Abstract
The effects of heavy-metal-containing sewage sludge on the soil microbial community were studied in two agricultural soils of different textures, which had been contaminated separately with three predominantly single metals (Cu, Zn, and Ni) at two different levels more than 20 years ago. We compared three community-based microbiological measurements, namely, phospholipid fatty acid (PLFA) analysis to reveal changes in species composition, the Biolog system to indicate metabolic fingerprints of microbial communities, and the thymidine incorporation technique to measure bacterial community tolerance. In the Luddington soil, bacterial community tolerance increased in all metal treatments compared to an unpolluted-sludge-treated control soil. Community tolerance to specific metals increased the most when the same metal was added to the soil; for example, tolerance to Cu increased most in Cu-polluted treatments. A dose-response effect was also evident. There were also indications of cotolerance to metals whose concentration had not been elevated by the sludge treatment. The PLFA pattern changed in all metal treatments, but the interpretation was complicated by the soil moisture content, which also affected the results. The Biolog measurements indicated similar effects of metals and moisture to the PLFA measurements, but due to high variation between replicates, no significant differences compared to the uncontaminated control were found. In the Lee Valley soil, significant increases in community tolerance were found for the high levels of Cu and Zn, while the PLFA pattern was significantly altered for the soils with high levels of Cu, Ni, and Zn. No effects on the Biolog measurements were found in this soil.
Ecotoxicological research usually involves investigations at either the single-species or ecosystem level. The latter is common in soil studies, e.g., measurements of different processes within the carbon or nitrogen cycle (for reviews, see references 2 and 8). In such studies, the microbial community is often considered a black box. However, studies within this black box, e.g., on species composition or level of tolerance of the community, can be useful indicators of toxic effects of pollutants since it is possible that the microbial community can be altered without resulting in changes in the overall performance of the soil system.
The effect of metal contamination of the soil on the species composition of soil microorganisms has been studied previously (2), but this is not a very common approach. Usually only standard techniques, such as measurements of different activities and biomass, are used (8). One reason for this is the amount of work involved in isolating and typing bacteria or fungi, which makes it difficult to process the large number of samples usually needed in ecological studies. There is thus a need for rapid techniques which give an indication of the composition of the microbial community. One such technique is the analysis of the phospholipid fatty acid (PLFA) pattern. Phospholipids are located in membranes of the cell. Since different subsets of microorganisms have different PLFA compositions, the PLFA pattern of a soil sample will reflect the microbial community composition. This technique has been used to detect changes in the soil microbial community structure due to metal pollution both in laboratory studies and in the field (12, 17–19, 27).
Another technique for monitoring changes in microbial communities is the use of sole-carbon-source tests. The method first described by Garland and Mills (22) involves a commercially available microtiter plate (Biolog), which can be used to simultaneously test the utilization of 95 substrates as sole carbon sources. Carbon source utilization is indicated by color development of a redox indicator dye, and changes in the overall patterns of carbon source utilization rates can be assessed by multivariate statistics. The technique has been used to detect differences between microbial communities in soil and the rhizosphere (16, 21, 23, 28, 29, 32, 34), but only in a few cases have the effects of metal pollution been studied (17, 25).
The presence of elevated metal concentrations can exert a selective pressure on the microbial community such that levels of metal-tolerant and -resistant species are increased. A simple way of measuring changes in the tolerance of microbial communities to metals is to use agar plates with different concentrations of the pollutant (1, 15, 24). However, this involves much work, and also only the culturable part of the microbial community is studied. A simple technique was devised by Bååth (4), who used thymidine incorporation of bacteria extracted from soil at different metal concentrations as a fast bioassay to determine the tolerance of bacterial communities. This technique has also been used to detect metal effects in both laboratory (13, 14) and field (12, 27) studies.
In the present study, the effects of metal-rich sludge amendments on these three different microbial community-based measurements have been compared. We used two different sludge-amended soils, in which the effects of metals on the microbial biomass were studied previously (11). Both these soils were contaminated over 20 years ago with either Cu, Zn, and Ni as the predominant metal, amended at two different concentrations. This would make it possible to test whether different metals induced different responses in the microbial community, as revealed by the three different methods used, as well as the sensitivity of each method in detecting the effects of heavy metals.
MATERIALS AND METHODS
Soils and pretreatments.
Soils from both the Lee Valley and Luddington long-term sewage sludge experiments were used and have been described in detail elsewhere (5–7, 10, 11). The Lee Valley experiment was set up in 1968 on a silty loam soil of the Hamble series (5). The Luddington soil belongs to the Wick series and has a sandy loam texture. The Luddington soil has a lower pH (5.8 and 6.5), a lower cation-exchange capacity (11.5 and 22.5 meq/100 g), and a lower organic-matter content (1.8 and 4.9%) than the Lee Valley soil (6).
The experiments consisted of control plots with no sludge added and plots amended with either uncontaminated sludge or sludge contaminated with one of either Cu, Ni, or Zn as the predominant metal. Each metal-rich sludge was applied at either a low or high rate of amendment, and the experiment had a replicated blocked factorial design (5).
Contaminated sludges were applied at rates of 16 g of Zn per kg, 8 g of Cu per kg, and 4 g of Ni kg for the high rate of application. The uncontaminated sludge was used to adjust the contaminated sludges to obtain the low rate of application at half of the high rate. Soils held in the Macaulay Land Use Research Institute archive store for blocks 2 and 4 of Lee Valley (10) and for blocks 1 to 4 of Luddington were used. The soils from Luddington and Lee Valley were sampled in 1990 from the 0- to 10-cm soil depth and in 1988 from the 0- to 15-cm soil depth, respectively, and were taken from the sides of profile pits dug in the center of each plot to avoid areas affected by soil mixing (11). The soils had been air dried, sieved, and stored in the dark at 10°C for 4 to 6 years.
For each soil sample, approximately 500 g of dry soil was wetted and allowed to drain freely in 7-cm-diameter plastic plant pots. The pots were then sown with white clover (Trifolium repens cv. Blanca) seeds and maintained at approximately 20°C in a greenhouse. The clover was cut at regular intervals of 14 days, and the accumulated dry-matter yield was recorded. After 3 months of growth, the clover was harvested and weighed.
Microbiological analysis.
Soil samples were sieved (mesh size, <2 mm) and sorted to remove plant debris and any soil animals and then allowed to stabilize for 7 days at 25°C before analysis of soil microbial biomass carbon by the fumigation-extraction procedure (30, 33) and basal respiration. Basal respiration (CO2 evolution) was measured in triplicate on 20-g samples of soil in 100-cm3 soil jars after 7 days by using a gas chromatograph to measure the headspace CO2 that accumulated over 6 h at 25°C. Thymidine incorporation was measured by the method of Bååth (3) (see below for further details).
Chemical analysis.
The moisture content was determined by drying at 105°C for 24 h. The soil pH was measured after equilibrating 10 g of soil with water. The total soil C and N were measured, after dry combustion, with an elemental analyzer (NA1500; Carlo-Erba, Milan, Italy). The soils were extracted with aqua regia (26), and all the metals in the extracts were analyzed by ICP optical emission spectroscopy, except for Cd, which was analyzed by graphite furnace atomic absorption spectroscopy. The metal content in the Lee Valley soil has been reported previously (10).
Tolerance of the bacterial community to metals.
The bacterial growth rate and heavy-metal tolerance of the bacterial community were estimated by the thymidine (TdR) incorporation technique (3, 4, 13) with the bacterial community extracted from soil by homogenization and centrifugation. The following metal salts were used: CuSO4, ZnSO4, Ni(NO3)2, and CdSO4. Each metal was added at five to seven different concentrations to the extracted bacterial solution before TdR was added. A control without any added metals was always included. TdR incorporation was expressed as a percentage of this control value, and the heavy-metal tolerance of the different bacterial communities was then estimated by calculating the concentration of added metals which resulted in 50% TdR incorporation (IC50) compared to the control value. The changes in the level of tolerance to different metals were expressed as ΔIC50, which was calculated by subtracting the IC50 for the unpolluted sludge treatments from that for the other treatments.
PLFA analysis.
The phospholipid extraction and PLFA analysis were performed as previously described by Frostegård et al. (18). Briefly, lipids from 3 g (fresh weight) of soil were extracted with a chloroform-methanol-citrate buffer mixture and the phospholipids were then separated from other lipids on a silicic acid column. The phospholipids were subjected to mild-alkali methanolysis, and the resulting fatty acid methyl esters were separated by gas chromatography. Fatty acids are designated in terms of the total number of carbon atoms: number of double bonds, followed by the position of the double bond from the methyl end of the molecule. The prefixes a and i indicate anteiso and iso branching; br indicates an unknown methyl branching position; and cy indicates a cyclopropane fatty acid. 10Me indicates a methyl group on the 10th carbon atom from the carboxyl end of the molecule.
Biolog method.
Fresh soil (10 g) was added to 90 ml of distilled water and shaken on a wrist action shaker at full speed for 10 min, and 10-fold serial dilutions were made. The 10−4 dilution, which was the lowest dilution with minimal background color, was used to inoculate the Biolog plates. The 10−4 dilution was centrifuged at 750 × g for 10 min to separate the soil, and 150 μl of supernatant was inoculated into each well of a GN type plate (Biolog Inc., Hayward, Calif.). The plates were incubated at 15°C for up to 4 days on an orbital shaker at 100 rpm. Color development was measured as absorbance at 590 nm on an automated plate reader (EMAX; Molecular Devices, Crawley, United Kingdom) after 24, 48, and 96 h, and the data were collected with Softmax (Molecular Devices) software. Because the plates were visibly colored by the addition of soil extract, the initial absorbances were measured immediately after inoculation and were subtracted from subsequent daily readings. The average well color development (AWCD) for all carbon sources (22) was calculated as being indicative of total activity.
Statistics.
The data was analyzed by one-way analysis of variance (ANOVA), with or without soil moisture as a covariate (see Results). Only differences between the uncontaminated-sludge treatment, which was considered to be the most proper control treatment, and the other treatments were assessed, with P < 0.05 (least significant difference) as the significance criteria.
Principal-components analyses were performed on the PLFA data and the Biolog data separately. The PLFA data was expressed as moles percent and logarithmically transformed before analyses. We used 29 (Luddington) or 30 PLFAs (Lee Valley) in the statistical analysis with Sirius software (Pattern Recognition Systems A/S, Bergen, Norway). The Biolog data was first transformed by being divided by the AWCD to avoid bias between samples with different numbers of culturable organisms (22) and was then analyzed by principal-component analysis with Genstat Rel 5.3 (NAG Ltd., Oxford, United Kingdom).
RESULTS
Luddington soil. (i) Metal levels.
The Cu content in soil subjected to the Cu-contaminated treatments was significantly higher than in soil given the uncontaminated-sludge treatment but did not increase much in the soils given the other treatments (Table 1). The Ni content was also significantly higher only in the Ni-contaminated plots, while the Zn level was high not only in the Zn-contaminated treatments but also in some of the treatments not intended to increase the Zn levels in soil. The Cd and Cr contents, although not studied specifically, were also significantly higher in the soils given the metal-polluted treatments (Table 1).
TABLE 1.
Mean metal concentrations in the Luddington soil
| Treatment | Mean concn (mg kg of dry soil−1) ofa:
|
||||
|---|---|---|---|---|---|
| Cu | Zn | Ni | Cd | Cr | |
| Uncontaminated sludge | 18 | 66 | 12 | 0.22 | 16 |
| No sludge | 24 | 67 | 12 | 0.23 | 21 |
| Low Cu | 125* | 145* | 17 | 0.54* | 48* |
| High Cu | 282* | 222* | 21 | 0.85* | 81* |
| Low Zn | 20 | 207* | 13 | 0.36* | 25* |
| High Zn | 26 | 359* | 13 | 0.56* | 38* |
| Low Ni | 30 | 150* | 69* | 0.55* | 51* |
| High Ni | 29 | 167* | 89* | 0.51* | 50* |
| Mean SE | 5.6 | 24.1 | 4.7 | 0.04 | 4.6 |
Asterisks indicate significant difference from the uncontaminated-sludge treatment (n = 4, P < 0.05; mean SE from ANOVA).
(ii) General biological and chemical effects.
The addition of uncontaminated or metal-contaminated sludge increased the carbon content of the soil compared to the no-sludge treatment (Table 2). This was usually also the case for the percent nitrogen, whereas the pH was unaffected by the different treatments. The microbial biomass carbon (C-mic) decreased in most of the metal-contaminated plots compared to the soil given the uncontaminated-sludge treatment, although this decrease was significant only for the high-Cu treatment (Table 2).
TABLE 2.
Effect of treatments on properties of the Luddington soila
| Treatment | %C | %N | pH | Moisture content (%) | C-mic (μg g−1) | Respiration (μg of C g−1 day−1) | TdR incorporation (10−13 mol ml−1 h−1) | Clover yield (g) | Principal-components analysis
|
|
|---|---|---|---|---|---|---|---|---|---|---|
| PC1 | PC2 | |||||||||
| Uncontaminated sludge | 2.00 | 0.16 | 5.1 | 7.90 | 117.7 | 1.78 (1.04) | 2.63 (2.04) | 2.72 | 0.166 | −0.230 |
| No sludge | 1.52 | 0.13 | 5.4 | 9.76 | 116.9 | 1.48 (1.46) | 2.60 (2.58) | 1.64* | −0.021 | −0.038* |
| Low Cu | 2.44 | 0.21 | 5.2 | 10.48* | 99.0 | 1.44 (1.70) | 1.07* (1.28) | 2.22 | −0.028 | −0.008* |
| High Cu | 2.64* | 0.23* | 5.4 | 14.15* | 55.4* | 0.74* (2.42) | 0.34* (1.68) | 1.50* | −0.587* | −0.027* |
| Low Zn | 2.25 | 0.16 | 5.4 | 7.94 | 124.2 | 1.71 (0.98) | 3.03 (2.39) | 3.06 | −0.155 | 0.063* |
| High Zn | 1.97 | 0.15 | 5.5 | 10.32* | 90.8 | 1.96 (2.16) | 1.72 (1.88) | 1.68* | 0.033 | 0.051* |
| Low Ni | 2.54 | 0.19 | 5.3 | 8.69 | 81.8 | 1.48 (1.04) | 2.64 (2.30) | 2.59 | 0.160 | 0.138* |
| High Ni | 1.73 | 0.13 | 5.4 | 9.25 | 85.0 | 1.34 (1.12) | 3.17 (2.99) | 2.02 | 0.121 | 0.117* |
| Mean SE | 0.19 | 0.02 | 0.11 | 0.82 | 19.5 | 0.19 (0.37) | 0.42 (0.49) | 0.33 | 0.088 | 0.042 |
Asterisks indicate significant difference from the uncontaminated sludge treatment (n = 4, P < 0.05, mean SE from ANOVA). For the respiration and TdR data, values are given after adjusting for covariance of the moisture content in the ANOVA, and the unadjusted values are given in parentheses.
The clover yield also decreased in the most highly contaminated plots, significantly so for the high-Cu and high-Zn treatments (Table 2). The difference in plant growth and subsequently greater evapotranspiration introduced a confounding variation in soil moisture content between treatments and replicates, in that there was a strong negative correlation (r = −0.75, P < 0.001, n = 32) between plant growth and moisture content. The effect of the moisture content could be seen for the respiration measurements, which were closely correlated with soil moisture (r = 0.86, P < 0.001). However, the high-Cu treatment led to significantly lower (P < 0.05) respiration rate than did all other treatments when the ANOVA was performed with soil moisture as a covariate (Table 2). The effect of moisture was also evident for the TdR incorporation rate. No significant differences due to the different metal sludge amendments were found, unless soil moisture was used as a covariate (Table 2), and then the high- and low-Cu treatments resulted in significantly lower TdR incorporation rates than did the uncontaminated sludge. The high-Zn treatment also led to lower TdR incorporation than did the uncontaminated sludge control.
(iii) Tolerance measurements.
The bacterial community tolerance, expressed as IC50s, increased in all the metal-polluted treatments compared to the uncontaminated sludge control treatment (Table 3). The effect was most pronounced for tolerance to the metal added to the soil, and a dose-response effect was also evident. Thus, community tolerance to Cu was highest for the high-Cu treatment followed by the low-Cu treatment, tolerance to Zn was highest for the bacterial community from the high-Zn treatment, and tolerance to Ni was highest for the community from the high-Ni treatment.
TABLE 3.
Bacterial community tolerance of metals in Luddington
| Treatment | Community tolerance (IC50) ofa:
|
|||
|---|---|---|---|---|
| Cu | Zn | Ni | Cd | |
| Uncontaminated sludge | −6.16 | −4.79 | −4.21 | −4.70 |
| No sludge | −5.83* (0.34) | −4.61 (0.18) | −4.21 (0.00) | −4.46 (0.24) |
| Low Cu | −5.48* (0.68) | ND | ND | ND |
| High Cu | −5.18* (0.98) | −4.10* (0.69) | −3.91 (0.30) | −4.10* (0.60) |
| Low Zn | ND | −4.32* (0.37) | ND | ND |
| High Zn | −5.55* (0.61) | −3.93* (0.86) | −3.77 (0.55) | −4.01* (0.69) |
| Low Ni | ND | ND | −3.82 (0.39) | ND |
| High Ni | −5.90 (0.26) | −4.15* (0.64) | −3.11* (1.10) | −4.35 (0.35) |
| Mean SE | 0.11 | 0.14 | 0.28 | 0.15 |
IC50s are expressed as log metal concentrations. Values in parentheses denote ΔIC50. Asterisks indicate significant difference from the uncontaminated-sludge treatment (n = 4, P < 0.05, mean SE from ANOVA). ND, not determined.
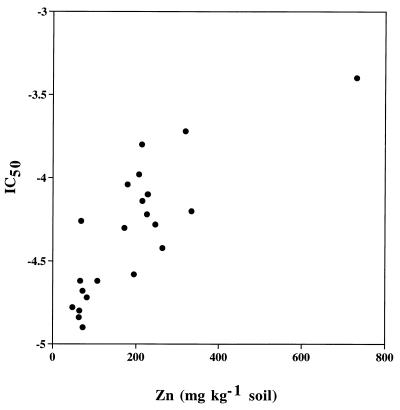
There were also indications of multiple heavy-metal tolerance, that is, tolerance to metals not added to the soil. Thus, the bacterial community from the soil given the high-Zn treatment was tolerant to both Cu and Cd, and the community from the soil given the high-Cu treatment was tolerant to Cd (Table 3). The extent of cross-tolerance for Zn could not be determined, since high levels of Zn could also be found in treatments not intended to increase Zn levels in soil. However, when all the treatments were included, a good linear regression between the Zn level in soil and the level of tolerance of the bacterial community to Zn was found (Fig. 1) (r = −0.81, P < 0.001, n = 24), indicating that increased tolerance of the bacterial community was found at least around 200 mg of Zn kg of soil−1.
FIG. 1.
Relationship between Zn concentrations in soil (aqua regia extracted) and bacterial community tolerance (IC50) for the Luddington soil. A high value indicates a high community tolerance. IC50s are expressed as log metal concentrations in the TdR incorporation bioassay.
The precision of determination of community tolerance differed for the different metals. The lowest mean standard error (SE) was found for Cu (0.11), with intermediate values for Zn and Cd, and the lowest precision was found for Ni (mean SE = 0.28) (Table 3).
(iv) PLFA.
The PLFA data was subjected to a principal-components analysis (Fig. 2A), where the first principal component explained 44.3% and the second component explained 15.6% of the variation of the data. The first component clearly differentiated between the high-Cu treatment and the other treatments, while for the second component all the metal treatments gave significantly different results from the uncontaminated sludge (Table 2). This might suggest that the high-Cu treatment had affected the microbial community the most. However, since the first component appeared to be related to the moisture content of the soil (Fig. 3, r = −0.87, P < 0.001, n = 24), it could not directly be concluded that the altered PLFA pattern was due to Cu or moisture content. However, with moisture as the covariate in the ANOVA of the first principal component, a significant difference between the high-Cu treatment and the uncontaminated-sludge treatment was found. The effect of the soil moisture content on the PLFA pattern could also be found in the no-sludge treatment, where two samples had high moisture contents and two had low moisture contents (Fig. 3). The variation in moisture content within treatments was the reason for the exceptionally high variation in the scores of the first principal component not only for the no-sludge treatment but also for the high-Zn treatment (Fig. 2A).
FIG. 2.
(A) Score plot from the principal-components analysis with PLFA data from the Luddington soil. Each value is the mean ± SE of four replicate determinations. US, uncontaminated sludge; NS, no sludge; LCu and HCu, low- and high-Cu-amended sludge, respectively; LNi and HNi, low- and high-Ni-amended sludge, respectively; LZn and HZn, low- and high-Zn-amended sludge, respectively. (B) Loading values for individual PLFAs from the principal-components analysis of the Luddington soil.
FIG. 3.
Relationship between the soil moisture content and the scores for the first principal component of the PLFA data from the Luddington soil. For abbreviations of the treatments, see the legend to Fig. 2A.
The second principal component can be regarded as a metal component, with the highest values being found for the two Ni treatments and the lowest values being found for the uncontaminated-sludge treatment (Fig. 2A). This component was characterized by relatively high proportions of the PLFAs il4:0 and br16:0 and relatively low proportions of the PLFAs cy19:0, 19:1a (branching position unknown), and 16:1ω7t compared to those for the uncontaminated-sludge treatment (Fig. 2B).
(v) Biolog.
The first principal component of the Biolog data accounted only for 18% of the variation, and the second accounted for 11% of the variation. The variation between replicates for the Biolog measurements was large, and thus no significant differences between treatments were found. However, the pattern found after the principal-components analysis was similar to that found for the PLFA pattern (Fig. 4; note that the axes 1 and 2 have been transposed to facilitate comparisons with Fig. 2A). The high-Cu treatment differed from the other treatments along one component, while the other component appeared to partly reflect heavy-metal pollution, in that the uncontaminated-sludge treatment had a low value and most of the metal-polluted soils had high values.
FIG. 4.
Score plot from the principal-components analysis with the Biolog data after 96 h of incubation for the Luddington soil. Note that axes 1 and 2 have been transposed to facilitate comparison with the PLFA data in Fig. 2A. Each value is the mean ± SE of four replicate determinations. For abbreviations of the treatments, see the legend to Fig. 2A.
Lee Valley soil.
The Lee Valley soil was used as a complement to the Luddington soil. Essentially, the findings for the two soils were similar. The bacterial community tolerance to Cu, Zn, and Ni increased the most in the Cu-polluted soils, the Zn-polluted soils, and the Ni-polluted soils, respectively. The ΔIC50 to Cu, Zn, and Ni was 0.38 for the high-Cu treatment, 0.85 for the high-Zn treatment, and 0.47 for the high-Ni treatment, respectively (the high-Cu and high-Zn treatments were significantly different from the uncontaminated sludge control). A dose-response effect was also found, since Zn tolerance was positively correlated with the amount of Zn in soil (r = 0.71, P < 0.05, n = 12). As with the Luddington soil, the greatest precision (judged from the mean SE) was for the tolerance measurements to Cu, and the least was for the tolerance measurements to Ni.
The Cu treatments were subjected to a principal-components analysis separately from the other metals, since the different metals appeared to have different effects on the PLFA pattern. The nonpolluted treatments differed from the metal-polluted treatments in both analyses (data not shown), and ANOVA of the scores showed a significant difference between the high levels of Cu, Zn, and Ni compared to the uncontaminated-sludge control. Zn and Ni pollution appeared to increase the relative amounts of PLFAs 18:2ω6,9, 20:4, and cy17:0, while the amounts of 16:1ω5 and several branched PLFAs decreased the most compared to the uncontaminated control treatments. In the Cu-polluted soil, the cy17:0 concentration increased and the concentrations of 18:1ω7, 16:1ω5, and several branched PLFAs decreased compared to the unpolluted controls. The Biolog measurements did not differentiate between the different treatments (data not shown).
DISCUSSION
Chander and Brookes (11), in their initial microbiological study of the Luddington sludge experiment, reported a reduced C-mic level for the low-Cu treatment and especially the high-Cu treatment, while no effects were found for the high-Ni and high-Zn treatments. In the present study, clover was grown to partly reconstitute the microbial populations in the stored and air-dried soils, since these soils had previously been planted with grass and clover pasture. The microbial biomass at the end of the plant growth period was still less than that found by Chander and Brookes (11) with fresh soils. Although the original microbial biomass was not restored fully, the same treatment effects were confirmed (Table 2), indicating that archived soils hitherto thought to be of little value for biological assessment may in fact prove worthy of further investigation.
It was clear that the high-Cu sludge amendment was most detrimental to the soil microorganisms. In these plots, not only C-mic but also soil respiration rate and bacterial activity (TdR incorporation) decreased the most compared to the values in the unpolluted-sludge treatment when the confounding effect of soil moisture was taken into consideration (Table 2). Also in this treatment, the community measurements were clearly affected by the toxic metals. There was some evidence that the microbial community was also affected by the metal additions in treatments where no effect on biomass and activity could be found. This was true, for example, for the low- and high-Zn treatments and the high-Ni treatments in the Luddington soil, which all had a significantly increased bacterial community tolerance (Table 3) and altered PLFA pattern compared to the uncontaminated sludge (Table 2; Fig. 2A). An altered microbial community composition, without any change in biomass, was also found in the Lee Valley soil, where the PLFA pattern was significantly altered for the high-Ni treatment, a treatment that did not alter C-mic (11). Thus, the microbial community-based techniques, measuring bacterial community tolerance and PLFA pattern, appeared at least as sensitive to the effects of heavy metals as did the biomass and activity measurements. This was also the case in earlier studies (12, 27).
The PLFA pattern can be affected by several environmental factors including the possible presence of toxic substances like heavy metals. This is also the case with the Biolog measurements, as well as most activity- and biomass-based measurements commonly used in soil ecotoxicological studies. In our study, it was exemplified by the effect of soil moisture, which affected not only the PLFA pattern (Fig. 2A and 3) and the Biolog results (Fig. 4) but also the soil respiration rate and bacterial TdR incorporation (Table 2). It can therefore be difficult, when using these nonspecific techniques, to conclude with certainty that a registered effect is due to toxicity and not to changes in other environmental variables. Measurement of bacterial community tolerance, on the other hand, can be a more direct way of detecting toxic effects in soil, since an altered tolerance should reflect a selection pressure due only to a toxic substance. This was also the case, since the moisture content had no effect on the level of community tolerance. Pennanen et al. (27) also found effects on the PLFA pattern which were not directly related to the heavy-metal pollution, since changes in the PLFA pattern did not always correlate with changes in bacterial community tolerance levels.
Pennanen et al. (27), who studied the effect of metals in forest soils around two smelters in Finland and Sweden, reported that metal toxicity always increased the relative abundance of some PLFAs and decreased the abundance of others. They therefore suggested that the PLFA pattern changed in a predictable way in coniferous-forest humus soils. However, such specific changes in the PLFA pattern appear not to be the case when other soil types are studied. This was found in a laboratory study using one forest humus and one agricultural soil (18), and in the present study the effect of metals on individual PLFAs was also different in the Luddington and the Lee Valley soils. For example, the concentration of the PLFA cy17:0 increased compared to that of the uncontaminated sludge in the high-Ni and high-Zn treatments at Lee Valley but decreased in the same treatments at Luddington. Similarly, concentration of the PLFA 16:1ω5, which increased with metal concentration at Luddington (Fig. 2B), decreased at Lee Valley.
Frostegård et al. (18) reported that within one soil, different metals (Cd, Ni, Pb, and Zn) induced similar changes in the PLFA pattern. In our study, the same alterations in the PLFA pattern were found for the Zn- and Ni-treated samples within both soils (Fig. 2 gives an example for the Luddington soil), thus providing additional evidence that these two metals appear to cause similar changes in the microbial community structure. The case with Cu was more complicated. For the Luddington soil, the high-Cu treatment differed from the other treatments along the first principal component (Fig. 2A), indicating that Cu had a different effect from the other metals on the PLFA pattern. One difference was, for example, that the concentration of 18:2ω6,9 decreased slightly in the soil given the high-Cu treatment but was unaffected or slightly higher in the soils given the other metal treatments compared to the soil used as the uncontaminated-sludge control (Fig. 2B). However, the first principal component was determined in large part by the soil moisture content (Fig. 3), making any conclusion about effects of Cu uncertain. However, in the Lee Valley soil, Cu clearly differed in its effect on the fungal PLFA 18:2ω6,9 (and 20:4, also common in some fungi) from the other metals. Cu appeared to decrease the relative amounts of these two PLFAs, while Ni and Zn increased them both. Thus, the present data support earlier laboratory results (18) that Cu appears to affect the fungal part of the microbial community differently from other metals in agricultural soils. Dahlin et al. (12) also reported that a high Cu content in soil was correlated with low concentrations of 18:2ω6,9 in an experiment in which sludge was added to agricultural soil.
Both the PLFA and the Biolog measurements are supposed to reflect the composition of the microbial community, although the former reflects mainly the structural aspect (species composition) and the latter might more closely indicate the potential metabolic capacity of the community. Although the Biolog method measures functional attributes (metabolic capacities) of the microbial community, it may also be yielding structural information, because the organisms express these different metabolic capacities only after they have grown in the Biolog microtiter plates. It is therefore also a potential measure rather than an actual functional expression. Although both techniques revealed the same pattern of changes due to metal pollution and soil moisture content in the Luddington soil (Fig. 2A and 4), the variation in the data was much smaller for the PLFA measurements. The large variation in the Biolog data was also evident by the low percent variation explained in the first two principal components. Thus, the PLFA technique appeared to be more sensitive in detecting metal pollution than the Biolog technique. This was also corroborated by the results with the Lee Valley soil, where Biolog found no significant differences between treatments but where the high Cu, Zn, and Ni treatments had a significantly altered PLFA pattern. Similar results indicating the PLFA technique to be more sensitive than Biolog were reported previously from a heavy-metal-polluted forest soil in Finland (17).
There could be several reasons for the difference in sensitivity between the Biolog and PLFA techniques. However, it is clear that the two techniques differ in the part of the microbial community that they measure. PLFA measurements include PLFAs from both eucaryotic and procaryotic organisms, while the Biolog technique probably reflects only the bacterial community. Furthermore, since the Biolog technique essentially consists of enrichment batch cultures for the different substrates in the 95 wells and considering that rather high substrate concentrations are used, only very fast-growing copiotrophs, probably initially nondormant, will be able to contribute to the color development in the wells. These bacteria will probably make up only a small fraction of the soil microbial community, since most bacteria in soil are considered oligotrophic and slow growing (31). The PLFA pattern, on the other hand, could be considered an integrated measure of all the microorganisms present in soil irrespective of the growth rate or metabolic capacity. However, in environments like the rhizosphere, where a larger part of the community is active compared to bulk soil, the Biolog technique might perform better. For example, this technique has been used to differentiate between rhizosphere communities of different plant species (21, 23). The use of a wider range of more ecologically relevant carbon sources can also increase the discriminating ability of this technique (9).
In contrast to us, Knight et al. (25), in a recent study of the combined effect of reduced pH and metal-rich sludge, claimed that the Biolog pattern showed significant effects due to metal concentrations similar to the ones reached in our soils. However, they found large differences in AWCD, which could be explained partly (but not entirely) by a lower biomass and activity in some of the treatments. They did not compensate for this variation, and therefore the separation of the Biolog data in their principal-components analysis could be explained entirely by the differences in AWCD. The importance of dividing by the AWCD before performing further analysis has been emphasized by Garland (20). In our study, only insignificant differences in AWCD between treatments were found, except for the high-Cu amendment in the Luddington soil, where AWCD was slightly lower than the other treatments (data not shown). Furthermore, we compensated for any variation by dividing our data set by the AWCD for each Biolog plate. Also, in contrast to our study, Knight et al. (25) reported a small variation between replicates within each treatment. However, they replicated their data by inoculating Biolog plates with the same soil suspension from only one soil sample. Since we used different soil samples as the basis for replication, we believe that our larger variations between replicates are more representative of the performance of the Biolog technique in soil and of the effect of heavy metal rich sludge on sole-carbon source profiles.
The precision in the community tolerance measurements differed between metals, being highest for Cu and lowest for Ni (Table 3). This was also found by Díaz-Raviña et al. (13). The precision of the IC50 estimate increases proportionally with the steepness of the slope of the dose-response curve. Cu produced dose-response curves with steep slopes in the TdR incorporation bioassay, whereas the slope of the curve for Ni toxicity was less steep. Zn and Cd had intermediate slopes for the dose-response curves. This difference in precision was a major reason for finding significant tolerance to Ni less often than finding significant tolerance to Cu, although the actual ΔIC50s were similar for the two metals in the Lee Valley soil, for example. One must therefore be cautious when comparing the tolerance changes for different toxic substances by the TdR incorporation approach.
The Luddington soil, with less clay and organic matter than the Lee Valley soil, should theoretically have a higher proportion of the total amount of metals available in soil. Thus, at similar total metal concentrations, the toxicity should be higher at Luddington than at Lee Valley. This was shown previously by comparing 50% effective concentrations for a protozoan growth bioassay (10) and the decrease in microbial biomass (11) in these two soils. The tolerance of the bacterial community also indicated this, since the ΔIC50s were higher at Luddington than at Lee Valley at comparable total metal concentrations. This might indicate that community tolerance can be used to compare actual metal toxicity between soil types, not only within one soil type.
One of our objectives was to study the extent to which metals added separately might cause multiple tolerance. This is difficult when using sludge contaminated from different sources. Our sludges were contaminated predominantly with a single metal but still had elevated levels of other elements. Thus, due to contamination of the Cu- and Ni-polluted soils with both Zn and Cd, no conclusions could be drawn about whether the increased community tolerances to Zn and Cd, found, for example, in the Cu treatments at Luddington, were due to multiple tolerance. However, in the case of Cu tolerance in the high-Zn treatment at Luddington (Table 3), this must clearly be due to multiple tolerance. The presence of multiple tolerance will obviously make it more difficult to elucidate from community tolerance measurements which metal is exerting the most toxic influence in a pollution situation with a mixture of metals. However, in all cases, the increase in tolerance was most pronounced for the metal added at the highest concentration, indicating that this can be used as a criterion for determining the most toxic metal. This was also indicated in both a field study (27) and a laboratory study (13).
A better criterion for estimating the most toxic metal is to use the dose-response relationship between the metal concentration in soil and the level of community tolerance to estimate a threshold metal concentration for toxic effects. This relationship was shown to be curvilinear (14) when the metal concentration was logarithmically transformed. When only a few data points are available, a linear approximation has to be used. This was done by Pennanen et al. (27) and indicated that Cu was the most toxic metal around a primary smelter that was also emitting large amounts of Cd and Zn. The use of only the low and high levels for the Luddington soil indicated that the threshold levels were 20 mg kg of soil−1 for increased Cu tolerance to Cu contamination, 60 mg kg−1 for Ni tolerance to Ni contamination, and 140 mg kg−1 for Zn tolerance to Zn contamination. These low values, although uncertain, still indicate that the use of community tolerance measurements can be a very sensitive technique to monitor the effects of heavy metals. This fact, together with the possibility of more directly indicating the exact cause of a toxic effect in a multiple-pollution situation, makes it an attractive technique to include in test batteries together with general measurements of biomass, activity, and community structure.
ACKNOWLEDGMENTS
This study was supported by grants from the European Environmental Research Organization (E.E.R.O.) and the Ministerio Español de Educación y Ciencia to M.D.-R., the Swedish Natural Science Research Council and the Swedish National Environment Protection Agency to E.B., and the Scottish Office Agricultural, Environment and Fisheries Department to C.D.C.
C.M. Cameron and J. Van Gelder are thanked for technical assistance.
REFERENCES
- 1.Arnebrant K, Bååth E, Nordgren A. Copper tolerance of microfungi isolated from polluted and unpolluted forest soil. Mycologia. 1987;79:890–895. [Google Scholar]
- 2.Bååth E. Effects of heavy metals in soil on microbial processes and populations: a review. Water Air Soil Pollut. 1989;47:335–379. [Google Scholar]
- 3.Bååth E. Thymidine incorporation into macromolecules of bacteria using thymidine incorporation into bacteria extracted after homogenization-centrifugation. Soil Biol Biochem. 1992;24:1157–1165. [Google Scholar]
- 4.Bååth E. Measurement of heavy metal tolerance of soil bacteria using thymidine incorporation into bacteria extracted after homogenization-centrifugation. Soil Biol Biochem. 1992;24:1167–1172. [Google Scholar]
- 5.Berrow M L, Burridge J C. Inorganic pollution and agriculture. MAFF reference book 326. London: Her Majesty’s Stationery Office; 1980. Trace element levels in soil: effects of sewage sludge; pp. 159–183. [Google Scholar]
- 6.Berrow M L, Burridge J C. Persistence of metals in sewage sludge treated soils. In: L’Hermite P, Ott H, editors. Processing and use of sewage sludge. Dordrecht, The Netherlands: Dordrecht Reidel Publishing Co.; 1984. pp. 418–421. [Google Scholar]
- 7.Berrow M L, Burridge J C. Persistence of metal residues in sewage-sludge treated soils over 17 years. Int J Environ Anal Chem. 1990;39:173–177. [Google Scholar]
- 8.Brookes P C. The use of microbial parameters in monitoring soil pollution by heavy metals. Biol Fertil Soils. 1995;19:269–279. [Google Scholar]
- 9.Campbell C D, Grayston S J, Hirst D J. Use of rhizosphere carbon sources in sole carbon source tests to discriminate soil microbial communities. J Microbiol Methods. 1997;30:33–41. [Google Scholar]
- 10.Campbell C D, Warren A, Cameron C M, Hope S J. Direct toxicity assessment of two soils amended with sewage sludge contaminated with heavy metals using a protozoan (Colpoda steinii) bioassay. Chemosphere. 1997;34:501–514. [Google Scholar]
- 11.Chander K, Brookes P C. Effects of heavy metals from past applications of sewage sludge on microbial biomass and organic matter accumulation in a sandy loam and silty loam U.K. soil. Soil Biol Biochem. 1991;23:927–932. [Google Scholar]
- 12.Dahlin S, Witter E, Mårtensson A, Turner A, Bååth E. Where’s the limit? Changes in the microbiological properties of agricultural soils at low levels of metal contamination. Soil Biol Biochem. 1997;29:1405–1415. [Google Scholar]
- 13.Díaz-Raviña M, Bååth E, Frostegård Å. Multiple heavy metal tolerance of soil bacterial communities and its measurement by a thymidine incorporation technique. Appl Environ Microbiol. 1994;60:2238–2247. doi: 10.1128/aem.60.7.2238-2247.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Díaz-Raviña M, Bååth E. Development of metal tolerance in soil bacterial communities exposed to experimentally increased metal levels. Appl Environ Microbiol. 1996;62:2970–2977. doi: 10.1128/aem.62.8.2970-2977.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doelman P, Jansen E, Michels M, van Til M. Effects of heavy metals in soil on microbial diversity and activity as shown by the sensitivity-resistance index, an ecologically relevant parameter. Biol Fertil Soils. 1994;17:177–184. [Google Scholar]
- 16.Ellis R J, Thompson I P, Bailey M J. Metabolic profiling as a means of characterizing plant-associated microbial communities. FEMS Microbiol Ecol. 1995;16:9–18. [Google Scholar]
- 17.Fritze H, Pennanen T, Vanhala P. Impact of fertilizers on the humus layer microbial community of Scots pine stands growing along a gradient of heavy metal pollution. In: Insam H, Rangger A, editors. Microbial communities: functional versus structural approaches. Berlin: Springer-Verlag KG; 1997. pp. 68–83. [Google Scholar]
- 18.Frostegård Å, Tunlid A, Bååth E. Phospholipid fatty acid composition, biomass, and activity of microbial communities from two soil types experimentally exposed to different metals. Appl Environ Microbiol. 1993;59:3605–3617. doi: 10.1128/aem.59.11.3605-3617.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frostegård Å, Tunlid A, Bååth E. Changes in microbial community structure during long-term incubation in two soils experimentally contaminated with metals. Soil Biol Biochem. 1996;28:55–63. [Google Scholar]
- 20.Garland J L. Analytical approaches to the characterization of samples of microbial communities using patterns of potential C source utilization. Soil Biol Biochem. 1996;28:213–221. [Google Scholar]
- 21.Garland J L. Patterns of potential C source utilization by rhizosphere communities. Soil Biol Biochem. 1996;28:223–230. [Google Scholar]
- 22.Garland J L, Mills A L. Classification and characterisation of heterotrophic microbial communities on the basis of patterns of community-level-sole-carbon-source-utilization. Appl Environ Microbiol. 1991;57:2351–2359. doi: 10.1128/aem.57.8.2351-2359.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grayston S J, Campbell C D. Functional biodiversity of microbial communities in the rhizospheres of hybrid larch (Larix eurolepis) and Sitka spruce (Picea sitchensis) Tree Physiol. 1996;16:1031–1038. doi: 10.1093/treephys/16.11-12.1031. [DOI] [PubMed] [Google Scholar]
- 24.Huysman F, Verstraete W, Brookes P C. Effect of manuring practices and increased copper concentrations on soil microbial populations. Soil Biol Biochem. 1994;26:103–110. [Google Scholar]
- 25.Knight B P, McGrath S P, Chaudri A M. Biomass carbon measurements and substrate utilization patterns of microbial populations from soils amended with cadmium, copper, or zinc. Appl Environ Microbiol. 1997;63:39–43. doi: 10.1128/aem.63.1.39-43.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGrath S P, Cunliffe C H. A simplified method for the extraction of the metals Fe, Zn, Cu, Ni, Cd, Pb, Cr, Co and Mn from soils and sewage sludges. J Sci Food Agric. 1985;36:794–798. [Google Scholar]
- 27.Pennanen T, Frostegård Å, Fritze H, Bååth E. Phospholipid fatty acid composition and heavy metal tolerance of soil microbial communities along two heavy metal-polluted gradients in coniferous forests. Appl Environ Microbiol. 1996;62:420–428. doi: 10.1128/aem.62.2.420-428.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfender W F, Fieland V P, Ganio L M, Seidler R J. Microbial community structure and activity in wheat straw after inoculation with biological control organisms. Appl Soil Ecol. 1996;3:69–78. [Google Scholar]
- 29.Vahjen W, Munch J-C, Tebbe C C. Carbon source utilization of soil extracted microorganisms as a tool to detect the effects of soil supplemented with genetically engineered and non-engineered Corynebacterium glutamicum and a recombinant peptide at the community level. FEMS Microbiol Ecol. 1995;18:317–328. [Google Scholar]
- 30.Vance E D, Brookes P C, Jenkinson D S. An extraction method for measuring microbial biomass C. Soil Biol Biochem. 1975;19:703–707. [Google Scholar]
- 31.Whang K, Hattori T. Oligotrophic bacteria from rendzina forest soil. Antonie Leeuwenhoek. 1988;54:19–36. doi: 10.1007/BF00393955. [DOI] [PubMed] [Google Scholar]
- 32.Winding A. Fingerprinting bacterial soil communities with Biolog microtitre plates. In: Ritz K, Dighton J, Giller K E, editors. Beyond the biomass: compositional and functional analysis of soil microbial communities. Chichester, United Kingdom: John Wiley & Sons, Inc.; 1994. pp. 85–94. [Google Scholar]
- 33.Wu J, Joergensen R G, Pommerening B, Chaussod R, Brookes P C. Measurement of soil microbial biomass C by fumigation-extraction—an automated procedure. Soil Biol Biochem. 1990;22:1167–1169. [Google Scholar]
- 34.Zak J C, Willig M R, Moorhead D L, Wildman H G. Functional diversity of microbial communities: a quantitative approach. Soil Biol Biochem. 1994;26:1101–1108. [Google Scholar]