid biosynthesis pathway in Pseudomonas syringae pv. syringae affects growth in Phaseolus vulgaris and syringomycin production. J Bacteriol. 1994;176:1374–1382. doi: 10.1128/jb.176.5.1374-1382.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quigley N B, Gross D C. Syringomycin production among strains of Pseudomonas syringae pv. syringae: conservation of the syrB and syrD genes and activation of phytotoxin production by plant signal molecules. Mol Plant-Microbe Interact. 1994;7:78–90. doi: 10.1094/mpmi-7-0078. [DOI] [PubMed] [Google Scholar]
- 28.Quigley N B, Mo Y-Y, Gross D C. SyrD is required for syringomycin production by Pseudomonas syringae pathovar syringae and is related to a family of ATP-binding secretion proteins. Mol Microbiol. 1993;9:787–801. doi: 10.1111/j.1365-2958.1993.tb01738.x. [DOI] [PubMed] [Google Scholar]
- 29.Raaijmakers J M, Weller D M, Thomashow L S. Frequency of antibiotic-producing Pseudomonas spp. in natural environments. Appl Environ Microbiol. 1997;63:881–887. doi: 10.1128/aem.63.3.881-887.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rich J J, Willis D K. Multiple loci of Pseudomonas syringae pv. syringae are involved in pathogenicity on bean: restoration of one lesion-deficient mutant requires two tRNA genes. J Bacteriol. 1997;179:2247–2258. doi: 10.1128/jb.179.7.2247-2258.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Segre A, Bachmann R C, Ballio A, Bossa F, Grgurina I, Iacobellis N S, Marino G, Pucci P, Simmaco M, Takemoto J Y. The structure of syringomycins A1, E and G. FEBS Lett. 1989;255:27–31. doi: 10.1016/0014-5793(89)81054-3. [DOI] [PubMed] [Google Scholar]
- 32.Sorensen, K. N. Unpublished data.
- 33.Sorensen K N, Kim K-H, Takemoto J Y. In vitro antifungal and fungicidal activities and erythrocyte toxicities of cyclic lipodepsinonapeptides produced by Pseudomonas syringae pv. syringae. Antimicrob Agents Chemother. 1996;40:2710–2713. doi: 10.1128/aac.40.12.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takemoto J Y. Bacterial phytotoxin syringomycin and its interaction with host membranes. In: Verma D P S, editor. Molecular signals in plant-microbe communications. Boca Raton, Fla: CRC Press; 1992. pp. 247–260. [Google Scholar]
- 35.Xu G-W, Gross D. Physical and functional analyses of the syrA and syrB genes involved in syringomycin production by Pseudomonas syringae pv. syringae. J Bacteriol. 1988;170:5680–5688. doi: 10.1128/jb.170.12.5680-5688.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J-H, Quigley N B, Gross D C. Analysis of the syrB and syrC genes of Pseudomonas syringae pv. syringae indicates that syringomycin is synthesized by a thiotemplate mechanism. J Bacteriol. 1995;177:4009–4020. doi: 10.1128/jb.177.14.4009-4020.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]