Abstract
We examined the genetic diversity of Nostoc symbionts in some lichens by using the tRNALeu (UAA) intron as a genetic marker. The nucleotide sequence was analyzed in the context of the secondary structure of the transcribed intron. Cyanobacterial tRNALeu (UAA) introns were specifically amplified from freshly collected lichen samples without previous DNA extraction. The lichen species used in the present study were Nephroma arcticum, Peltigera aphthosa, P. membranacea, and P. canina. Introns with different sizes around 300 bp were consistently obtained. Multiple clones from single PCRs were screened by using their single-stranded conformational polymorphism pattern, and the nucleotide sequence was determined. No evidence for sample heterogenity was found. This implies that the symbiont in situ is not a diverse community of cyanobionts but, rather, one Nostoc strain. Furthermore, each lichen thallus contained only one intron type, indicating that each thallus is colonized only once or that there is a high degree of specificity. The same cyanobacterial intron sequence was also found in samples of one lichen species from different localities. In a phylogenetic analysis, the cyanobacterial lichen sequences grouped together with the sequences from two free-living Nostoc strains. The size differences in the intron were due to insertions and deletions in highly variable regions. The sequence data were used in discussions concerning specificity and biology of the lichen symbiosis. It is concluded that the tRNALeu (UAA) intron can be of great value when examining cyanobacterial diversity.
Cyanobacteria of the genus Nostoc can form symbiotic associations with a wide range of organisms such as bryophytes, the small water fern (pteridophyte) Azolla, gymnosperms (cycads), the angiosperm Gunnera, and fungi (lichens) (1, 26). The taxonomic identity of the host and the structure in which the symbiont is housed show great diversity. However, the systematic and genetic diversity of the Nostoc symbionts is largely unknown (1, 26).
Lichens are symbiotic associations between a mycobiont (fungus) and a photobiont (green alga and/or cyanobacterium). Estimates of the number of lichen species range from 13,000 to 17,000, of which about 10% contain a cyanobacterium (24). The most frequent cyanobacterial genus in lichens is Nostoc (11). The present knowledge about the identity of the cyanobacterial symbionts in lichens does not extend beyond the genus level and is based on microscopy and biochemical and physiological experiments on freshly isolated symbionts and free-living isolates (26, 27).
The examination of cyanobacterial diversity and genetic variation within one lichen species, one population, one individual, or one lichen thallus has been complicated for several reasons (5, 11, 15). (i) The plastic morphology of the cyanobacterium, especially in the symbiotic condition, makes in situ identification of the cyanobiont difficult. (ii) The difficulties in isolating the primary symbiont, as opposed to a fast-growing minor biont or contaminant, makes results obtained with isolates equivocal. For the Azolla symbioses, this problem is well known; isolates previously thought to be the primary symbionts proved after closer examination not to be the same cyanobacterial strains as those detected in the leaf cavities of the symbioses (12). (iii) The fact that very few characters exist for differentiating among cyanobacterial strains and species have further complicated taxonomic studies.
An earlier study on the lichen Nephroma laevigatum, using Southern hybridizations with probes for conserved regions, showed different patterns for the cultured cyanobiont and for freshly isolated cyanobionts from the same lichen (20). This study indicates both that one lichen species may be fairly restricted in its cyanobiont diversity and that culturing the primary symbiotic cyanobacterium is difficult.
To investigate differences between closely related organisms, highly variable parts of the genome are often examined. The intron in the tRNALeu (UAA) gene is a suitable candidate for studies concerning cyanobacterial strains. This intron, the first to be discovered in eubacteria (33), has been shown to be highly variable among different cyanobacteria of the Nostocaceae family (filamentous heterocystous cyanobacteria, e.g., Anabaena and Nostoc [28]), ranging in size from 249 bp in Anabaena strain PCC 7120 to 291 bp in Anabaena azollae (33). At the nucleotide level, these two cyanobacteria show considerable differences for this intron. Other sequenced cyanobacterial tRNALeu introns include Scytonema strain PCC 7110, Anacystis strain R2, and Phormidium strain N182 (23). The position of this intron, interrupting the anticodon of the tRNALeu (UAA) gene, is conserved from cyanobacteria to plant chloroplasts (23). In plants, this intron has been used in both systematic studies where high resolution is needed and for population genetics (10, 13, 14, 21). It has, for example, been used to resolve phylogenetic relations when the sequence for the rbcL gene showed too little variation (13). Showing such great differences between close relatives and being present in only one copy in the genome, it is a suitable region for comparing closely related cyanobacteria and for examining the genetic diversity and specificity of cyanobacterial symbioses.
We examined the genetic variability of cyanobacterial tRNALeu (UAA) introns in some symbiotic lichen associations containing Nostoc symbionts. A method to directly amplify the tRNALeu (UAA) intron from small pieces of crude lichen material without any DNA extraction was developed. We used lichens from different genera (Nephroma and Peltigera), lichens in which the cyanobacterium plays different physiological roles (either photosynthesis plus nitrogen fixation [as in the bipartite lichens, where the cyanobiont is the only photobiont] or primarily nitrogen fixation [as in the tripartite lichens, where also a green alga is a part in the symbiosis]), and lichens in which the cyanobacteria are housed in different morphological structures (internal/external cephalodia [tripartite lichens] and throughout the thallus [bipartite lichens]). The intron was studied after direct amplification from crude lichen material by using size variation of obtained PCR fragments, the single-stranded conformational polymorphism (SSCP) pattern of both PCR products and cloned fragments, and nucleotide sequences from cloned introns.
Since so little is known about the diversity of the cyanobionts, one task in this study was to examine the level at which the diversity is found: within one cephalodium/thallus piece, within one lobe, within one individual, within one population, or between different lichen species — all fundamental questions that until now have not been addressed (27).
MATERIALS AND METHODS
Biological material.
Based on morphological characters, all lichens used in this study have been reported to have Nostoc as their cyanobacterial symbionts (26). The lichens belong to four different species; two are tripartite cephalodiate lichens (Nephroma arcticum [with internal cephalodia] and Peltigera aphthosa [with external cephalodia]), and two are bipartite lichens (P. membranacea and P. canina). All the lichens were freshly collected from Särna in the Dalecarlia area in Sweden. Thalli were taken from three different localities: near the Näcksjö mountain (locality 1; thallus samples A, D, J, and K), along the river Fulan (locality 2; thallus samples B, C, E, H, and I), and by the river Yxningån (locality 3; thallus samples G and F). These three localities are situated about 30 km from each other. In total, 11 thalli, all from different stands, were examined. From each thallus, three samples were prepared for PCR. Two of the products of these PCR samples were further characterized by cloning, SSCP screening, and sequencing. The laboratory strains used included Nostoc strain PCC 73102, N. muscorum CCAP 1453/12, and Anabaena/Nostoc strain PCC 7120 (referred to in the text as Anabaena strain PCC 7120). These nitrogen-fixing strains were grown in BG110 medium (29) in continuous light at 26°C. The equivalent of Nostoc strain PCC 73102 in the American Type Culture Collection, Nostoc strain ATCC 29133, was also analyzed in this study. These two strains were shown to have identical sequences for the tRNALeu (UAA) intron (data not shown).
Preparation of biological material for PCR.
To obtain biological material for PCR, a small fragment (less than 1 mm3) of the cephalodium/thallus was removed and placed in a microcentrifuge tube. Water to a volume of 40 μl was added, and the lichen piece was ground in a mortar (prepared by melting the tip of a 200-μl pipette tip). Aliquots of 2 μl from this slurry were used directly in the PCR without any DNA extraction. At least three separate subsamples were taken from each sample (thallus) and used for PCR.
When strains from pure cultures were used, either whole cells (Nostoc strain PCC 73102 and Nostoc muscorum) or purified DNA (Anabaena strain PCC 7120) was used in the PCRs.
Primers used in PCR.
Two primer pairs (outer and inner, respectively) were designed for specific amplification of cyanobacterial tRNALeu (UAA) introns by nested PCR. Both the specificity and the increased sensitivity obtained with nested PCR were necessary for good amplification from the biologically complex lichen samples containing only small amounts of cyanobiont cells. All primers contained a restriction site (lowercase letters) to facilitate cloning. The outer primers were 5′ggaattcGGGGRTRTGGYGRAAT3′ and 5′tcccGGGGRYRGRGGGACTT3′, and the inner primers were 5′agaattcGGTAGACGCWRCGGACTT3′ and 5′acccgggTWTACARTCRACGGATTTT3′. The primers were designed by Jeff Elhai, Department of Biology, University of Richmond, Richmond, Va.
PCR.
PCRs were performed on a model 2400 Perkin-Elmer thermocycler. Reaction mixtures of 20 μl contained 2 μl of cells or DNA (as described above), each deoxynucleoside triphosphate at 0.2 mM, 1.5× Taq buffer, 1 U of Taq DNA polymerase, and each primer at 1 μM. All PCRs were performed with 30 cycles at an anealing temperature of 55°C (first 4 cycles) and 60°C (next 26 cycles). The PCR products were diluted 1,000-fold in water between the first reaction (outer primers) and the second reaction (inner primers). At least three PCRs were performed for each thallus with different parts of the thallus.
Cloning.
Two of the PCR products from each thallus were used for subcloning. PCR-generated tRNALeu (UAA) introns were digested with the restriction enzymes SmaI and EcoRI (Pharmacia) and ligated into the vector pBluescript II SK+ (Stratagene). Transformation into Escherichia coli (Epicurian Coli XL-1 blue; Stratagene) was performed as described by the manufacturer.
SSCP analysis.
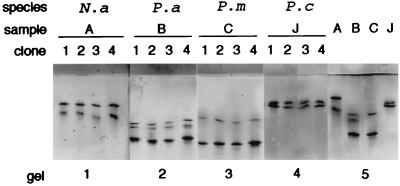
SSCP is a method which can detect small differences between PCR products. Double-stranded fragments are denatured, and the single-stranded products are run on a polyacrylamide gel under nondenaturing conditions. The single-stranded DNA molecules will fold in a sequence-dependent manner, and separation occurs based on the conformation of the molecule, which may allow the detection of differences as small as single mutations (25). PCR products from both crude lichen samples and plasmids with cloned introns were used for the SSCP analysis. The PCR products were precipitated and resuspended in water to the same volume as before. The samples were prepared by mixing equal amounts of sufficiently diluted PCR product and denaturing solution (formamide containing 2% glycerol, 0.05% bromophenol blue, and 0.05% xylene cyanol). The PCR products were made single stranded by being heated for 5 min in 95°C and then quickly cooled on ice. The samples were separated on a homogeneous 12.5% polyacrylamide gel with a native buffer system and a PhastSystem (Pharmacia Biotech) before being silver stained (Pharmacia Biotech) as recommended by the manufacturer. From two PCRs for each lichen species, at least five clones were screened by this method (four clones from each species shown in Fig. 2).
FIG. 2.
SSCP pattern of four clones from each of four samples (gels 1 to 4) compared to four different samples from the four different thalli run on the same gel (gel 5). The fact that all clones show a similar pattern indicates that each PCR product contains only one intron type. Samples B and C (P. aphthosa and P. membranacea living in close contact in the field) resulted in PCR products of very similar size, but they could be shown to be different by SSCP. N.a, N. arcticum; P.a, P. aphthosa; P.m, P. membranacea; P.c, P. canina.
Sequencing.
Both strands of the cloned fragments were sequenced with the PRISM Ready Reaction Dye Deoxy Terminator cycle-sequencing kit (Perkin-Elmer) with 1 μg of purified plasmid and T3 and T7 primers (Stratagene). The samples were run and analyzed on a 373A automated DNA sequencer (Applied Biosystems).
Intron folding.
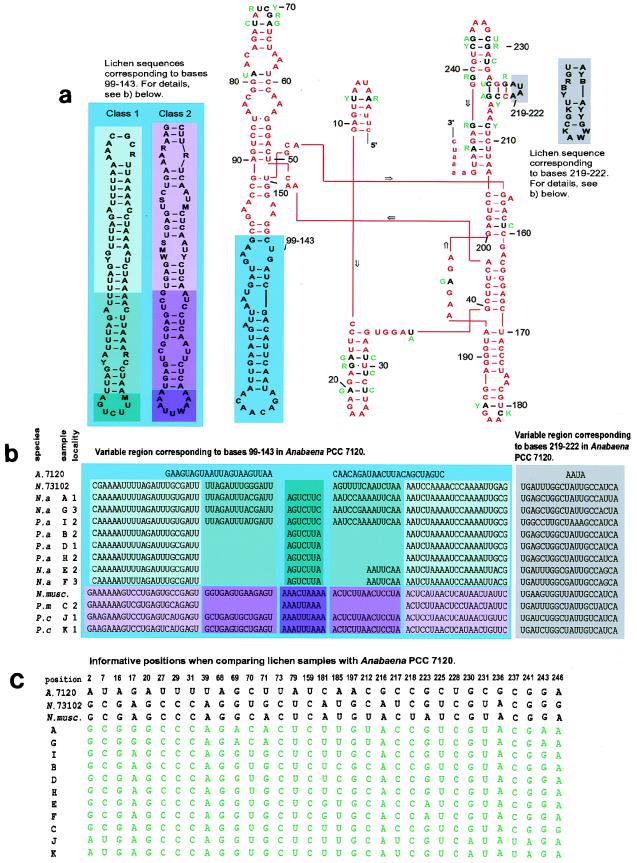
The intron sequences were used to construct a model for the secondary structure of the transcribed intron. As a reference, the model suggested by Cech et al. (4) for Anabaena strain PCC 7120 was used. Where the lichen sequences were not alignable with the Anabaena strain PCC 7120 sequence (the regions corresponding to bases 99 to 143 and bases 219 to 222) the program MulFold (17, 18, 34) was used to predict the stem-loop structure. The large region (corresponding to bases 99 to 143) could be divided into two classes to which all sequences obtained from this study were aligned (see Fig. 3). The two free-living Nostoc strains also used in this study could each be aligned with one of these two classes.
FIG. 3.
(a) Two-dimensional representation of the tRNALeu (UAA) intron from Anabaena strain PCC 7120 demonstrating the secondary structure of the intron as suggested by Cech et al. (4). The intron is shown in capital letters. Comparisons with the lichen cyanobiont sequences from this study are made in the model. Identity between Anabaena strain PCC 7120 and all lichen sequences is shown in red. Where differences occur, these are indicated either in the colored boxes on the sides (further explained for panel b) or in green next to their respective position (further explained for panel c). The fact that the intron has a secondary structure clearly restricts evolutionary changes to certain positions and regions. Compensating base pair changes and base pairing of the G · U type retain the secondary structure. (b) Size differences found by comparing Anabaena strain PCC 7120 with sequences in this study occur in two regions corresponding to positions 99 to 143 and 219 to 222 in the Anabaena strain PCC 7120 sequence. In one of these regions (corresponding to bases 99 to 143), the lichen sequences can be aligned to either of two classes. Size differences within each class occur due to insertions and deletions. Note that some samples (E and F) do not readily fit into the folding of this region as suggested in the legend to panel a. Sequences from free-living strains also obtained in this study are included. (c) Informative positions when comparing alignable regions (all bases except those corresponding to the variable regions shown in panel b) of the tRNALeu intron from Anabaena strain PCC 7120 and of all sequences obtained in the present study. N.a, N. arcticum; P.a, P. aphthosa; P.c, P. canina; P.m, P. membranacea; A. 7120 Anabaena strain PCC 7120; N.73102, Nostoc strain PCC 73102; N.musc, N. muscorum.
Phylogenetic analysis.
All published tRNALeu (UAA) intron sequences from cyanobacteria belonging to the order Nostocales (16), section IV (28), were used for phylogenetic analysis. This included the lichen samples used in the present study, as well as the sequences from N. muscorum (this study), Nostoc strain PCC 73102 (this study), Anabaena strain PCC 7120 (33), A. azollae (33), and Scytonema strain PCC 7110 (23). The sequences were aligned by using the conserved secondary structure, and all the information corresponding to bases 1 to 98, 144 to 218, and 223 to 249 in Anabaena strain PCC 7120 was used in the phylogenetic analysis. These parts of the sequences correspond to the entire intron, with the exception of the two variable regions (corresponding to bases 99 to 143 and 219 to 222), which could not be readily aligned (see Fig. 3). The data matrices were analyzed with Phylogenetic Analysis Using Parsimony (PAUP) software, version 4.0d49_noFPU (32), on an Apple Macintosh LC630 with 20 MB of physical RAM. An exact search was performed with the branch-and-bound algorithm as implemented in PAUP. Initially, all characters were given equal weight and treated as unordered. All uninformative characters were excluded from the analysis. With the results obtained from the first analysis, a successive-approximations reweighting by the method of Farris (7) was performed, reweighting characters according to their rescaled consistency index. This quickly resulted in a stable solution giving 90 trees. Following the first analysis, a bootstrap analysis (9) and a parsimony jackknifing analysis (8) as implemented in PAUP were performed.
Image processing.
Photographs (Fig. 1) and gels (Fig. 2) were scanned and digitalized with a Studio Scan II si scanner and Fotolook PS 2.07.2 (Agfa Gevaert N.V.). The brightness and contrast were adjusted with Adobe Photoshop 3.0 (AdobeSystems Europe Ltd.). The figures were then completed with Adobe Illustrator 5.5 (AdobeSystems Europe Ltd.) and printed with a Fujix Pictography 3000 (Fuji Photo Film Co.).
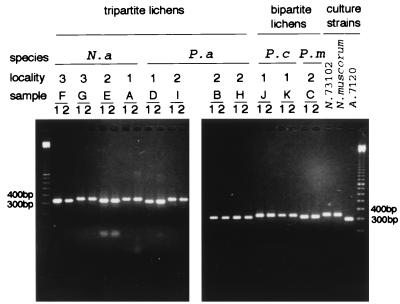
FIG. 1.
PCR products after amplification of the tRNALeu (UAA) intron from lichen samples and three free-living strains from culture collections analyzed by agarose gel electrophoresis. Different samples from one thallus never showed size variation (subsamples 1 and 2). All samples have an intron which is larger than that of Anabaena strain PCC 7120 (A. 7120). N.a, N. arcticum; P.a, P. aphthosa, P.c, P. canina; P.m, P. membranacea; N.73102, Nostoc strain PCC 73102. Size markers (100-bp ladder) are shown to the left and to the right.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the cyanobiont sequences are AF019912 to AF019922 for lichen samples A to K, and those for the free-living sequences are AF019924 and AF019925 for Nostoc strain PCC 73102 and N. muscorum, respectively.
RESULTS
PCR.
One DNA fragment of 300 to 400 bp was consistently obtained from the different biological samples (Fig. 1) after nested PCR. All examined samples had an intron larger than that of Anabaena strain PCC 7120. No size variation among the three samples from a single lichen thallus was observed. The sizes of the introns were not strictly correlated to the taxonomic or geographical origin of the lichen.
SSCP analysis.
At least five clones from two PCRs for each species were further investigated by using the SSCP pattern. All the clones from a single PCR consistently showed an identical pattern (Fig. 2). Direct SSCP analysis of the PCR products showed the same SSCP pattern as did analysis of their cloned counterparts. The fact that more than two bands can be seen in some samples is accounted for by the presence of metastable bands. These have the advantage of further increasing resolution, but their presence is sensitive to any changes in the sample preparation and/or running conditions. SSCP analysis in this study is not, as often is the case, used for the detection of single mutations but, rather, is used as a screening method for the examination of the diversity of different intron types in one single sample. Biologically different samples, with amplified introns which could not be separated on an agarose gel, showed differences in their SSCP pattern. For example, it was possible to discriminate between two of the Peltigera species (P. aphthosa and P. membranacae) living close to each other in the same locality and giving PCR bands of very similar sizes (compare samples B and C in Fig. 1 and Fig. 2).
Sequences.
For each sample (thallus), clones from at least two PCRs performed on different parts of the thallus were completely sequenced from both directions. The intron sequences from these lichens show strong similarities to the previously published sequences from the free-living strains Anabaena strain PCC 7120 and A. azollae (33), as well as to the sequences from Nostoc strain PCC 73102 and N. muscorum (Fig. 3). However, substantial differences did occur in two regions when the cyanobiont sequences were compared with that of Anabaena strain PCC 7120, and differences occurred one region when the lichen sequences were compared with each other (Fig. 3a and b). These regions also account for the observed size variations of the cyanobacterial tRNALeu (UAA) introns. The large, highly variable region (corresponding to bases 99 to 143) in all lichen samples exhibit either of two separate themes. Thus, this highly variable region in the introns can be divided into two classes (Fig. 3). The two free-living Nostoc strains used in this study can also be aligned with these classes, whereas Anabaena strain PCC 7120 shows a third pattern for this region. In this region, insertions and deletions give rise to structural differences rather than variations on the nucleotide level.
The stability of the secondary structure is often retained in areas where changes in the sequence have occurred. This can be accomplished by a compensating base change on the base-pairing strand in the secondary structure or by changes which allow base pairing of the G · U type. Different parts of the intron also have different degrees of variability, with the catalytic core (3) remaining virtually invariable while some peripheral parts show more changes (such as the highly variable regions discussed above and the loop corresponding to bases 68 to 73 in Anabaena strain PCC 7120).
Phylogenetic analysis.
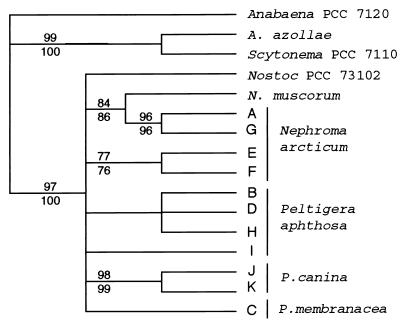
The result of the analysis consists of 90 equally parsimonious trees with a length of 47 steps. The trees were rooted by defining Anabaena strain PCC 7120, Scytonema strain PCC 7110, and A. azollae as outgroups. The retention index (RI) (6) and consistency index (CI) (22) were calculated to be 0.708 (RI) and 0.702 (CI), respectively. The combinable-component (2) consensus tree from the analysis is shown in Fig. 4. Supportive indices (bootstrap values and jackknife fractions) are displayed along the branches in Fig. 4.
FIG. 4.
Phylogenetic tree including all published tRNALeu intron sequences from cyanobacteria of the order Nostocales (section IV). Only the conserved, readily alignable parts of the intron have been used in this analysis (for details, see the text). Bootstrap values are shown above branches, and jackknife fractions are shown under branches. The intron sequences of all examined lichen cyanobionts are grouped with the sequences of the free-living Nostoc strains also used in this study. In the phylogenetic analysis, only few characters are informative within this group (Fig. 3c). The additional information present in the variable parts of the intron is difficult to use in this kind of phylogenetic analysis because they are not readily alignable, but it can be of great value when analyzing closely related cyanobacteria such as the lichen cyanobionts.
The analysis supports a close relationship between the intron sequences of all cyanobionts used in this study and the sequences from the free-living Nostoc strains also obtained in this study. The highly variable region corresponding to bases 99 to 143 in Anabaena strain PCC 7120, which is responsible for the division of the lichen introns into two classes in Fig. 3, is left out in this analysis. There is no support for this division in the analyzed conserved parts of the intron.
DISCUSSION
The cyanobacterial symbionts of the examined lichens seem not to consist of a community of different strains but, rather, of one cyanobacterium in each thallus. Although different cyanobacterial intron types were found in this study, even in lichens living close to each other, no variation was ever observed within a sample or within a single lichen thallus. That a minor intron type could have avoided detection is possible, but there is substantial evidence in favor of only one cyanobiont in each thallus. Five clones from two PCRs for each species were screened by SSCP analysis, and two clones from independent samples from each thallus were sequenced (in total, at least two times 11 sequences) without any differences being detected within one thallus. That identical intron sequences could be derived from different Nostoc strains is possible, but they should nevertheless be more closely related to each other than to cyanobionts with different sequences. For a biological discussion, it is thus reasonable to consider each intron type to be derived from one Nostoc strain.
This apparent lack of variation within one thallus could have different biological reasons. It is possible that it is the result of high specificity, which allow only one or a few cyanobacterial strains to be accepted by one fungal species. Another (or an additional) reason could be that cyanobacteria efficiently spread to younger parts of the thallus either internally or by external migration and reinfection. In the bipartite lichens, where the cyanobiont layer is continuous, it is relatively easy for the cyanobacterium to divide and grow together with the thallus margin. Hence, there is no need for more than one infection, which could result in several Nostoc strains in one thallus. It is more difficult to determine how this is accomplished in a tripartite lichen, where the cyanobacterial population is confined to small cephalodia separated by cyanobacterium-free areas of only fungus and green alga. Several authors have described, by observation of field material (19) and in resynthesis studies (30, 31), how each cephalodium is the product of a new infection. However, at least in the case of P. aphthosa, which has external cephalodia situated on the upper side of the thallus, the older cephalodia have been reported to release hormogonia, motile Nostoc filaments (30).
The highly variable region of the intron corresponding to bases 99 to 143 in Anabaena PCC 7120 shows two basic patterns to which all lichen sequences can be aligned (Fig. 3). One is shared by the bipartite lichens examined (P. membanacea and P. canina) and the free-living N. muscorum. The cyanobacteria of these lichens are both the photobionts of the symbiosis and the nitrogen-fixing bionts, and the N. muscorum strain is a phototrophic free-living organism. All other lichen samples, as well as Nostoc strain PCC 73102 (originally isolated from symbiosis with the cycad Macrozamia), contain another theme in this highly variable region. These are all cyanobacteria with nitrogen fixation as their primary task in their respective symbiosis. Thus, a functional difference exists between our samples regarding their role in symbiosis, with the cyanobiont of the tripartite lichens being involved primarily in nitrogen fixation and the cyanobiont of the bipartite lichen having both photosynthesis and nitrogen fixation as its roles in the symbiosis. However, there are not enough differences in the conserved parts of the intron, which were used in the phylogenetic analysis, to be able to support or reject this division. From a symbiotic perspective, this division of the samples in the present study into two types of introns correlated to two different physiological roles for the cyanobiont is interesting and should be investigated further.
The two different classes of variable loop also have some features in common. They are both composed of several repeats of the same or very similar sequences (see, for example, the base pairing GAUUU/AAAUC for class 1 and GAGU/ACUC for class 2, which are repeated several times). Different numbers of these repeats yield the size differences within the classes. It can also be noted that samples E and F do not readily fit into the secondary-structure model in Fig. 3 but do provide a possible form between a shorter and a longer stem-loop where insertion or deletion has occurred on only one side of the structure.
Given the high degree of identity between the symbionts examined in this study, it appears that the presently studied lichen symbioses are restricted in their choice of cyanobiont to a relatively small number of Nostoc strains. The fact that the same cyanobacterial intron type also can be found in different samples from the same lichen species (see, for example, samples A and G or samples B, D, and H) is also an indication of specificity. To determine to what extent the fungus really discriminates between cyanobacterial strains when establishing the symbiosis, we would need a much better knowledge of the diversity of Nostoc strains in the soil.
This study is the first step in a more extensive survey of the genetic diversity of symbiotic cyanobacteria. We have addressed several important questions concerning lichen biology and presented a method which opens up the possibility of studying these, and other, symbiotic systems in more detail. Examining other symbiotic systems by the same approach will not only provide information concerning the biology of these systems but will also reveal similarities and differences between cyanobacterial symbioses in general.
ACKNOWLEDGMENTS
We thank Anna och Gunnar Vidfelts fond, Karin och Axel Binnings fond, and the Swedish Natural Science Research Council for supporting this project.
Jeff Elhai (Department of Biology, University of Richmond, Richmond, Va.) is gratefully acknowledged for designing the primers and for valuable discussions. Anders Backlund (Department of Systematic Botany, Uppsala University, Uppsala, Sweden) is thanked for help with the phylogenetic analysis.
REFERENCES
- 1.Bergman B, Matveyev A, Rasmusson U. Chemical signaling in cyanobacterial-plant symbioses. Trends Plant Sci. 1996;1:191–197. [Google Scholar]
- 2.Bremer K. The limits of amino acid sequence data in angiosperm phylogenetic reconstruction. Evolution. 1988;42:795–803. doi: 10.1111/j.1558-5646.1988.tb02497.x. [DOI] [PubMed] [Google Scholar]
- 3.Cate J H, Gooding A R, Podell E, Zhou K, Golden B L, Kundrot C E, Cech T R, Doudna J A. Crystal structure of a group 1 ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 4.Cech T R, Damberger S H, Gutell R R. Representation of the secondary and tertiary structure of group I introns. Nat Struct Biol. 1994;1:273–280. doi: 10.1038/nsb0594-273. [DOI] [PubMed] [Google Scholar]
- 5.Fahselt D. Individuals, populations and population ecology. In: Nash T H, editor. Lichen biology. Cambridge, United Kingdom: Cambridge University Press; 1996. pp. 181–198. [Google Scholar]
- 6.Farris J S. The retention index and the rescaled consistency index. Cladistics. 1989;5:417–419. doi: 10.1111/j.1096-0031.1989.tb00573.x. [DOI] [PubMed] [Google Scholar]
- 7.Farris J S. A successive approximations approach to character weighting. Syst Zool. 1969;18:374–385. [Google Scholar]
- 8.Farris J S, Albert V A, Källersjö M, Lipscomb D, Kluge A G. Parsimony jackknifing outperforms neighbor-joining. Cladistics. 1996;12:99–124. doi: 10.1111/j.1096-0031.1996.tb00196.x. [DOI] [PubMed] [Google Scholar]
- 9.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 10.Ferris C, Oliver R P, Davy A J, Hewitt G M. Using chloroplast DNA to trace postglacial migration routes of oaks into Britain. Mol Ecol. 1995;4:731–738. doi: 10.1111/j.1365-294x.1995.tb00273.x. [DOI] [PubMed] [Google Scholar]
- 11.Friedl T, Büdel B. Photobionts. In: Nash T H, editor. Lichen biology. Cambridge, United Kingdom: Cambridge University Press; 1996. pp. 8–23. [Google Scholar]
- 12.Gebhardt J S, Nierzwicki Bauer S A. Identification of a common cyanobacterial symbiont associated with Azolla spp. through molecular and morphological characterization of free-living and symbiotic cyanobacteria. Appl Environ Microbiol. 1991;57:2141–2146. doi: 10.1128/aem.57.8.2141-2146.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gielly L, Taberlet P. The use of chloroplast DNA to resolve plant phylogenies: noncoding versus rbcL sequences. Mol Biol Evol. 1994;11:769–777. doi: 10.1093/oxfordjournals.molbev.a040157. [DOI] [PubMed] [Google Scholar]
- 14.Gielly L, Taberlet P. A phylogeny of the European gentians inferred from chloroplast trnL (UAA) intron sequences. Bot J Linn Soc. 1996;120:57–75. [Google Scholar]
- 15.Hill D J. The nature of the symbiotic relationship in lichens. Endeavour. 1994;18:96–103. [Google Scholar]
- 16.Holt J G, Krieg N R, Sneath P H A, Staley J T, Williams S T. Bergey’s manual of determinative bacteriology. 9th ed. Baltimore, Md: The Williams & Wilkins Co.; 1994. [Google Scholar]
- 17.Jaeger J A, Turner D H, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaeger J A, Turner D H, Zuker M. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 1989;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- 19.Jordan W P, Rickson F R. Cyanophyte cephalodia in the lichen genus Nephroma. Am J Bot. 1971;58:562–568. [Google Scholar]
- 20.Kardish N, Rotem Abarbanell D, Zilberstein A, Galun M. Comparison between the symbiotic Nostoc of the lichen Nephroma laevigatum Ach. and its cultured, isolated Nostoc by recombinant DNA. Symbiosis. 1990;8:135–146. [Google Scholar]
- 21.Kita Y, Ueda K, Kadota Y. Molecular phylogeny and evaluation of the Asian Aconitum subgenus Aconitum (Ranunculaceae) J Plant Res. 1995;108:429–442. [Google Scholar]
- 22.Kluge A G, Farris J S. Quantitative phyletics and the evolution of the anurans. Syst Zool. 1969;18:1–32. [Google Scholar]
- 23.Kuhsel M G, Strickland R, Palmer J D. An ancient group I intron shared by eubacteria and chloroplasts. Science. 1990;250:1570–1573. doi: 10.1126/science.2125748. [DOI] [PubMed] [Google Scholar]
- 24.Nash T H. Nitrogen, its metabolism and potential contribution to ecosystems. In: Nash T H, editor. Lichen biology. Cambridge, United Kingdom: Cambridge University Press; 1996. pp. 121–135. [Google Scholar]
- 25.Pharmacia LKB Biotechnology. PCR-SSCP analysis, separation technique. File 131. Pharmacia LKB Biotechnology, Uppsala, Sweden.
- 26.Rai A N. Cyanobacterial-fungal symbioses: the cyanolichens. In: Rai A N, editor. Handbook of symbiotic cyanobacteria. Boca Raton, Fla: CRC Press, Inc.; 1990. pp. 9–41. [Google Scholar]
- 27.Rikkinen J. What’s behind the pretty colours? A study on the photobiology of lichens. Bryobrothera. 1995;4:1–239. [Google Scholar]
- 28.Rippka R, Herdman M. Pasteur culture collection of cyanobacterial strains in axenic culture. Vol. 1. Paris, France: Institute Pasteur; 1992. [Google Scholar]
- 29.Stanier R Y, Kunisawa R, Mandel M, Cohen-Bazire G. Purification and properties of unicellular blue-green algae (Order Chrococcales) Bacteriol Rev. 1971;35:171–205. doi: 10.1128/br.35.2.171-205.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stocker-Woergoetter E. Experimental cultivation of lichens and lichen symbionts. Can J Bot. 1995;73:S579–S589. [Google Scholar]
- 31.Stocker Woergoetter E, Tuerk R. Artificial resynthesis of the photosymbiodeme Peltigera leucophlebia under laboratory conditions. Cryptogam Bot. 1994;4:300–308. [Google Scholar]
- 32.Swofford D L. PAUP 4.0. Phylogenetic Analysis Using Parsimony, version 4.0. Sunderland, Mass: Sinauer Associates; 1996. [Google Scholar]
- 33.Xu M Q, Kathe S D, Goodrich Blair H, Nierzwicki Bauer S A, Shub D A. Bacterial origin of a chloroplast intron: Conserved self-splicing group I introns in cyanobacteria. Science. 1990;250:1566–1570. doi: 10.1126/science.2125747. [DOI] [PubMed] [Google Scholar]
- 34.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]