Abstract
This study focuses on the different efficiencies of secretion of two fungal cutinases by Saccharomyces cerevisiae, a wild-type cutinase (CY000) and a hydrophobic mutant cutinase (CY028). Both cutinases are placed under control of the GAL7 promoter, by which the expression levels can be regulated. Wild-type cutinase was secreted at up to 25 mg per g (dry weight), while CY028 was secreted at a level of 2 mg per g (dry weight); this difference is nearly independent of the expression level. Pulse-chase experiments revealed that whereas CY000 cutinase is secreted, CY028 is irreversibly retained in the cell. Immunogold labelling followed by electron microscopy revealed colocalization of CY028 with immunoglobulin heavy-chain binding protein (BiP) in the endoplasmic reticulum (ER). The increase of wild-type cutinase expression did not result in higher levels of the molecular chaperone BiP, but BiP levels are raised by increased induction of the hydrophobic mutant cutinase. Immunoprecipitation studies showed that in contrast to the wild-type cutinase, the hydrophobic mutant cutinase interacts with BiP. These results indicate that the introduction of two exposed hydrophobic patches in cutinase results in a higher affinity for BiP which might cause the retention of this mutant cutinase in the ER.
Cutinase from Fusarium solani pisi is a lipase with a molecular mass of 21.6 kDa containing two disulfide bridges (14). This enzyme degrades the cutin layer of plants, enabling penetration by the fungus. Cutinase is active in aqueous solutions, without need of interfacial activation (32), and is therefore potentially suitable for lipid stain removal applications in the detergent industry (5, 6). However, the natural cutinase has two clear shortcomings: a low level of effective interaction with lipid substrate (both on the molecular and the micellar levels) and sensitivity to anionic detergents. Cutinase lacks a large hydrophobic surface around the active site, in contrast to other lipases (18). To improve the interaction with lipid substrates, a large series of cutinase mutants has been constructed by using a synthetic cutinase gene (30) in which the hydrophobic surface around the active site has been increased (around amino acids 80 and 185). Some of the designed cutinase mutants indeed exhibit improved wash performance, making them interesting for use in detergents. In order to obtain an efficient and low-cost production system for cutinase, this enzyme was overproduced in Saccharomyces cerevisiae (30). However, some of the mutant cutinases with increased wash performance were significantly impaired in secretion compared to the wild-type enzyme. Because CY028 cutinase was the best in performance but was secreted at a very low level, we studied this mutant in more detail.
Secretion efficiency is dependent on proper intracellular sorting and folding of the heterologous protein (13, 21, 24). Molecular chaperones play an important role in these processes. The hsp 70 protein chaperone BiP (immunoglobulin heavy-chain binding protein) was originally identified as an endoplasmic reticulum (ER) protein (20, 22) found in association with unassembled antibody heavy chains (10), thereby preventing their premature secretion. It is now clear that BiP interacts with exposed hydrophobic patches of various newly synthesized translocating proteins which are entering the ER lumen, preventing aggregation of these proteins and accompanying the process of folding of these polypeptides (9).
The aim of this study was to identify the cause of the low level of secretion of a hydrophobic mutant cutinase by S. cerevisiae. Here, we report the interaction of a hydrophobic mutant cutinase with BiP during the secretion process, which could be a factor in the ER retention of this hydrophobic cutinase as observed by immunoelectron microscopy.
MATERIALS AND METHODS
Strains and genetic constructs.
S. cerevisiae SU50 (MEL− his4 leu2 cir0) (31) was used as a host for cutinase production. The cutinase gene was cloned behind the invertase signal sequence, placed under control of the GAL7 promoter, and integrated on the chromosomal ribosomal DNA locus; the construct contained a leu2 gene which enabled selection on leu-lacking agar plates (30). The cutinase expression strains and the mutant cutinases (see Table 1) were constructed and provided by C. Visser, Unilever Research Laboratory, Vlaardingen, The Netherlands.
TABLE 1.
Specific activities and production of designed cutinase mutants
| Mutant | Mutation(s) | Sp act (SLU/mg)a | Extracellular production (%)b |
|---|---|---|---|
| CY000 | 700 | 100 | |
| CY004 | A85F | 945 | 34 |
| CY008 | A185L | 735 | 120 |
| CY009 | L189F | 847 | 183 |
| CY026 | G82A, A85F | 812 | 120 |
| CY027 | V184I, A185L, L189F | 483 | 83 |
| CY028 | G82A, A85F, V184I, A185L, L189F | 1,085 | 22 |
SLU, specific lipase units (1 SLU corresponds to 1 μmol of free fatty acid release per min).
Production levels were determined as described in Materials and Methods.
Media and growth conditions.
Defined Egli medium was used in a glucose-limited chemostat (27) supplemented with 200 mg of histidine per liter. The yeast was grown at 30°C in a Bioflow III fermentor (New Brunswick Scientific) essentially as described by Silljé et al. (28). After the batch phase, a continuous feed was connected with 20 g of glucose per liter at a dilution rate of 0.07 h−1. When a steady state was reached, samples were taken, as indicated below. The amount of galactose in the feed was increased after each steady state, thereby increasing the level of induction of the cutinase gene.
Fed-batch fermentations.
Fed-batch fermentations were performed in a bioreactor with a working volume from 5 to 7.5 liters. The pH and dissolved-oxygen tension were measured with Ingold probes; exhaust gas was analyzed on-line with a Prima 600 mass spectrometer (VG Gas Analysis Systems Limited). All fermentations were done under standard conditions; the pH was maintained at 5.0 with 12.5% ammonia, and the dissolved-oxygen tension was kept above 15% by adjusting the stirrer speed. After the batch phase, a fed batch with an exponential feed strategy was used to increase biomass and to induce the cutinase gene. The batch phase medium (5 liters) was composed of the following (in grams per liter): glucose, 22; galactose, 7.4; yeast extract (Difco), 10; Trusoy, 5; histidine, 0.05; MgSO4 · 7H2O, 0.05; KH2PO4, 2.1; and Egli trace elements, 10. The feed medium (2.5 liters) was composed of the following (in grams per liter): glucose, 393; galactose, 53.6; yeast extract (Difco), 22.3; KH2PO4, 7.4; Egli trace elements, 17.86; and histidine, 2.25. After sterilization, 5 ml of 1,000× Egli vitamin solution was added. Cutinase production was determined at the end of the feed phase.
Western blotting.
From continuous cultures, 1 ml of culture was diluted to an optical density at 600 nm (OD600) of 10, washed with phosphate-buffered saline (PBS), and stored at −80°C for Western blot analysis. The cells were disrupted by five cycles of vortexing in the presence of glass beads (0.45- to 0.6-mm diameter; Sigma). The equivalent of 32 μg (dry weight) of cells was mixed with 10 μl of sample buffer (23), boiled for 3 min, loaded on a 12% polyacrylamide–sodium dodecyl sulfate (SDS) gel (16), and blotted onto an Immobilon membrane (Millipore). The blots were blocked with 3% skim milk powder in PBS for 15 min. Cutinase antibody (30) was diluted 1,000-fold in 0.3% skim milk powder, and the blot was incubated for 45 min. Subsequently, the blot was incubated with goat anti-rabbit immunoglobulin G coupled to horseradish peroxidase (Jackson Immunoresearch), which was diluted 10,000-fold in the same solution and was also incubated for 45 min. Between the two antibody incubations, the blots were washed with 0.3% skim milk powder in PBS and H2O separately (twice, 10 min each). The resulting immune complexes were detected by chemiluminescence (NEN Du Pont). The results were quantified with a personal densitometer (Molecular Dynamics).
Coimmunoprecipitation.
From continuous cultures, cells were diluted to an OD600 of 10 with radioimmunoprecipitation assay (RIPA) buffer (1% SDS, 20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.5% Triton X-100, 1 mM EDTA), centrifuged for 30 s at 12,000 × g, and resuspended in 1 ml of RIPA buffer. The cells were disrupted by five cycles of vortexing with glass beads. Samples (100 μl) of cell lysates were centrifuged at 14,000 × g in an Eppendorf table centrifuge for 1 min. The supernatant was separated from the pellet, which was resuspended in 100 μl of RIPA buffer. These fractions were precleared with 25 μl of protein A-Sepharose CL4B (Pharmacia) (0.07 g in 0.5 ml of RIPA buffer) for 1 h at 4°C and centrifuged for 5 s at 1,000 × g, and the precleared supernatant was incubated with cutinase antibody (5 μl) for 2 h at 4°C. Protein A-Sepharose (25 μl) was added, and the mixture was incubated for 1 h at 4°C. The suspension was centrifuged for 5 s at 1,000 × g, and the pellets were washed twice with wash buffer (20 mM Tris-HCl [pH 7.4], 0.15 mM NaCl, 0.1% Triton X-100) at the end of the process. Between the washing steps, the suspensions were centrifuged at 1,000 × g for 5 s. The pellets were resuspended in 10 μl of SDS sample buffer, and after being boiled for 5 min, the suspension was centrifuged for 5 s at 1,000 × g and the supernatant was loaded on an SDS-polyacrylamide gel.
Enzyme assays.
One milliliter of culture was centrifuged for 1 min at 14,000 × g in a table centrifuge (Eppendorf), and the supernatant was stored at −80°C for further analysis. Extracellular cutinase was determined by activity assays (30) with p-nitrophenyl butyrate (Sigma) as a substrate.
KMnO4 fixation and transmission electron microscopy.
S. cerevisiae cells were grown in yeast-peptone-glucose to an OD600 of 0.5, harvested, washed twice with distilled H2O, and fixed in 1.5% KMnO4 for 20 min at room temperature. After dehydration in acetone, the samples were infiltrated and embedded with Spurr’s resin. After 24 h of polymerization at 60°C, 80-nm-thick sections were cut with a diamond knife on an ultramicrotome (Reichert-Jung). The sections were mounted on 0.7% pioloform (Polaron Equipment Ltd., Watford, England)-coated, carbon-evaporated one-hole copper grids and dried for 16 h. Subsequently, the sections were viewed on a Philips EM420 electron microscope at an operating voltage of 80 kV.
Immunogold labelling and transmission electron microscopy.
Samples of S. cerevisiae wild-type CY000 and mutant CY028 cutinase-producing cells were taken from continuous cultures with 4 g of galactose per liter and 20 g of glucose per liter in the feed, which results in full induction of the cells. The samples were cryofixed in liquid propane by means of a double-jet freeze device (JFD 030; Baltec) and were freeze-substituted in a mixture of 0.3% uranyl acetate and 0.01% glutaraldehyde in methanol at −90°C for 2 days (11). The samples were subsequently warmed to −45°C at a rate of 5°C/h. Then the specimens were rinsed with methanol and infiltrated with Lowicryl HM20. After 16 h, the specimens embedded with Lowicryl HM20 at −45°C. Polymerization at −45°C for 48 h was carried out in a freeze-substitution apparatus (cryo-substitution auto; Reichert-Jung) with a UV light source (360-nm) attachment (29) followed by a 2-day curing by UV light at room temperature. After ultramicrotomy with an Ultracut E (Reichert-Jung), the 80-nm Lowicryl HM20 sections of the yeast cells were mounted on nickel grids. The nickel grids were coated with 1% Formvar and carbon. To detect cutinase, HM20 sections were incubated with anticutinase polyclonal antibodies (1:250). The antigen-antibody complex was visualized with secondary goat anti-rabbit antibodies (1:10) conjugated with 10-nm-diameter gold particles (Aurion, Wageningen, The Netherlands) (19). To detect both cutinase and BiP, first Lowicryl HM20 sections were incubated with anti-BiP polyclonal antibodies (1:200). The anti-BiP antibody complex was visualized with secondary goat anti-rabbit antibodies (1:10) conjugated with 10-nm-diameter gold particles, and subsequently the Lowicryl HM20 sections were incubated with anticutinase polyclonal antibodies (1:250). The anticutinase antibody complex was visualized with secondary goat anti-rabbit antibodies (1:10) conjugated with 6-nm-diameter gold particles. The labelled ultrathin sections were viewed in a Philips EM420 electron microscope, and micrographs were taken at an acceleration voltage of 80 kV.
Pulse-chase.
Yeast cells were grown in shake flasks to the mid-logarithmic phase in YPG (1% yeast extract, 2% Bacto Peptone, 2% galactose). The cells were resuspended in 1 ml of YNB (0.67% Yeast Nitrogen Base without amino acids) supplemented with the appropriate amino acids (20 mg/liter each) and with 2% galactose as a carbon source, to an OD600 of 10. After incubation for 30 min at 30°C, the cells were labelled for 10 min with 15 μCi of [35S]methionine-cysteine (2.5 μCi/μl) (Amersham). Chase of radioactivity was done by washing the cells in 1 ml of YNB–2% galactose, resuspending the cells in YNB–2% galactose, and adding 200 μl of a nonradioactive mixture of methionine (100 mM) and cysteine (50 mM). At the indicated times, 200 μl of cells was lysed by addition of 120 μl of 1.85 M NaOH–7.5% β-mercaptoethanol and then incubated on ice for 10 min. The labelled proteins were precipitated by an incubation on ice for 10 min in the presence of 120 μl of trichloroacetic acid. The precipitated proteins were pelleted by centrifugation at 13,000 × g for 10 min. The pellet was washed with 500 μl of 1 M Tris and resuspended in 400 μl of 25 mM imidazole–2.5 mM EDTA–2% SDS (pH 6.8). For solubilization, the samples were boiled for 7 min and diluted in 1 ml of INET (50 mM imidazole, 140 mM NaCl, 5 mM EDTA, 1% Triton X-100 [pH 8.0]). After centrifugation at 13,000 × g for 10 min, the supernatant was isolated and diluted in INET to a final volume of 5 ml for immunoprecipitation.
Immunoprecipitation.
A 10-μl volume of rabbit anticutinase serum was added to the labelled proteins, and overnight incubation was carried out at 4°C in an orbital shaker. A 25-μl volume of protein A coupled to Sepharose (protein A-Sepharose CL4B; Pharmacia) in RIPA buffer (0.07 mg in 500 μl) was added, and this mixture was incubated at 4°C for 2 h. The immunocomplexes were washed three times with INET, and 30 μl of sample buffer (23) was added. The samples were boiled for 5 min and centrifuged for 1 min at 13,000 × g. A 12.5% polyacrylamide–SDS gel was loaded with 20 μl of the supernatant. After amplification (Amersham Amplify), the gels were dried and autoradiography was performed.
RESULTS
Specific activity and production of designed cutinase mutants.
Mutants were designed with the aid of molecular dynamics based on the X-ray structure of cutinase (18) in an attempt to increase the affinity of cutinase for lipid substrates. These mutated cutinase genes were expressed in S. cerevisiae. The transformed yeasts were grown in fed-batch cultures under standard conditions as described in Materials and Methods. Table 1 clearly shows that most of the designed mutants have an increased activity caused by the introduction of hydrophobic amino acids around the active site which probably results in a better interaction with the substrate. The mutant cutinases are produced at different levels, ranging from 22 to 183% of wild-type production.
The hampered secretion of CY028 cutinase is a major problem in the large-scale production of this improved cutinase. Previously, it has been reported that production of heterologous proteins in yeast may result in an overflow of the secretory and/or folding pathway, resulting in an impaired production level (21); this relationship is protein dependent. To investigate the relationship between the level of induction of the cutinase gene and the production level, a method by which the induction level of the cutinase gene was regulated independently of other parameters was developed.
Cutinase production as a function of induction level.
S. cerevisiae SU50 transformed with expression vectors for wild-type cutinase or CY028 cutinase was grown in continuous cultures at a dilution rate of 0.07 h−1 under glucose limitation (see Materials and Methods). By addition of different amounts of galactose to the feed, the induction of the cutinase gene under control of the GAL7 promoter was regulated. Stability of the cutinase integrations was confirmed by Southern blotting. By using a constant glucose concentration of 20 g/liter and increasing the galactose concentration from 0 to 7 g/liter, the biomass increased from 8.5 to 11 g/liter. No residual galactose and glucose were observed, apart from when 7 g of galactose per liter was used in the feed (the residual galactose concentration was 1.8 g/liter). This is probably due to a limitation in the galactose uptake rate of SU50. The wild-type and CY028 transformants exhibit identical growth characteristics in terms of yield, CO2 production, and O2 consumption, and under all conditions growth was respiratory (data not shown).
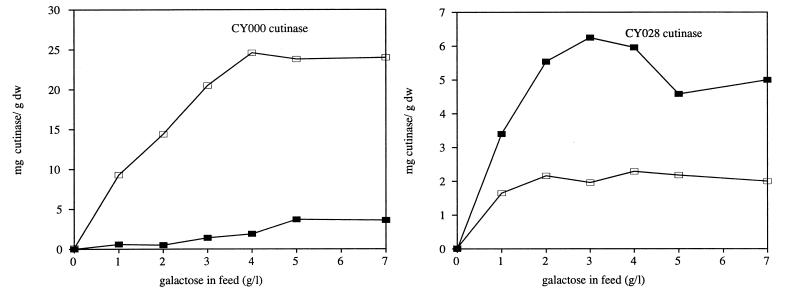
Figure 1 shows the secretion and intracellular levels of wild-type and CY028 cutinases. The secretion of wild-type cutinase increased with the concentration of galactose in the feed. When more than 4 g of galactose per liter was added, the amount of secreted CY000 cutinase did not increase but remained constant at 25 mg/g (dry weight) of cells. The intracellular amount of CY000 cutinase increased slightly with increasing amounts of galactose in the feed. The intracellular level of CY028 cutinase increased also with higher concentrations of galactose in the feed but more rapidly and to a higher level than that of wild-type cutinase. The maximal intracellular levels of wild-type and CY028 cutinases were 4 and 6 mg/g (dry weight), respectively. However, CY028 cutinase increased only to 2.2 mg/g (dry weight) extracellularly.
FIG. 1.
Wild-type and CY028 cutinase production in a continuous culture by S. cerevisiae SU50 with increasing amounts of galactose in the feed. At all points, 20 g of glucose per liter was present in the feed. Intracellular cutinase (▪) was determined by Western blotting and the amount of extracellular cutinase (□) was determined by activity assay, as described in Materials and Methods. dw, dry weight.
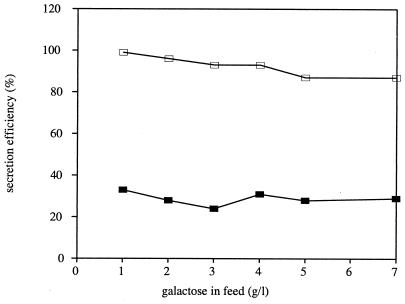
These observations clearly demonstrate that the mutant cutinase CY028 is secreted poorly in S. cerevisiae compared to the wild-type cutinase. The low level of CY028 secretion could be due to either a low rate of CY028 cutinase synthesis or a low efficiency of CY028 cutinase secretion. In the latter case, the secretion efficiency of mutant cutinase is low compared to that of the wild-type cutinase, while in the former case the secretion efficiencies of wild-type and mutant cutinases are comparable. Secretion efficiency is defined as the ratio of extracellular cutinase produced per hour to the total amount of cutinase produced per hour multiplied by 100.
As shown in Fig. 2, the secretion efficiency of CY000 cutinase is 87 to 97%, and CY028 is secreted with an efficiency of 24 to 33%, nearly independently of the expression level. Therefore, it is concluded that the low level of extracellular CY028 cutinase is due to a low efficiency of secretion.
FIG. 2.
Secretion efficiencies of CY000 (□) and CY028 (▪) cutinases produced by S. cerevisiae SU50, calculated from data presented in Fig. 1 as extracellular cutinase produced per hour divided by the total amount of cutinase produced per hour multiplied by 100.
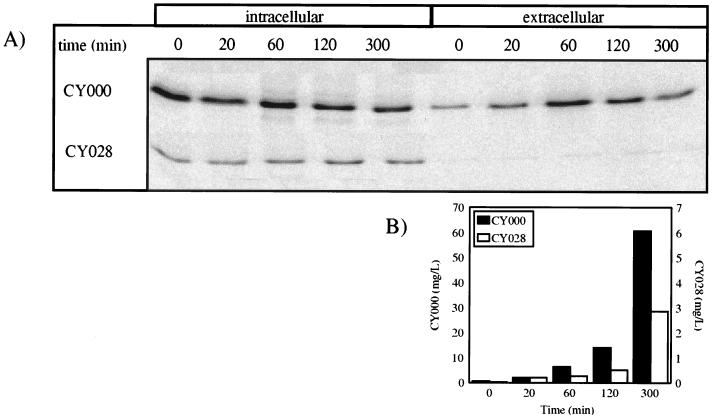
Pulse-chase experiments with CY000 and CY028 cutinases.
In order to investigate if intracellular wild-type or CY028 cutinase is degraded, we performed pulse-chase experiments. As shown in Fig. 3A, the intracellular wild-type cutinase decreased. In contrast, intracellular CY028 cutinase remained constant, at least until 300 min after the pulse. This indicates that most CY028 cutinase is neither degraded nor secreted but is retained inside the cell. The extracellular amount of labelled wild-type cutinase increases in time; however, slight decreases are observed after 120 and 300 min. However, if the extracellular amount of total cutinase is determined with the activity assay as depicted in Fig. 3B, an increase during the chase period is observed. From Fig. 3B, it can be concluded that the decrease in the amount of immunoprecipitated labelled extracellular wild-type cutinase (Fig. 3A) is due to competition between labelled cutinase and unlabelled cutinase which is produced during the chase period.
FIG. 3.
Pulse-chase analysis of CY000 and CY028 cutinases. Cells were labelled with 15 μCi of [35S]methionine-cysteine for 10 min. After the pulse, samples were taken at the times indicated above the lanes. Cutinase was immunoprecipitated and loaded on an SDS-polyacrylamide gel, and autoradiography was performed. (A) Autoradiograph; (B) amount of extracellular cutinase during the chase period, determined by activity assay as described in Materials and Methods.
From the pulse-chase experiments, it can be concluded that the signal sequence is cleaved off efficiently, because even at the shortest chase time, no unprocessed cutinase was detected. Therefore, the translocation at the ER membrane of wild-type and mutant cutinases and the processing of these cutinases are rapid processes. This indicates that the secretion of CY028 cutinase is hampered downstream of the processing of the translocating nascent cutinase polypeptide chain. In order to trace the bottleneck in the secretion of CY028 cutinase, localization studies were performed.
Localization of intracellular CY000 and CY028 cutinases.
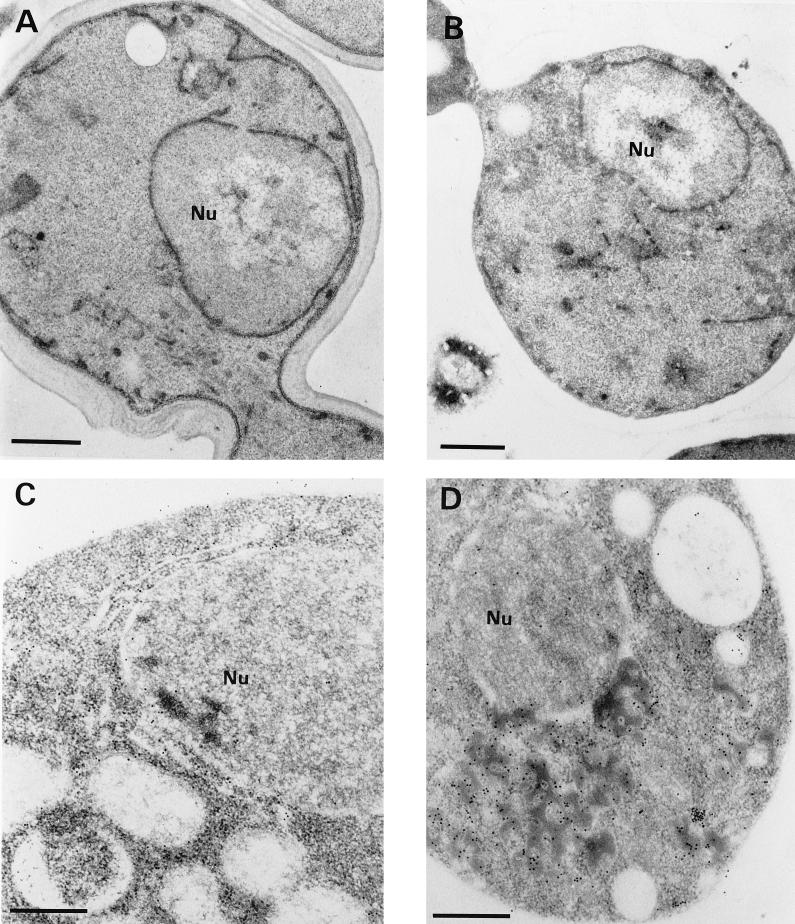
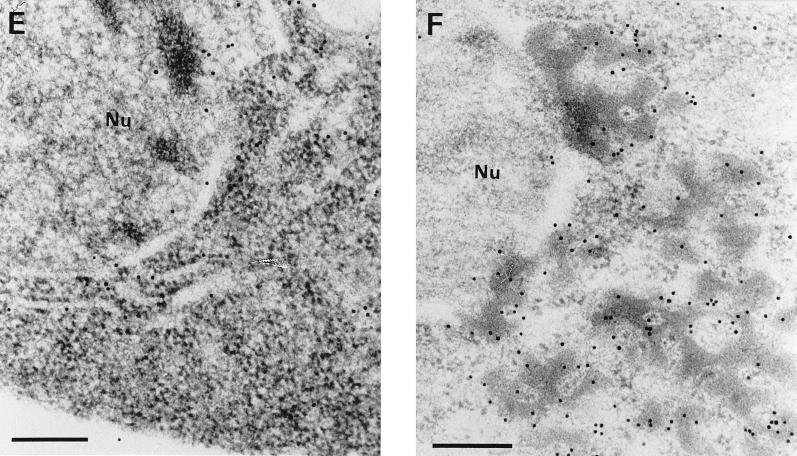
Ultrathin sections of yeast cells grown under noninducing conditions (glucose as the sole carbon source) were chemically fixed with KMnO4. After ultramicrotomy and transmission electron microscopy, the morphologies of CY000- and CY028-transformed yeast cells were found to be similar (Fig. 4A and B). To preserve both the ultrastructure of the yeast cells and antigenicity, we have used a combination of cryofixation, freeze-substitution, and low-temperature embedding. Under inducing conditions, cutinase was localized in both CY000 and mutant CY028 cutinase-producing cells by immunogold labelling as described in Materials and Methods. The ER in CY000 cutinase-producing cells (Fig. 4C and E) showed gold particles and had a morphology comparable to that under noninducing conditions (Fig. 4A). In contrast, CY028 cutinase-producing cells (Fig. 4D and F) showed electron-dense structures 200 to 700 nm in diameter which were labelled with anticutinase. These electron-dense structures, possibly residing in the ER, were found only in mutant CY028-producing cells (Fig. 4D and F).
FIG. 4.
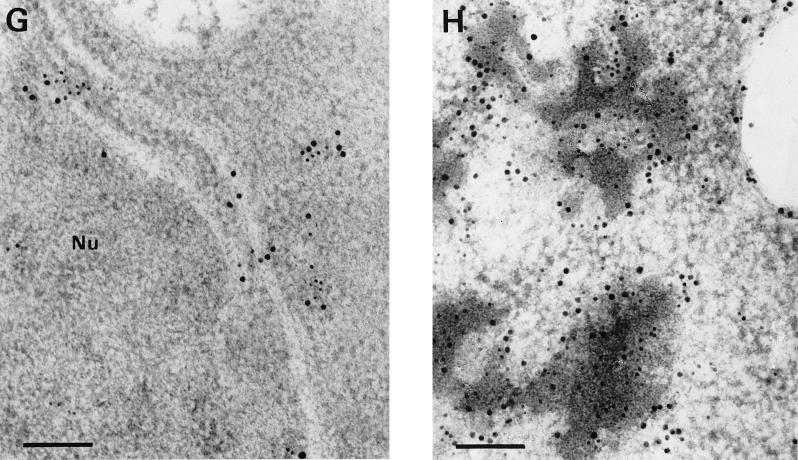
(A and B) KMnO4-fixed S. cerevisiae under noninducing conditions. The CY000-transformed (A) and the CY028-transformed (B) cells show no marked ultrastructural differences. The ERs of both cells exhibit a plate-like morphology. Bars = 1 μm. (C to F) Cryofixed and freeze-substituted S. cerevisiae cells under inducing conditions. After immunolabelling with anticutinase (1:250) and goat anti-rabbit antibodies conjugated with 10-nm-diameter gold (1:10), CY000 cutinase resides in plate-like ERs (C and E). In contrast, CY028 cutinase locates predominantly in electron-dense structures, which results in an abnormal ER morphology (D and F). Bars = 1 μm in panels C and D and 500 nm in panels E and F. (G and H) Double immunolabelling of cutinase and BiP of cryofixed and freeze-substituted S. cerevisiae cells under inducing conditions. The cutinase was located with anticutinase (1:250) and goat anti-rabbit antibodies conjugated with 6-nm-diameter gold (1:10), and BiP was located with anti-BiP (1:200) and goat anti-rabbit antibodies conjugated with 10-nm diameter gold (1:10). The plate-like ER of the CY000-producing cells contains both CY000 cutinase and BiP (G). In CY028-producing cells, the abnormal ER confines electron-dense structures containing CY028 cutinase and BiP (H). Bars = 250 nm. Nu, nucleus.
The results of the pulse-chase experiments suggested that the hampered secretion of CY028 cutinase occurs after the cotranslational translocation over the ER membrane. To support the suggestion that CY028 cutinase is located in the ER, we used the ER-localized chaperone BiP (21) as an ER marker. After double labelling with anticutinase and anti-BiP, the ER in CY000-producing cells exhibited cutinase (6-nm-diameter gold) and BiP (10-nm-diameter gold) (Fig. 4G). In CY028 cutinase-producing cells, more gold-labelled cutinase and BiP were found than in the CY000-producing cells, in electron-dense structures. The ultrastructural data clearly showed that the ER in CY000-producing cells had a normal plate-like morphology without electron-dense structures. In contrast, the CY028-producing cells exhibited pronounced electron-dense structures resulting in totally different ER morphology.
Induction of BiP during CY028 cutinase production.
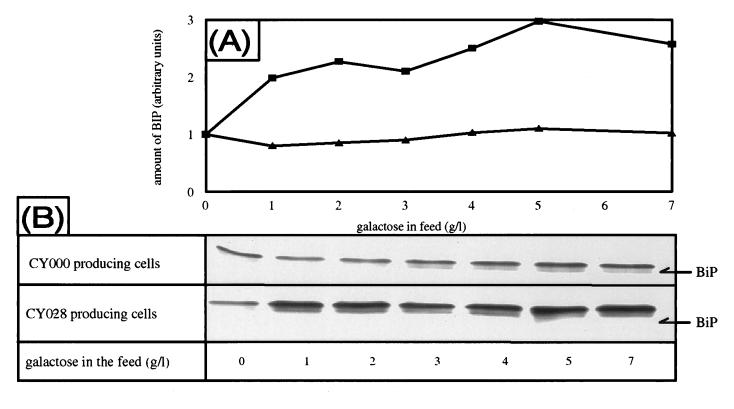
When misfolded proteins accumulate in the ER, the cell reacts by inducing BiP (8, 21, 22, 25). This phenomenon is known as the unfolded-protein response. To determine whether the accumulation of CY028 cutinase in structures which contain BiP also leads to increased BiP production, we performed Western blotting using antibodies against BiP. BiP is increased threefold compared with the amount under noninducing conditions (Fig. 5) in the CY028-expressing strain. No induction of BiP was observed with expression of CY000 cutinase. When the cutinase gene was not induced (0 g of galactose per liter), CY000- and CY028-transformed cells exhibited similar levels of BiP. The level of induction of BiP increases with the level of intracellular CY028 cutinase (Fig. 2 and 6).
FIG. 5.
(A) Quantification of Western blot analysis of BiP levels during increased expression of CY000 (▴) and CY028 (▪) cutinases. Quantification was performed with a personal densitometer from Molecular Dynamics. (B) Western blot analysis of cutinase-producing cells with anti-BiP antibody. Samples were taken from a continuous culture with increasing induction levels and prepared for Western blotting as described in Materials and Methods. All amounts of BiP are normalized against the level with no cutinase induction (0 g of galactose per liter).
FIG. 6.
Immunoprecipitation of BiP followed by detection with anticutinase antibody. Samples were taken from the fermentor at maximal induction (4 g of galactose/liter) and were corrected for the amount of cutinase so that CY000 and CY028 cutinases were present in the same amounts. The anti-BiP-immunoprecipitated complexes were subjected to Western blotting and detected with anticutinase antibody as described in Materials and Methods. The total cell lysates were not immunoprecipitated. A 100-ng sample of cutinase was used as a marker. sup, supernatant; pel, pellet.
Association of the chaperone BiP with the hydrophobic mutant cutinase.
As described previously, BiP exhibits a stable interaction with proteins which are misfolded and retained in the lumen of the ER (13). To determine whether BiP is associated with CY028 cutinase, we immunoprecipitated extracts from cutinase-producing cells with anti-BiP antibody and determined by Western blotting whether cutinase was present in these fractions.
As shown in Fig. 6, CY028 cutinase is present in the anti-BiP-immunoprecipitated insoluble fraction. The anti-BiP-immunoprecipitated soluble fraction of the CY028-expressing cells did not contain CY028 cutinase. The CY000-expressing strain did not contain cutinase in anti-BiP-immunoprecipitated fractions. It is concluded that association of BiP with the hydrophobic mutant cutinase occurred in the pellet fraction (Fig. 6). This association is in agreement with the observation by electron microscopy that CY028 cutinase colocalizes with BiP. As expected from the electron microscopy studies, no stable association between CY000 cutinase and BiP was found.
DISCUSSION
When heterologous proteins are expressed in yeast, the high level of expression due to the use of strong inducible promoters and high gene copy numbers may lead to an overflow of the secretory pathway of the yeast cell, resulting in a decreased secretion efficiency (21). To obtain insight into the relationship between secretion efficiency and the level of expression of the cutinase gene, we used a continuous-culture system. In this system, it is possible to regulate the cutinase flux through the secretion pathway by adjusting the induction level of the cutinase gene while keeping all other parameters constant. We observed a lower secretion efficiency of the hydrophobic mutant cutinase than of the wild-type cutinase which was nearly independent of the induction level.
For various induction levels, the secretion efficiency of CY028 varied between 24 and 33% and the wild-type cutinase secretion efficiency ranged from 87 to 97%. This implies that the nature of the cutinase which has to be transported through the secretion pathway, rather than the amount of cutinase to be transported by the secretion route, determines the secretion efficiency. The pulse-chase experiments revealed that most of the CY028 cutinase is retained irreversibly inside the cell and therefore not secreted. Neither wild-type nor mutant cutinase is degraded inside the cell. The synthesis of CY028 cutinase is impaired compared to that of wild-type cutinase (Fig. 1 and 3), probably because of the intracellular accumulation of CY028 cutinase as aggregates in ER-derived structures. The exact mechanism of this feedback system is not known.
Poor secretion of heterologous proteins in yeast because of retention in the ER has been reported before (2, 26). Various findings revealed that CY028 cutinase is retained in the ER. First, CY028 cutinase colocalizes with BiP. Second, an association between CY028 cutinase and the ER-localized chaperone BiP is observed (Fig. 6). Third, BiP is induced when CY028 cutinase is expressed, a reaction of the cell to the accumulation of proteins in the ER (15, 22).
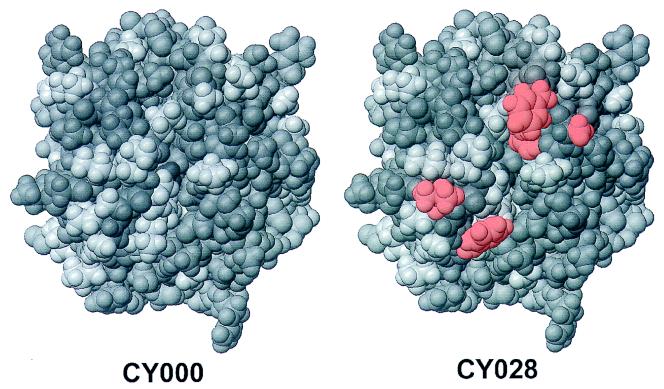
The stable association of BiP with misfolded proteins has been described previously (4, 7, 12). These misfolded proteins are thought to have exposed hydrophobic stretches which would normally be buried in the interior of the protein. Putative binding sites for BiP have been determined with a library of random peptides displayed on bacteriophages. This affinity panning technique revealed specific, 7-residue-long hydrophobic stretches which can form a BiP binding site (3). The mutant tested in this study has two exposed hydrophobic stretches, which results in a more hydrophobic surface than that of wild-type cutinase, as shown in Fig. 7. This could mean that these mutations cause the stable association of CY028 cutinase with BiP. Due to the position of these mutations, this would be possible even if CY028 is folded into the correct conformation. As a result of this stable association with BiP, the CY028 cutinase can reach a high concentration in the ER. It has been determined in vitro that CY028 cutinase aggregates at a lower concentration than CY000 cutinase (data not shown). This high concentration may lead to aggregation, as shown in Fig. 4D, F, and H.
FIG. 7.
Predicted positions of the mutations made in CY028 cutinase, obtained by molecular modelling. The top view of wild-type cutinase (left) and CY028 cutinase (right) is shown, with the introduced hydrophobic amino acids (red) positioned at the periphery around the enzymatic cleft.
In cells expressing CY028 cutinase, the aggregates are 200 to 700 nm in diameter. Normally, the secretion vesicles are small vesicular coated carriers 50 to 80 nm in diameter (1). Presumably, the CY028 cutinase aggregates are too large to be transported through the secretion pathway and are therefore retained in ER-derived structures.
In the recent past, the formation of homologous prolamine bodies in rice, triggered by stable association of prolamine with BiP and resulting in aggregation, has been reported (17). We extrapolate this mechanism to retention of hydrophobic mutated cutinase in yeast. In this model, BiP is thought to screen cutinase for hydrophobic residues, as shown in Fig. 7, and to bind to these residues. Therefore, BiP is part of a control system which prevents proteins with exposed hydrophobic residues from leaving the ER. CY028 is nearly incapable of passing this control system in the ER. This leads to high concentrations of CY028 cutinase in the ER, resulting in large aggregates which are too large to be transported through the secretion pathway.
ACKNOWLEDGMENTS
Chris Visser is gratefully acknowledged for construction of the cutinase mutants. John Chapman is acknowledged for critically reading the manuscript.
REFERENCES
- 1.Aridor M, Balch W E. Principles of selective transport: coat complexes hold the key. Trends Cell Biol. 1996;6:315–320. doi: 10.1016/0962-8924(96)10027-1. [DOI] [PubMed] [Google Scholar]
- 2.Biemans R, Thines D, Rutgers T, DeWilde M, Cabezon T. The large surface protein of hepatitis B virus is retained in the yeast endoplasmic reticulum and provokes its unique enlargement. DNA Cell Biol. 1991;10:191–200. doi: 10.1089/dna.1991.10.191. [DOI] [PubMed] [Google Scholar]
- 3.Blond-Elguindi S, Cwirla S E, Dower W J, Lipshutz R J, Sprang S R, Sambrook J F, Gething M J H. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993;75:717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- 4.Bole D G, Hendershot L, Kearney J F. Posttranslational association of immunoglobulin heavy chain binding protein with nascent heavy chain in nonsecreting and secreting hybridomas. J Cell Biol. 1986;102:1558–1566. doi: 10.1083/jcb.102.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egmond, M. R., H. W. T. M. van der Hijden, W. Musters, H. Peters, and C. T. Verrips. 1994. Patent WO 94/14963.
- 6.Egmond, M. R., H. W. T. M. van der Hijden, W. Musters, H. Peters, and C. T. Verrips. 1994. Patent WO 94/14964.
- 7.Gething M J, McCammon K, Sambrook J. Expression of wild-type and mutant forms of influenza hemagglutinin: the role of folding and intracellular transport. Cell. 1986;46:939–950. doi: 10.1016/0092-8674(86)90076-0. [DOI] [PubMed] [Google Scholar]
- 8.Gething M J, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 9.Haas I G. BiP—a heat shock protein involved in immunoglobulin chain assembly. Curr Top Microbiol Immunol. 1991;167:71–82. doi: 10.1007/978-3-642-75875-1_4. [DOI] [PubMed] [Google Scholar]
- 10.Haas I G, Wabl M. Immunoglobulin heavy chain binding protein. Nature. 1983;306:387–389. doi: 10.1038/306387a0. [DOI] [PubMed] [Google Scholar]
- 11.Humbel B M, Schwartz H. Freeze-substitution for immunochemistry. In: Verkleij A J, Leunissen J L M, editors. Immuno-gold labelling in cell biology. Boca Raton, Fla: CRC Press, Inc.; 1989. pp. 115–134. [Google Scholar]
- 12.Hurtley S M, Bole D G, Hoover L H, Helenius A, Copeland C S. Interactions of misfolded influenza virus hemagglutinin with binding protein (BiP) J Cell Biol. 1989;108:2117–2126. doi: 10.1083/jcb.108.6.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim P S, Bole D, Arvan P. Transient aggregation of nascent thyroglobulin in the endoplasmic reticulum: relationship to the molecular chaperone, BiP. J Cell Biol. 1992;118:541–549. doi: 10.1083/jcb.118.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolattukudy P E. Cutinases from fungi and pollen. In: Borgstrom B, Brockman H, editors. Lipases. Amsterdam, The Netherlands: Elsevier Science; 1984. pp. 471–504. [Google Scholar]
- 15.Kozutsumi Y, Segal M, Normington K, Gething M J, Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose regulated proteins. Nature. 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Wu Y, Zhang D, Gillikin J W, Boston R S, Franceschi V R, Okita T W. Rice prolamine protein body biogenesis: a BiP-mediated process. Science. 1993;262:1054–1056. doi: 10.1126/science.8235623. [DOI] [PubMed] [Google Scholar]
- 18.Martinez C, de Geus P, Lauwereys M, Matthysens G, Cambillau C. Fusarium solani cutinase is a lipolytic enzyme with a catalytic serine accessible to solvent. Nature. 1992;356:615–618. doi: 10.1038/356615a0. [DOI] [PubMed] [Google Scholar]
- 19.Müller W H, van der Krift T P, Knoll G, Smaal E B, Verkleij A J. A preparation method of specimens of the fungus Penicillium chrysogenum for ultrastructural and immuno-electron microscopical studies. J Microsc. 1991;169:29–41. doi: 10.1111/j.1365-2818.1991.tb03189.x. [DOI] [PubMed] [Google Scholar]
- 20.Normington K, Kohno K, Kozutsumi Y, Gething M J, Sambrook J. S. cerevisiae encodes an essential protein homologous in sequence and function to mammalian BiP. Cell. 1989;57:1223–1236. doi: 10.1016/0092-8674(89)90059-7. [DOI] [PubMed] [Google Scholar]
- 21.Parekh R, Forrester K, Wittrup D. Multicopy overexpression of bovine pancreatic trypsin inhibitor saturates the protein folding and secretory capacity of Saccharomyces cerevisiae. Protein Expr Purif. 1995;6:537–545. doi: 10.1006/prep.1995.1071. [DOI] [PubMed] [Google Scholar]
- 22.Rose M D, Misra L M, Vogel J P. Kar2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Sawa T, Imamura T, Haruta T, Sasaoka T, Ishiki M, Takata Y, Takada Y, Morioka H, Ishihara H, Usui I, Kobayashi M. Hsp70 family molecular chaperones and mutant insulin receptor: differential binding specificities of BiP and Hsp70/Hsc70 determine accumulation or degradation of insulin receptor. Biochem Biophys Res Commun. 1996;218:449–453. doi: 10.1006/bbrc.1996.0080. [DOI] [PubMed] [Google Scholar]
- 25.Shamu C E, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signalling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- 26.Shuster J R. Gene expression in yeast: protein secretion. Curr Opin Biotechnol. 1991;2:685–690. doi: 10.1016/0958-1669(91)90035-4. [DOI] [PubMed] [Google Scholar]
- 27.Sierkstra L N, Verbakel J M A, Verrips C T. Analysis of transcription and translation of glycolytic enzymes in glucose limited continuous cultures of S. cerevisiae. J Gen Microbiol. 1992;138:2559–2566. doi: 10.1099/00221287-138-12-2559. [DOI] [PubMed] [Google Scholar]
- 28.Silljé H H W, ter Schure E G, Verkleij A J, Boonstra J, Verrips C T. The Cdc25 protein of Saccharomyces cerevisiae is required for normal glucose transport. Microbiology. 1996;142:1765–1773. doi: 10.1099/13500872-142-7-1765. [DOI] [PubMed] [Google Scholar]
- 29.Sitte H, Neumann K, Edelmann L. Cryofixation and cryosubstitution for routine work in transmission electron microscopy. In: Müller M, Becker R P, Boyde A, Wolosewick J J, editors. Science of biological specimen preparation. Chicago, Ill: SEM Inc., AMF O’Hare; 1985. pp. 103–118. [Google Scholar]
- 30.van Gemeren I A, Musters W, van den Hondel C A M J J, Verrips C T. Construction and heterologous expression of a synthetic copy of the cutinase cDNA from Fusarium solani pisi. J Biotechnol. 1995;40:155–162. doi: 10.1016/0168-1656(95)00041-n. [DOI] [PubMed] [Google Scholar]
- 31.Verbakel J M A. Heterologous gene expression in the yeast Saccharomyces cerevisiae. Ph.D. thesis. Utrecht, The Netherlands: Utrecht University; 1991. [Google Scholar]
- 32.Verger R, de Haas G H. Interfacial enzyme kinetics of lipolysis. Annu Rev Biophys Bioeng. 1979;5:77–117. doi: 10.1146/annurev.bb.05.060176.000453. [DOI] [PubMed] [Google Scholar]