Abstract
The scope of marine phytoplankton diversity is uncertain in many respects because, like bacteria, these organisms sometimes lack defining morphological characteristics and can be a challenge to grow in culture. Here, we report the recovery of phylogenetically diverse plastid small-subunit (SSU) rRNA gene (rDNA) clones from natural plankton populations collected in the Pacific Ocean off the mouth of Yaquina Bay, Oreg. (OCS clones), and from the eastern continental shelf of the United States off Cape Hatteras, N.C. (OM clones). SSU rRNA gene clone libraries were prepared by amplifying rDNAs from nucleic acids isolated from plankton samples and cloning them into plasmid vectors. The PCR primers used for amplification reactions were designed to be specific for bacterial SSU rRNA genes; however, plastid genes have a common phylogenetic origin with bacteria and were common in both SSU rRNA gene clone libraries. A combination of restriction fragment length polymorphism analyses, nucleic acid sequencing, and taxon-specific oligonucleotide probe hybridizations revealed that 54 of the 116 OCS gene clones were of plastid origin. Collectively, clones from the OCS and OM libraries formed at least eight unique lineages within the plastid radiation, including gene lineages related to the classes Bacillariophyceae, Cryptophyceae, Prymnesiophyceae, Chrysophyceae, and Prasinophyceae; for a number of unique clones, no close phylogenetic neighbors could be identified with confidence. Only a group of two OCS rRNA gene clones showed close identity to the plastid SSU rRNA gene sequence of a cultured organism [Emiliania huxleyi (Lohmann) Hay and Mohler; 99.8% similar]. The remaining clones could not be identified to the genus or species level. Although cryptic species are not as prevalent among phytoplankton as they are among their bacterial counterparts, this genetic survey nonetheless uncovered significant new information about phytoplankton diversity.
Eukaryotic phytoplankton in the ultraplankton size range (cells of <5 to 10 μm in diameter) are recognized as important members of the photosynthetic assemblage in the world’s oceans (23, 28, 42). These organisms are also commonly referred to as nanoplankton (cells of <2 to 20 μm in diameter) and make up a majority of the eukaryotic phytoplankton cells present in many, if not most, pelagic marine habitats, including oligotrophic open-ocean and ocean margin environments (8).
The identification of ultraphytoplankton, either in natural communities or clonal cultures, is often hindered by their size and lack of taxonomically informative morphological features. In addition, many of the smaller plankton cells are difficult to culture. The taxonomic identification of small eukaryotic phytoplankton often involves the techniques of electron microscopy (23, 28), analysis of photosynthetic pigments by high-performance liquid chromatography (16, 41), or immunological cross-reactivity (6, 50). The identification of individual cells at the class level is difficult at best and is rarely accomplished with the ease and speed necessary to analyze large numbers of field samples.
The identification of small eukaryotic phytoplankton has benefited from the application of molecular phylogenetic techniques. Recent descriptions of phytoplankton taxa include sequence analyses of the nucleus-encoded 18S rRNA gene (2). As well as aiding in the placement of the newly described taxon in a phylogenetic framework, the use of conserved gene sequences has provided information for the design of phylogenetically specific oligonucleotide hybridization probes (32, 35, 40).
Nucleic acid probing has become a common approach for assessing the diversity of microorganisms in natural communities (1). In pelagic marine environments, the focus has been on prokaryotic plankton, including organisms from the domains Bacteria and Archaea. The findings have been dramatic; many novel bacterioplankton and archaea have been described only by their small-subunit (SSU) rRNA gene (rDNA) sequences, with most having thus far evaded culture in the laboratory. Oligonucleotide probes based on SSU rRNA sequence signatures are being designed for widespread application to ecological problems, including determining the distribution of bacterial species in their natural habitats (11, 20, 21). rRNA probes have also previously been constructed for cultured ultraphytoplankton with the intention of identifying cells rapidly in field samples (51).
This study was undertaken with the intent of examining the diversity of bacterioplankton in Oregon coastal seawater. The Oregon coast is an area of high productivity. Wind-driven coastal upwelling processes seasonally dominate the region’s oceanography (26, 27). Similar hydrographical events occur in both the northern and southern hemispheres along eastern ocean boundaries (3). The high nutrient availability in these regions results in high ecosystem productivity; they may be responsible for half the world’s catch of fish, even though they represent only 0.1% of the world’s surface area (47). High phytoplankton abundance and elevated levels of primary production are characteristic of the Oregon coast during upwelling events (24, 52).
In this study, the fortuitous recovery of plastid-derived SSU rRNA gene clones in high abundance from two clone libraries prepared from natural plankton populations off the Pacific and Atlantic coasts of the United States allowed us to assess phytoplankton diversity. Most of the plastid-derived rDNA clones recovered belonged to several classes of eukaryotic algae which are known to contain marine phytoplankton members (7). Several groups of rRNA gene clones did not affiliate with any currently available plastid SSU rRNA gene sequence and may represent cryptic species previously unrecognized because of their lack of distinguishing characters.
MATERIALS AND METHODS
Construction of the Oregon coast study (OCS) rRNA gene clone library has previously been partially described (55). Construction of the ocean margins (OM) rRNA gene clone library has previously been described (44, 45). Here, we provide details of the construction and analysis of the OCS clone library not provided elsewhere.
Sample collection and nucleic acid isolation.
On 28 April 1993, a 16-liter plankton sample was collected from a depth of 10 m at a station located 8 km west of Yaquina Bay, Oregon (44°39.1′N, 124°10.6′W). The water depth was ca. 50 m at the sampling site. The water was prescreened through 10-μm-pore-size nylon mesh and immediately transported in autoclaved polyethylene carboys to the laboratory, where the plankton sample was collected by filtration onto 0.2-μm-pore-size polysulfone filters (Supor-200; Gelman, Inc., Ann Arbor, Mich.). For subsample 2, which was used for length heterogeneity-PCR (LH-PCR), water was prefiltered through a 0.8-μm-pore-size polycarbonate membrane (Poretics, Livermore, Calif.) and cells were collected on 0.2-μm-pore-size filters as described above. Total cellular nucleic acids were isolated by cell lysis with proteinase K and sodium dodecyl sulfate (SDS) and phenol-chloroform extraction as previously described (19, 55).
Clone library construction.
SSU rRNA genes were amplified from the environmental sample of genomic DNAs by PCR (48) with two general bacterial SSU rDNA primers and Pfu DNA polymerase as previously described (55). The amplification products from six reactions were pooled, inserted into the SmaI restriction site of phagemid vector pBluescript KSII− (Stratagene) by blunt-end ligation as previously described (19, 55), and used to transform competent Escherichia coli XL1-Blue cells (Stratagene, La Jolla, Calif.). Positive (white-colony morphotype) transformants were streaked for isolation and stored in both stab cultures and dimethyl sulfoxide. Clones were numbered discontinuously from 1 to 182 and assigned the prefix OCS (for Oregon coast study).
RFLP analysis.
All gene clones containing full-length inserts were characterized by HaeIII restriction fragment length polymorphisms (RFLPs) as previously described (55). Briefly, insert-bearing plasmids, isolated by alkaline lysis, were used as templates in the amplification of environmental clone rDNAs by PCR with the same primers previously used to amplify rRNA genes. Since all of the PCRs yielded similar amounts of product, 7 μl of nonpurified PCR products was digested with 3 U of restriction endonuclease HaeIII (Promega, Madison, Wis.) for 2 h. Restriction fragments were resolved by gel electrophoresis on 3% NuSieve low-melting-point agarose (FMC, Rockland, Maine) in 1× Tris-acetate-EDTA (TAE) buffer and stained with ethidium bromide (0.5 μg · ml−1).
rDNA sequencing and phylogenetic analyses.
Template plasmid DNAs for sequencing were prepared by alkaline lysis as previously described (55). Clone sequences submitted to GenBank were sequenced on both strands with an ABI 377 or 373a automated sequencer (Applied Biosystems Inc., Foster City, Calif.), dye-terminator chemistry, and conserved bacterial SSU rDNA primers (34). DNA sequences were manually aligned to bacterial and plastid sequences obtained from GenBank and the Ribosomal Database Project (RDP) (37) by using the Genetic Data Environment (version 2.2) sequence analysis software package (provided by Steve Smith).
Evolutionary trees were constructed by distance, maximum-parsimony, and maximum-likelihood methods. Each phylogenetic analysis employed conservative phylogenetic masks which included only regions of unambiguous alignment. In addition, all phylogenetic analyses were repeated multiple times, with different taxa employed as outgroups. The phylogeny inference package (PHYLIP version 3.5c [14]) was used for phylogenies constructed from evolutionary distance matrices and by the maximum-parsimony method. Evolutionary distances were calculated from pairwise sequence similarities with the Kimura two-parameter model for nucleotide change and a transition/transversion ratio of 2.0 (31) by using the DNADIST program. The NEIGHBOR program was used to reconstruct phylogenetic trees from evolutionary distance matrices by the neighbor-joining method (49). Parsimony trees were reconstructed by using the DNAPARS program. Maximum-likelihood analyses were performed with the fastDNAml program distributed by RDP (37). The relative confidence in monophyletic groups within each phylogenetic analysis was estimated by bootstrap analysis, which included 100 replicate resampled data sets with random sequence addition and global rearrangement (13). Test version 4.0.0d53 of PAUP∗ was used for all LogDet phylogenetic analyses (56).
Secondary-structure analyses and identification of chimeras.
Several methods were employed to verify the integrity of sequence data and to aid in the detection of chimeric gene artifacts. First, the gRNAid (version 1.4) program (57) was used to construct secondary structural models for all sequenced SSU rDNAs. These models were manipulated manually for optimal agreement with published SSU rRNA secondary structures (22), and the natures and locations of mutations were analyzed. Second, all gene sequences were submitted to the RDP CHECK_CHIMERA program (37), which is useful for detecting chimeric gene artifacts created from parent sequences that are no greater than ca. 84% similar (46). Phylogenetic trees were also constructed and compared from different regions within single rRNA gene clones, under the assumption that if two regions of an rRNA gene clone originate from different parental rRNA genes, they separate into different lineages that reflect the phylogenies of their different parent rRNA gene sequences (15, 46).
Oligonucleotide probe hybridizations.
To help in identifying unique SSU rDNA clones, taxon-specific oligonucleotide probes were used to screen a dot blot of 32 clones, representing 23 RFLP patterns. For the selected clones, PCR amplicons from the RFLP analysis were purified with a Qiaquick-spin PCR purification kit (Qiagen, Chatsworth, Calif.). For each amplicon, 30 ng of product was resuspended in 180 μl of Tris-EDTA buffer. Products were denatured by the addition of 20 μl of 2.0 N NaOH and incubated for 10 min at room temperature prior to being blotted on a Zetaprobe nylon membrane (Bio-Rad, Hercules, Calif.). Each well was washed with 200 μl of 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4 [pH 7.2], 1 mM EDTA) to neutralize NaOH. The membrane was dried under vacuum at 80°C for 15 min, exposed to 260-nm UV radiation at 12 J/m2, and stored (desiccated) before being probed.
The following oligonucleotide probes were hybridized to the OCS unknown dot blot: SAR83R (probe hybridization stringency temperature [TH] = 45°C), SAR11A1 (TH = 37°C) (15), SAR86 (TH = 45°C) (45), CHRYS1 (TH = 45°C) (44), and HAP1 (TH = 45°C) (44). The TH for each probe was determined empirically by washing at successively increasing temperatures. All probes used in this study were constructed with an automated DNA synthesizer (Applied Biosystems Inc.) and were 32P labeled on their 5′ termini with T4 polynucleotide kinase as previously described (18).
Prior to each probe hybridization, the OCS unknown blot was prehybridized in 15 ml of Z-hyb buffer (1.0 mM EDTA, 0.50 mM NaH2PO4 [pH 7.2], 7.0% SDS) for 45 to 60 min at room temperature. After the prehybridization buffer was decanted, the blot was hybridized in 6.0 ml of Z-hyb buffer containing 200 μl of 32P-labeled oligonucleotide (25 to 50 ng of oligonucleotide) for 8 to 16 h at room temperature. The blot was washed three times for 15 min each in 25 ml of wash buffer (0.2× SSPE, 0.1% SDS) at room temperature and one time for 30 min at the probe TH. The blot was stripped of probe by being washed three times in 25 to 50 ml of wash buffer at 70°C for 10 min each. Individual probe hybridizations were visualized with a PhosphorImager SI (Molecular Dynamics, Sunnyvale, Calif.) and autoradiographically with X-ray film.
LH-PCR.
Ten nanograms of purified genomic DNA from each subsample was used as a template for PCR. The forward primer, 27F (5′-AGAGTTTGATCMTGGCTCAG-3′), was labeled at the 5′ terminus with the dye 6-FAM (graciously supplied by Applied Biosystems Inc.). General bacterial primer 338R (5′-GCTGCCTCCCGTAGGAGT-3′) was employed as the reverse primer. Labeled PCR products were purified by using Qiaquick-spin columns. Ten femtomoles of DNA was electrophoretically separated by polyacrylamide gel electrophoresis with Long Ranger acrylamide (FMC) in an ABI 377 automated DNA sequencer (Applied Biosystems Inc.) in Genescan mode. Genescan measures both the sizes of molecules by electrophoretic mobilities and integrated fluorescence emissions by bands. The output is an electropherogram in which bands are represented by peaks and the integrated fluorescence of each band is the area under the peak. Since the integrated fluorescence increased linearly with concentrations of up to 50 fmol of PCR products (data not shown), the relative proportion of the integrated fluorescence of each peak was observed to correspond to the proportion of each amplicon in the PCR product.
Nucleotide sequence accession numbers.
Nucleotide sequences were filed in the GenBank database under the following accession numbers: OCS20, AF001654; OCS31, AF001655; OCS50, AF001656; OCS54, AF001657; OCS56, AF001658; OCS162, AF001659; OCS182, AF001660; OM134, AF001661. GenBank accession numbers were obtained previously for the following environmental clone rDNAs used in this study: OM20 (U32670) and OM21 (U32671) (44); OM5 (U70715), OM39 (U70716), OM81 (U70717), OM111 (U70718), OM125 (U70719), OM153 (U70720), OM164 (U70721), OM255 (U70722), OM270 (U70723), and OM283 (U70724) (45).
RESULTS
OCS clone library.
Of 182 putative positive transformants (white colonies) in the OCS library, 116 clones were shown to have inserts of the correct size (1.5 kb) by gel electrophoresis of both plasmids and amplicons derived from plasmids. HaeIII RFLP analyses revealed 37 banding patterns distributed among OCS clones, with 19 OCS clones containing unique HaeIII RFLP patterns. Except where noted below, a minimum of two clones from all RFLP types containing two or more clones and all clones with unique RFLP banding patterns were sequenced. Cursory phylogenetic analyses identified 53 OCS SSU rDNA clones (seven RFLP banding patterns) as members of the plastid line of descent. An additional clone with a unique RFLP banding pattern was identified as plastid related, based on its hybridization with a taxon-specific oligonucleotide probe.
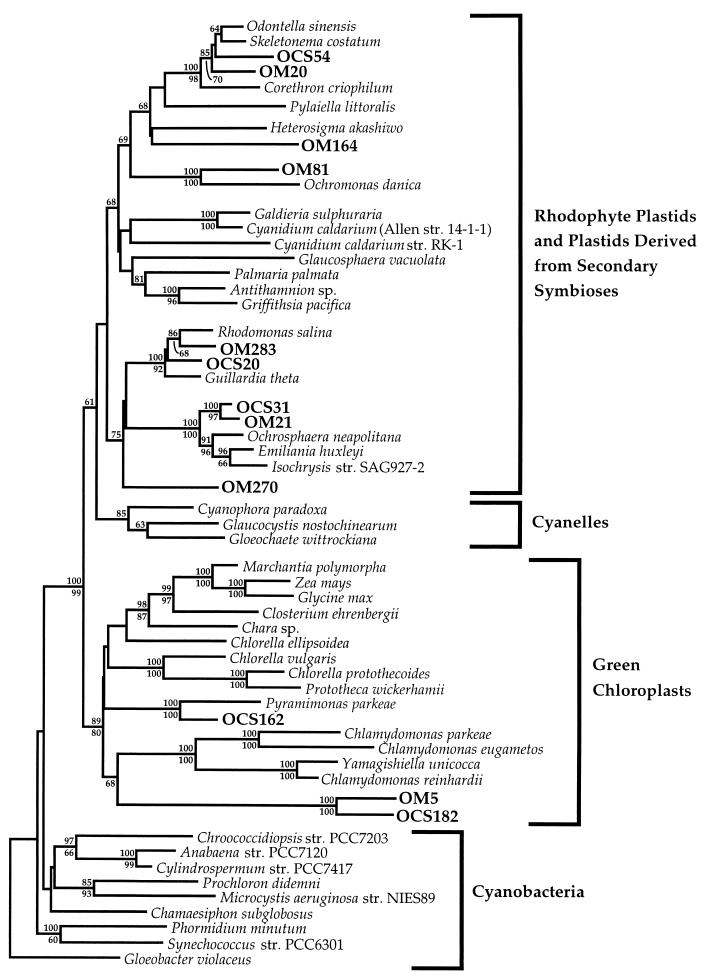
An overview of the phylogenetic diversity among plastid SSU rRNA genes recovered in the OCS clone library is shown in Fig. 1. Of the eight plastid-related RFLP types recovered (Table 1), five were included in the phylogenetic dendrogram depicted in Fig. 1, which employed a sequence spanning the entire SSU rRNA gene. Complete SSU rRNA gene clone sequences were not obtained from representative clones possessing RFLP types IX and XI (Table 1); thus, they were not included in the analysis depicted in Fig. 1. OCS129, representing RFLP type XXX, also was not included in the analysis depicted in Fig. 1 because no gene sequence was determined for this clone. Clones representing the remaining five RFLP types appeared to originate from the plastids of five distinct classes of eukaryotic algae, including three within the major plastid lineage defined by rhodophyte plastids (and plastids derived by secondary symbioses) and two within the major lineage defined by chloroplasts of green algae (algae with chloroplasts possessing chlorophylls a and b) (Fig. 1) (4, 39).
FIG. 1.
Phylogenetic relationships among OCS and OM plastid rDNA clones and plastid SSU rRNA gene sequences from cultured organisms. This phylogenetic tree was constructed by the neighbor-joining method with a LogDet matrix as the input (36). A conserved mask of 987 nucleotides, spanning the entire length of the plastid SSU rRNA gene, was included. Bootstrap values (100 replications) generated by the neighbor-joining method and the Kimura 2-parameter model for nucleotide change are shown above relevant nodes, and bootstrap values generated by maximum-parsimony analysis are shown below relevant nodes. All the sequences used, except for those of Corethron criophilum (38), Cyanidium caldarium (17), Closterium ehrenbergii, Chlamydomonas parkeae, Yamagishiella unicocca, and P. parkeae (30), are available from the GenBank or RDP database. str., strain.
TABLE 1.
Summary data for plastid gene clones reported here
| RFLP type | Phylogenetic affiliation | No. of clones recovered | Repre- sentative clonea | Sequence length (nt)b | GenBank accession no. |
|---|---|---|---|---|---|
| I | Prymnesiophyceae | 30 | OCS31 | 1,472 | AF001655 |
| IX | 3 | OCS50 | 956 | AF001656 | |
| III | Bacillariophyceae | 9 | OCS54 | 1,478 | AF001657 |
| XI | 2 | OCS56 | 554 | AF001658 | |
| XXX | 1 | OCS129 | NDc | ||
| V | Cryptophyceae | 6 | OCS20 | 1,479 | AF001654 |
| XVII | Prasinophyceae | 2 | OCS162 | 1,490 | AF001659 |
| XXXVI | 1 | OCS182 | 1,482 | AF001660 |
Identical sequences were not filed in the GenBank database.
Includes the sequences of primers used in the amplification of environmental nucleic acids. nt, nucleotides.
ND, no sequence was determined.
Figure 1 illustrates the fundamental dichotomy between green chloroplasts and other plastids, as well as the extraordinary diversity encompassed by the entire plastid clade. The large evolutionary distances between many of the OM and OCS plastid genes and their nearest neighbors (Fig. 1) underscore the unusual results reported in further detail below; the natural diversity of plastid genes from unidentified organisms adds significantly to the overall diversity within this clade.
Prymnesiophyceae.
SSU rRNA gene sequences from prymnesiophyte alga plastids formed a monophyletic cluster, strongly supported by both neighbor-joining (100 of 100 replicates) and maximum-parsimony (100 of 100 replicates) bootstrap analyses, within the line of descent that includes rhodophyte plastids and plastids derived from secondary symbioses (Fig. 1). Sequenced clones from two RFLP types within the OCS library (RFLP types I and IX) were found to be related to prymnesiophyte alga plastid SSU rRNA genes (Fig. 1 and 2; Table 1).
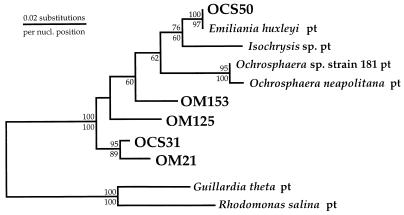
FIG. 2.
Phylogenetic positions of OCS and OM plastid rDNA clone sequences related to members of the class Prymnesiophyceae. This phylogenetic tree was constructed by the maximum-likelihood method (fastDNAml [43]). Bootstrap values (100 replicates) from both distance (above each node) and maximum-parsimony (below each node) analyses are shown. A mask of 750 nucleotides (nucl.), corresponding to the 5′ end of the plastid SSU rRNA gene, was employed. The plastid (pt) SSU rRNA gene sequences from cryptophyte algae G. theta and R. salina were used to root the phylogenetic tree.
RFLP type I, represented by OCS31 (Fig. 1 and 2), consisted of 30 clones, or 56% of the plastid SSU rDNA clones recovered in the OCS clone library. OCS31 was most closely related to OM21, an SSU rDNA clone recovered off the coast of Cape Hatteras, N.C. (97.6% similar for 1,470 nucleotide positions). These two environmental clones formed a monophyletic cluster in the phylogenies depicted in Fig. 1 and 2, although bootstrap support for this relationship was low for full-length sequences under maximum-parsimony criteria (69 of 100 replicates).
RFLP type IX consisted of three clones in the OCS library (6% of plastid clones [Table 1]) and was represented by OCS50 (Fig. 2). OCS50 was closely related to the plastid SSU rRNA gene sequence available for the prymnesiophyte Emiliania huxleyi (Lohmann) Hay and Mohler (99.8% similar for 955 nucleotide positions). Bootstrap support for the monophyletic clustering of OCS50 and E. huxleyi was high in both neighbor-joining (100 of 100 replicates) and maximum-parsimony (97 of 100 replicates) bootstrap analyses (Fig. 2).
Bacillariophyceae.
SSU rRNA gene sequences from bacillariophyte alga plastids formed a monophyletic cluster within the heterokont alga line of descent that was strongly supported by both neighbor-joining (100 of 100 replicates) and maximum-parsimony (98 of 100 replicates) bootstrap analyses (Fig. 1). Three RFLP types in the OCS library (RFLP types III, XI, and XXX) were related to bacillariophyte plastid SSU rRNA genes (Table 1). RFLP type III consisted of nine clones (17% of OCS plastid clones) and was represented by OCS54 (Fig. 1). Though not closely affiliated with any plastid SSU rRNA gene sequence available from cultured diatoms, OCS54 clearly emerged within the bacillariophyte SSU rDNA clade (Fig. 1). OCS56, representing RFLP type XI (two clones [4% of OCS plastid clones]), also did not specifically affiliate with any currently available bacillariophyte plastid SSU rRNA gene sequence but clearly branched within the bacillariophyte SSU rDNA clade as well (data not shown). OCS129, a unique clone in the OCS library (RFLP type XXX), was identified as being related to bacillariophyte plastid SSU rRNA genes by its hybridization with the bacillariophyte plastid-specific probe CHRYS1 (data not shown) and was not sequenced.
Cryptophyceae.
One RFLP type within the OCS library (RFLP type V; six clones [11% of plastid clones recovered]) was related to cryptophyte alga SSU rRNA genes (Table 1). Represented by OCS20, RFLP type V clearly branched within the cryptophyte plastid SSU rRNA gene clade, specifically affiliating with the cryptophyte Rhodomonas salina (95.9% similar for 1,479 positions) (Fig. 1). Together with OM283, a unique clone from the OM SSU rDNA clone library, OCS20 formed a monophyletic clade with the R. salina and Guillardia theta plastid SSU rRNA genes within the line of descent of rhodophyte plastids and plastids derived from secondary symbioses in the plastid SSU rRNA gene tree (Fig. 1). This clade received high bootstrap support in both neighbor-joining (100 of 100 replications) and maximum-parsimony (92 of 100 replications) analyses (Fig. 1).
Prasinophyceae.
Three clones representing two RFLP types in the OCS clone library formed two separate lineages within the chlorophyte (green chloroplast) line of descent (RFLP types XVII and XXXVI) (Table 1). Two clones with RFLP type XVII were represented by OCS162 (prasinophyte I lineage) (Fig. 1 and 3). OCS162 showed a close phylogenetic affiliation with the plastid SSU rRNA gene sequence from prasinophyte alga Pyramimonas parkeae (30), which was well supported by both neighbor-joining (100 of 100 replicates) and maximum-parsimony (100 of 100 replicates) bootstrap analyses of full-length SSU rRNA gene sequences (92.8% similar for 1,414 positions) (Fig. 1). Phylogenetic positioning of the prasinophyte clade represented by OCS162 and P. parkeae within the green-chloroplast line of descent could not be determined with confidence from the available green-plastid SSU rRNA data set (Fig. 3). Instead, this lineage appeared to diverge very early in the evolution of the green-chloroplast line of descent under a wide range of phylogenetic reconstructions with the full-length SSU rRNA gene sequence, including maximum-likelihood (Fig. 3), LogDet (Fig. 1), distance (Kimura two-parameter model with neighbor joining) (Fig. 1 and 3), and maximum-parsimony (Fig. 1 and 3) analyses.
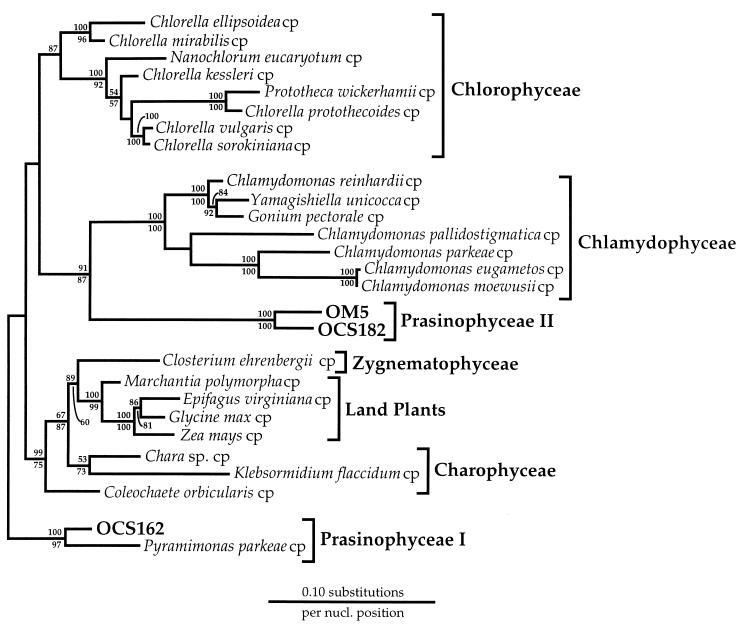
FIG. 3.
Phylogenetic positions of representative OCS and OM rDNA clones related to the chlorophyte line of descent. This phylogenetic tree was constructed by the maximum-likelihood method (fastDNAml [43]). Bootstrap values (100 replicates) from both neighbor-joining (above each node) and maximum-parsimony (below each node) analyses are shown. A conserved mask of 1,133 nucleotides (nucl.), spanning the entire length of the plastid SSU rRNA gene, was included. The gene sequences of Cyanophora paradoxa, Ochrosphaera neapolitana, E. huxleyi, and an Isochrysis sp. were used to root the phylogenetic tree. All the sequences used, except for those of Closterium ehrenbergii, Chlamydomonas parkeae, Yamagishiella unicocca, Gonium pectorale, and P. parkeae (30), are available from the GenBank or RDP database. cp, chloroplast.
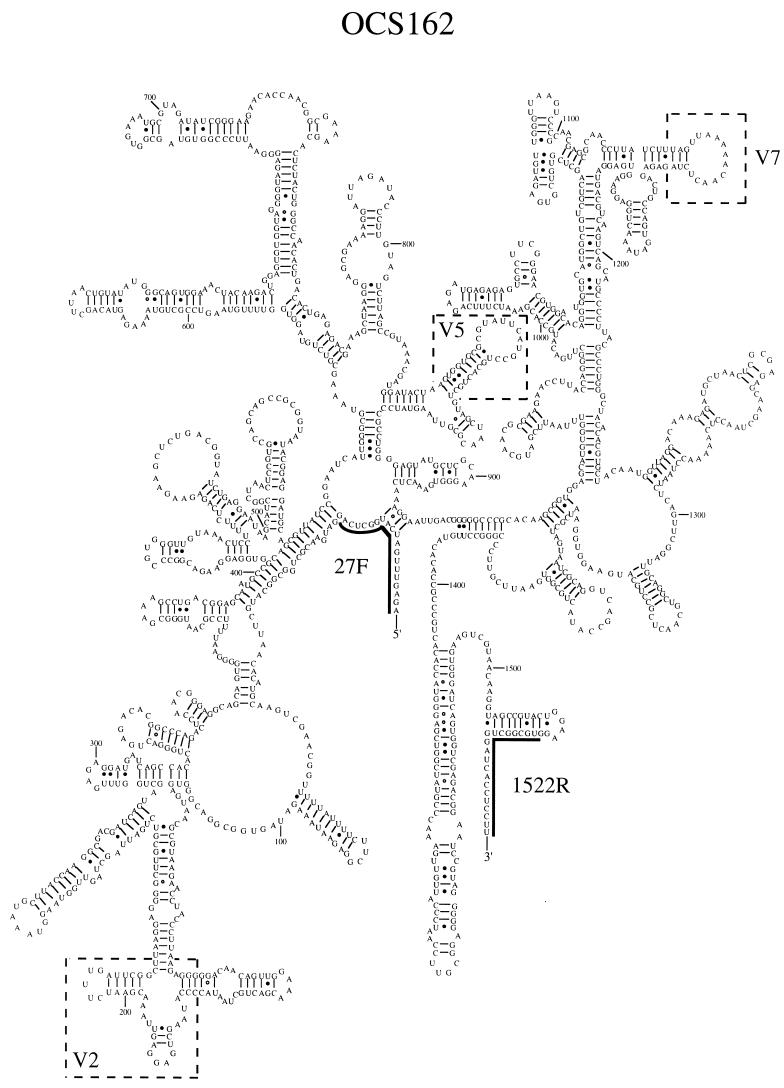
A secondary structural model for the SSU rRNA of OCS162 was constructed to check for base pairing idosyncrasies due to sequencing errors and chimeric gene formation (33) and to identify signatures unique to the prasinophyte lineage, as defined by OCS162 and P. parkeae (Fig. 4). The majority of nucleotide substitutions between OCS162 and P. parkeae were compensatory base changes that preserved the integrity of the SSU rRNA secondary structure across regions of double-stranded base pairing or occurred in highly variable loop regions (Fig. 4). Though clearly related phylogenetically, OCS162 and P. parkeae showed significant structural variation in two hypervariable regions of the bacterial SSU rRNA secondary structure (variable regions previously described [9]). Variable region two of the P. parkeae plastid SSU rRNA secondary structural model contained an insertion of 27 nucleotides relative to that of OCS162. In addition, the secondary structural model for OCS162 contained a 9-nucleotide deletion in variable region seven relative to that of P. parkeae. The structural variation observed in variable region two for these two related SSU rRNA genes is consistent with the structural variations observed in other groups of closely related bacterial rRNA genes (20, 59). However, structural variation in variable region seven of the bacterial SSU rRNA gene has previously been observed in only a limited number of major bacterial phyla (58). Deletions in this region of the bacterial SSU rRNA secondary structure appear to be common in the plastid SSU rRNA gene lineages we have examined.
FIG. 4.
Proposed secondary structural model for clone OCS162 SSU rRNA. Boxed areas indicate variable regions in the SSU rRNA gene product that distinguish OCS162 from P. parkeae. Variable regions of the bacterial SSU rRNA are numbered as previously described (9). Numbers refer to nucleotide positions in the E. coli SSU rRNA gene (5). The target sites for primers 27F and 1522R are indicated.
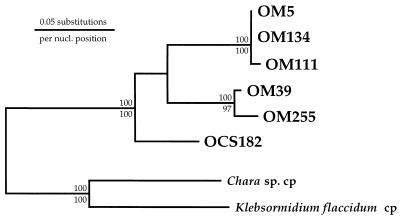
A unique clone in the OCS library, OCS182 (RFLP type XXXVI), was phylogenetically affiliated with OM5, an SSU rRNA gene clone recovered in the OM SSU rDNA clone library (Fig. 1 and 3). LogDet analysis (Fig. 1) revealed that the clade defined by OCS182 and OM5 was a deeply branching member of the green-plastid line of descent. A maximum-likelihood phylogenetic analysis which included full-length sequences and more members of the green-plastid line of descent showed the OCS182 and OM5 lineage to affiliate with the Chlamydophyceae class of single-celled algae (prasinophyte II lineage) (Fig. 3). This affiliation was supported only weakly by both neighbor-joining (<60 of 100 replicates) and maximum-parsimony (68 of 100 replicates) bootstrap analyses (Fig. 3), though it was also recovered in a LogDet analysis of the green-plastid data set (data not shown). As in the prasinophyte I lineage described above, the prasinophyte II lineage appeared to diverge early in the evolutionary history of green chloroplasts. Phylogenetic analyses of six members of the OCS182 and OM5 clade revealed that these clones fell into three closely related but well-defined lineages (Fig. 5). One lineage consisted of an Oregon coast clone (OCS182) alone, whereas the other two lineages consisted of only OM clones (Fig. 5). It was determined that the OCS182 and OM5 clade was closely related to prasinophyte alga plastids by phylogenetic analyses with an unpublished prasinophyte plastid SSU rRNA gene sequence (25).
FIG. 5.
Phylogenetic relationships among OCS and OM clones related to the Prasinophyceae II lineage. The phylogenetic tree was constructed by the maximum-likelihood method (fastDNAml [43]). This analysis included a mask of 382 nucleotides (nucl.), corresponding to the 5′ end of the plastid SSU rRNA gene. Bootstrap replications (100 resamplings) were performed by distance (above each node) and maximum-parsimony (below each node) methods. Bootstrap values of <50% are not shown. Chloroplast (cp) SSU rRNA sequences from charophytes, a Chara sp. and Klebsormidium flaccidum, were used to root the tree.
OM clone library.
In addition to the OM plastid rDNA clones described above and previously (44), three clones in the OM SSU rDNA clone library, OM81, OM164, and OM270, formed unique lineages within the plastid radiation defined by rhodophyte plastids and plastids derived from secondary symbioses. In LogDet analysis (Fig. 1), OM164 did not specifically affiliate with any class of algae for which plastid SSU rRNA gene sequences were available. OM81 associated specifically with the chrysophyte alga Ochromonas danica (91.4% similar for 1,272 nucleotide positions), a relationship supported by both neighbor-joining (100 of 100 replicates) and maximum-parsimony (100 of 100 replicates) bootstrap proportions, employing the entire length of the SSU rRNA gene (Fig. 1).
The third clone (OM270) did not specifically affiliate with any class of algae for which plastid SSU rRNA gene sequences were available (Fig. 1). In LogDet analysis (Fig. 1), OM270 occupied one lineage in a polytomy which also included the prymnesiophyte and cryptophyte plastid SSU rRNA lines of descent.
Chimeric gene product.
One OCS rDNA clone, preliminarily identified as plastid related, was identified as a chimeric gene product when the sequences obtained from both the 5′ and 3′ regions of the clone were examined. A phylogenetic analysis of ca. 400 nucleotides on the 3′ end of clone OCS22 revealed that this portion of the clone was closely related to bacillariophyte plastid SSU rRNA genes (data not shown). The sequence obtained from the 5′ end of OCS22 (ca. 400 nucleotides) was identical to an OCS rRNA gene clone (OCS126) related to the alpha subdivision of proteobacteria (data not shown).
LH-PCR.
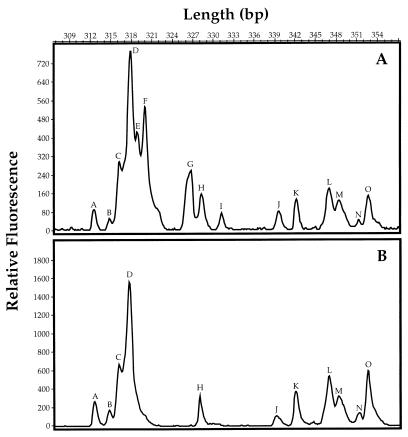
The identifications of plastid genes were supported by data showing that cells containing these genes could be selectively excluded from seawater samples by filtration through a 0.8-μm-pore-size polycarbonate filter. Electropherograms that chromatographically separated rDNA amplicons by molecular weight showed that major peaks corresponding to the sizes of prymnesiophyte, bacillariophyte, and prasinophyte genes (318, 319, and 326 bp) were present in a sample of coastal seawater (Fig. 6; Table 2) that had been collected on a 0.2-μm-pore-size polysulfone filter. These peaks were absent in samples of the same water that had been prefiltered through a 0.8-μm-pore-size polycarbonate filter, showing that eukaryotic phytoplankton cells could be selectively removed by size. The peak of 317 bp (Fig. 6) corresponds to the sizes of amplicons from both alpha proteobacteria and plastids (Table 2).
FIG. 6.
Electropherograms of bacterial and plastid rDNA diversity in unfiltered (A) and filtered (B) seawater at a depth of 10 m from 8 km off the coast of Yaquina Head, Oreg. SSU rRNA genes were amplified with the 27F and 338R primer pair, and amplicons were separated by natural length polymorphisms. Peaks E, F, G, and I were removed by filtration through a 0.8-μm-pore-size polycarbonate filter. Peaks E, F, and G have been identified as plastid genes (prymnesiophytes, bacillariophytes, and prasinophytes, respectively). Peak I is unidentified.
TABLE 2.
Length heterogeneity between SSU rRNA gene primers 27F and 338R for selected plastid and environmental clone sequences
| Organism or clone(s) | Class | Length (bp) | GenBank accession no. |
|---|---|---|---|
| OCS31 | Prymnesiophyceae | 317 | |
| OM21 | Prymnesiophyceae | U32671 | |
| OM13 | Bacillariophyceae | U32667 | |
| OM81 | Chrysophyceae | U70717 | |
| R. salina | Cryptophyceae | 318 | X55015 |
| OCS20 | Cryptophyceae | ||
| OM283 | Cryptophyceae | U70724 | |
| E. huxleyi | Prymnesiophyceae | 319 | X82156 |
| Isochrysis str. SAG927-2 | Prymnesiophyceae | X75518 | |
| Ochrosphaera neapolitana | Prymnesiophyceae | X80390 | |
| Ochrosphaera str. 181 | Prymnesiophyceae | X65101 | |
| OCS50 | Prymnesiophyceae | ||
| OM125 | Prymnesiophyceae | U70719 | |
| OM153 | Prymnesiophyceae | U70720 | |
| Odontella sinensis | Bacillariophyceae | ||
| Corethron cryophillum | Bacillariophyceae | ||
| OCS56 | Bacillariophyceae | ||
| OM19, OM20, OM22 | Bacillariophyceae | U32668, U32669, U32670 | |
| G. theta | Cryptophyceae | ||
| OM270 | Unidentified | U70723 | |
| Glaucosphaera vacuolata | Glaucosophyceae | 321 | |
| OCS54 | Bacillariophyceae | ||
| OM164 | Unidentified | U70721 | |
| Skeletonema costatum | Bacillariophyceae | 325 | X82154 |
| Skeletonema pseudocostatum | Bacillariophyceae | X82155 | |
| OM5, OM39, OM111, OM255 | Prasinophyceae II | 326 | U70715, U70716, U70718, U70722 |
| OCS182 | Prasinophyceae II | 327 | |
| OCS162 | Prasinophyceae I | 336 | |
| Nanochlorum eukaryotum | Chlorophyceae | 337 | X76084 |
| Heterosigma akashiwo | Chrysophyceae | 354 | M82860, M34370 |
| P. parkeae | Prasinophyceae | 360 |
DISCUSSION
The plastid genes that we describe here and those that we have described previously were discovered in investigations that had been designed to survey bacterioplankton diversity. Other studies with similar experimental designs have previously reported the presence of diatom plastid 16S rRNA genes recovered from phytodetrietal aggregates (10). Nonetheless, we were surprised to find so many plastid genes since numerous previous phylogenetic surveys of bacterioplankton diversity have reported relatively few plastid genes. In the two clone libraries we describe here, we identified 109 plastid genes belonging to five major classes of algae, including bacillariophytes (diatoms), prymnesiophytes (coccolithophorids), cryptophytes, chrysophytes, and prasinophytes. Although many examples of identical genes were found, in other cases the data support the presence of gene clusters, which most probably represent assemblages of related phytoplankton species.
A combination of several factors may have contributed to the contrast between the relative absence of plastid genes from clone libraries obtained from open-ocean samples and the large number of such genes found in these environmental rDNA clone libraries from coastal sites. Continental shelf regions are known to be areas of high nutrient availability and high primary productivity. Although marine Synechococcus spp. and prochlorophytes are common on continental shelves, the ratio of these unicellular prokaryotic phytoplankton species to their eukaryotic counterparts generally decreases as nutrient levels and primary production increase. Thus, the observation of large numbers of phytoplankton plastids in these clone libraries may result directly from the higher proportion of phytoplankton nucleic acids in samples from continental shelf regions. A second factor which may contribute to large numbers of phytoplankton genes is the high copy numbers of plastid organelle genomes. Some phytoplankton plastids contain up to 650 copies of small circular genomes per plastid, with each usually containing two ribosomal operons per genome (12). Prokaryotes frequently possess only 1 to 10 copies of their ribosomal operons. Although it appears likely that the large numbers of plastid genes observed in these clone libraries are in part due to the higher abundances of phytoplankton on continental shelves, the ratios between the observed numbers of plastid genes and bacterial genes may be skewed toward the former as a consequence of these differences in gene copy number.
As with phylogenetic surveys of bacterial diversity, this analysis of phytoplankton diversity uncovered evidence of novel groups of phytoplankton that had not been recognized previously on the basis of culture and morphological studies. We observed two phylogenetically distinct Prasinophyceae lineages. Prasinophytes are small flagellated cells that possess chlorophylls a and b. These organisms are thought to be primitive species because of their simple cell morphology, a feature that also makes them relatively difficult to identify in seawater samples, with the consequence that the marine members of this group are not routinely identified below the family level.
This report is the first to show the positions of two prasinophyte plastid SSU rRNA gene lineages in phylogenetic trees. In our analysis, which was based upon complete prasinophyte SSU rRNA genes, these plastids appeared to be most closely allied with the class Chlamydophyceae. However, the divergence between members of Prasinophyceae group I and Prasinophyceae group II was so deep that they appeared to be polyphyletic in Fig. 3, a result which is consistent with those of phylogenetic studies of nucleus-encoded rRNA gene sequence data (29, 53). An estimate of statistical confidence by bootstrap replication failed to resolve relationships between the two prasinophyte groups; however, clearly these are highly divergent lineages which warrant independent study.
Clone OM270 was unique among the genes investigated; it was phylogenetically allied to rhodophyte plastids and plastids derived by secondary symbioses, although bootstrap replicates provided only modest support for its position in the plastid phylogenetic tree. The phylogenetic position of this deeply branching clone has interesting evolutionary implications. In Fig. 1 and other analyses, OM270 appeared as an outgroup to prymnesiophytes and cryptophytes. Prymnesiophytes are flagellates that exhibit the photosynthetic pigments chlorophylls a and c. Members of the class Cryptophyceae are an enigmatic group; they appear to be the result of a sequential series of symbioses involving the capture of a rhodophyte phytoplankton cell by a eukaryotic cell, resulting in a cell configuration that includes a eukaryotic nucleus and a nucleomorph. The nucleomorph resides within an internal membrane that also encloses a plastid containing chlorophylls a and c and phycobilliproteins. Given this complicated phylogenetic picture, it would be of considerable interest to examine the phenotype of the lineage represented by OM270. An analysis of the predicted OM270 rRNA secondary structure failed to uncover any evidence that this unusual gene was a result of cloning or sequencing artifacts (data not shown).
An analysis of prymnesiophyte clones suggested that the prymnesiophyte E. huxleyi is not the most common prymnesiophyte in coastal seawater, at least in the environments we examined. E. huxleyi is a cultured species of marine prymnesiophyte, which is thought to represent the large blooms of coccolithophorids that are abundant primary producers in many regions of the world’s oceans. We observed a single gene, OCS50, that allied very closely with the published SSU rRNA gene sequence from the E. huxleyi plastid. Published studies suggest that the various morphotypes of E. huxleyii have identical plastid rRNA gene sequences. In addition to OCS50, we observed multiple plastid sequences that were related to prymnesiophytes but clearly distinct from Emiliania and Ochrosphaera species.
The view of phytoplankton diversity emerging from this study is that phytoplankton are better understood than are their prokaryotic counterparts; nonetheless, many members have not been identified previously by scientific investigations. There are several alternative explanations for these observations, the most likely of which is that some phytoplankton may be very difficult to identify by either morphology or cultivation. Alternatively, some of the novel phytoplankton lineages we describe here may correspond to cultured phytoplankton that have not been studied by chloroplast rRNA gene sequencing.
In the eyes of many practitioners, a survey of novel microbes by rRNA gene cloning and sequencing is not an ultimate goal but instead a stage of progress that may ultimately lead to the understanding of species that have not yielded to other means of study. The electropherogram in Fig. 6 is an example of the utility of plastid rDNA sequence data for the identification of plastid groups in a complex community. It shows the distribution of three major phytoplankton groups in rDNA amplicons from a coastal seawater sample. The different plastid types were resolved by LH-PCR in the 5′ regions of their plastid SSU rDNAs. Such measurements are highly reproducible and make it possible to rapidly survey the microbial diversities within samples for comparative purposes (54). rRNA gene sequences also provide the information needed to design molecular probes that can be used to study the ecological distributions of organisms by hybridization of probes to community nucleic acid samples or by microscopic observation of cells that have been hybridized to fluorescently labeled DNA probes.
ACKNOWLEDGMENTS
We are grateful to Katharine Field, Ena Urbach, Doug Gordon, Brian Lanoil, and Terah Wright for many useful comments regarding the manuscript. In addition, we thank Lee Kerkoff and Paul Kemp, who provided us with the nucleic acid sample used in the construction of this clone library. We are also indebted to Volker Huss and Linda Medlin for comparisons of our plastid clones to a database of unpublished plastid rRNA gene sequences.
This work was supported by Department of Energy grant FG0693ER61697.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen R A, Saunders G W, Paskind M P, Sexton J. Ultrastructure and 18S rRNA gene sequence for Pelagomonas calceolata gen. et sp. nov. and the description of a new algal class, the Pelagophyceae classis nov. J Phycol. 1993;29:701–715. [Google Scholar]
- 3.Barber R T, Smith R L. Coastal upwelling ecosystems. In: Longhurst A R, editor. Analysis of marine ecosystems. New York, N.Y: Academic Press; 1981. pp. 31–68. [Google Scholar]
- 4.Bhattacharya D, Medlin L. The phylogeny of plastids: a review based on comparisons of small-subunit ribosomal coding regions. J Phycol. 1995;31:489–498. [Google Scholar]
- 5.Brosius J, Palmer M L, Kennedy P J, Noller H F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell L, Shapiro L P, Haugen E. Immunochemical characterization of eukaryotic ultraplankton from the Atlantic and Pacific oceans. J Plankton Res. 1994;16:35–51. [Google Scholar]
- 7.Chrétiennot-Dinet M-J, Sournia A, Ricard M, Billard C. A classification of the marine phytoplankton of the world from class to genus. Phycologia. 1993;32:159–179. [Google Scholar]
- 8.Courties C, Vaquer A, Troussellier M, Lautier J, Chrétiennot-Dinet M J, Neveux J, Machado C, Claustre H. Smallest eukaryotic organism. Nature. 1994;370:255. [Google Scholar]
- 9.Dams E, Hendriks L, Van de Peer Y, Neefs J-M, Smits G, Vandenbempt I, De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1988;16:r87–r173. doi: 10.1093/nar/16.suppl.r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLong E F, Franks D G, Alldredge A L. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 11.DeLong E F, Wu K Y, Prézelin B B, Jovine R V M. High abundance of Archaea in Antarctic marine picoplankton. Nature. 1994;371:695–697. doi: 10.1038/371695a0. [DOI] [PubMed] [Google Scholar]
- 12.Ersland D R, Aldrich J, Cattolico R A. Kinetic complexity, homogeneity, and copy number of chloroplast DNA from the marine alga Olisthodiscus luteus. Plant Physiol. 1981;68:1468–1473. doi: 10.1104/pp.68.6.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein J. PHYLIP—phylogeny inference package (v3.5) Cladistics. 1989;5:164–166. [Google Scholar]
- 15.Field K G, Gordon D, Wright T, Rappé M, Urbach E, Vergin K, Giovannoni S J. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl Environ Microbiol. 1997;63:63–70. doi: 10.1128/aem.63.1.63-70.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gieskes W W C. Algal pigment fingerprints: clue to taxon-specific abundance, productivity and degradation of phytoplankton in seas and oceans. In: Demers S, editor. Particle analysis in oceanography. Berlin, Germany: Springer-Verlag; 1991. pp. 62–99. [Google Scholar]
- 17.Giovannoni, S. J. Unpublished observations.
- 18.Giovannoni S J, DeLong E F, Olsen G J, Pace N R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988;170:720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giovannoni S J, DeLong E F, Schmidt T M, Pace N R. Tangential flow filtration and preliminary phylogenetic analysis of marine picoplankton. Appl Environ Microbiol. 1990;56:2572–2575. doi: 10.1128/aem.56.8.2572-2575.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giovannoni S J, Rappé M S, Vergin K, Adair N. 16S rRNA genes reveal stratified open ocean bacterioplankton populations related to the green non-sulfur bacteria. Proc Natl Acad Sci USA. 1996;93:7979–7984. doi: 10.1073/pnas.93.15.7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon D A, Giovannoni S J. Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic and Pacific oceans. Appl Environ Microbiol. 1996;62:1171–1177. doi: 10.1128/aem.62.4.1171-1177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutell R R. Collection of small subunit (16S and 16S-like) ribosomal RNA structures. Nucleic Acids Res. 1994;22:3502–3507. doi: 10.1093/nar/22.17.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoepffner N, Haas L W. Electron microscopy of nanoplankton from the North Pacific central gyre. J Phycol. 1990;26:421–439. [Google Scholar]
- 24.Hood R R, Neuer S, Cowles T J. Autotrophic production, biomass and species composition at two stations across an upwelling front. Mar Ecol Prog Ser. 1992;83:221–232. [Google Scholar]
- 25.Huss, A. R. Personal communication.
- 26.Huyer A. Seasonal variation in temperature, salinity, and density over the continental shelf off Oregon. Limnol Oceanogr. 1977;22:442–453. [Google Scholar]
- 27.Huyer A. Coastal upwelling in the California current system. Prog Oceanogr. 1983;12:259–284. [Google Scholar]
- 28.Johnson P W, Sieburth J M. In-situ morphology and occurrence of eucaryotic phototrophs of bacterial size in the picoplankton of estuarine and oceanic waters. J Phycol. 1982;18:318–327. [Google Scholar]
- 29.Kantz T S, Theriot E C, Zimmer E A, Chapman R L. The Pleurastrophyceae and Micromonadophyceae: a cladistic analysis of nuclear rRNA sequence data. J Phycol. 1990;26:711–721. [Google Scholar]
- 30.Kim Y-S, Oyaizu H, Matsumoto S, Watanabe M M, Nozaki H. Chloroplast small-subunit ribosomal RNA gene sequence from Chlamydomonas parkeae (Chlorophyta): molecular phylogeny of a green alga with a peculiar pigment composition. Eur J Phycol. 1994;29:213–217. [Google Scholar]
- 31.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 32.Knauber D C, Berry E S, Fawley M W. Ribosomal RNA-based oligonucleotide probes to identify marine green ultraphytoplankton. J Eukaryot Microbiol. 1996;43:89–94. doi: 10.1111/j.1550-7408.1996.tb04486.x. [DOI] [PubMed] [Google Scholar]
- 33.Kopczynski E D, Bateson M M, Ward D M. Recognition of chimeric small-subunit ribosomal DNAs composed of genes from uncultivated microorganisms. Appl Environ Microbiol. 1994;60:746–748. doi: 10.1128/aem.60.2.746-748.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley and Sons; 1991. pp. 115–148. [Google Scholar]
- 35.Lange M, Guillou L, Vaulot D, Simon N, Amann R I, Ludwig W, Medlin L K. Identification of the class Prymnesiophyceae and the genus Phaeocystis with ribosomal RNA-targeted nucleic acid probes detected by flow cytometry. J Phycol. 1996;32:858–868. [Google Scholar]
- 36.Lockhart P J, Steel M A, Hendy M D, Penny D. Recovering evolutionary trees under a more realistic model of sequence evolution. Mol Biol Evol. 1994;11:605–612. doi: 10.1093/oxfordjournals.molbev.a040136. [DOI] [PubMed] [Google Scholar]
- 37.Maidak B L, Larsen N, McCaughey M J, Overbeek R, Olsen G J, Fogel K, Blandy J, Woese C R. The ribosomal database project. Nucleic Acids Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Medlin, L. K. Personal communication.
- 39.Medlin L K, Cooper A, Hill C, Wrieden S, Wellbrock U. Phylogenetic position of the chromista plastids based on small subunit rRNA coding regions. Curr Genet. 1995;28:560–565. doi: 10.1007/BF00518169. [DOI] [PubMed] [Google Scholar]
- 40.Miller P E, Scholin C A. Identification of cultured Pseudo-Nitzschia (Bacillariophyceae) using species-specific LSU rRNA-targeted fluorescent probes. J Phycol. 1996;32:646–655. [Google Scholar]
- 41.Millie D F, Paerl H W, Hurley J P. Microalgal pigment assessments using high-performance liquid chromatography: a synopsis of organismal and ecological applications. Can J Fish Aquat Sci. 1993;50:2513–2527. [Google Scholar]
- 42.Murphy L S, Haugen E M. The distribution and abundance of phototrophic ultraplankton in the North Atlantic. Limnol Oceanogr. 1985;30:47–58. [Google Scholar]
- 43.Olsen G J, Matsuda H, Hagstrom R, Overbeek R. fastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 44.Rappé M S, Kemp P F, Giovannoni S J. Chromophyte plastid 16S ribosomal RNA genes found in a clone library from Atlantic Ocean seawater. J Phycol. 1995;31:979–988. [Google Scholar]
- 45.Rappé, M. S., P. F. Kemp, and S. J. Giovannoni. Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol. Oceanogr., in press.
- 46.Robinson-Cox J F, Bateson M M, Ward D M. Evaluation of nearest-neighbor methods for detection of chimeric small-subunit rRNA sequences. Appl Environ Microbiol. 1995;61:1240–1245. doi: 10.1128/aem.61.4.1240-1245.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ryther J H. Photosynthesis and fish production in the sea. Science. 1969;166:72–76. doi: 10.1126/science.166.3901.72. [DOI] [PubMed] [Google Scholar]
- 48.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 49.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 50.Shapiro L P, Haugen E M, Keller M D, Bidigare R R, Campbell L, Guillard R R L. Taxonomic affinities of marine coccoid ultraphytoplankton: a comparison of immunochemical surface antigen cross-reactions and HPLC chloroplast pigment signatures. J Phycol. 1989;25:794–797. [Google Scholar]
- 51.Simon N, LeBot N, Marie D, Partensky F, Vaulot D. Fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes to identify small phytoplankton by flow cytometry. Appl Environ Microbiol. 1995;61:2506–2513. doi: 10.1128/aem.61.7.2506-2513.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Small L F, Menzies D W. Patterns of primary productivity and biomass in a coastal upwelling region. Deep Sea Res. 1981;28:123–149. [Google Scholar]
- 53.Steinkötter J, Bhattacharya D, Semmelroth I, Bibeau C, Melkonian M. Prasinophytes form independent lineages within the Chlorophyta: evidence from ribosomal RNA sequence comparisons. J Phycol. 1994;30:340–345. [Google Scholar]
- 54.Suzuki, M., M. S. Rappé, and S. J. Giovannoni. Unpublished observations.
- 55.Suzuki M T, Rappé M S, Haimberger Z W, Winfield H, Adair N, Ströbel J, Giovannoni S J. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swofford, D. Personal communication.
- 57.Whitmore S. gRNAid: an interactive graphics program for predicting the secondary structures of ribonucleic acid molecules. M.S. thesis. Corvallis: Oregon State University; 1992. [Google Scholar]
- 58.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wright T D, Vergin K, Boyd P W, Giovannoni S J. A novel δ-subdivision proteobacterial lineage stratified in the lower ocean surface layer. Appl Environ Microbiol. 1997;63:1441–1448. doi: 10.1128/aem.63.4.1441-1448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]