Abstract
The term microsporidia is used to describe several species of opportunistic protozoan parasites. Encephalitozoon intestinalis and Enterocytozoon bieneusi have been found in stools of more than 40% of AIDS patients with diarrhea. Diagnosis of infection with these small protozoans has been difficult, and until recently their occurrence has not been well documented. Formalin is widely used to preserve clinical specimens, but due to the nature of the fixation process, subsequent analysis, especially analysis by the PCR, is difficult. This study evaluated methods used to prepare formalin-fixed fecal specimens for PCR amplification of microsporidial DNA. Two methods were devised to allow PCR detection and subsequent identification of microsporidia in formalin-fixed fecal specimens to the species level. One method involved immunomagnetic separation to concentrate microsporidial spores from fecal specimens. In the second method Chelex resin (Bio-Rad, Hercules, Calif.) was used to remove inhibitory substances, followed by a DNA concentration step. Both methods resulted in reproducible, confirmed detection of microsporidia in formalinized fecal specimens and subsequent species determination by PCR sequencing. The detection sensitivity was two in vitro culture-derived spores (Encephalitozoon intestinalis) for the direct PCR. The reproducible detection sensitivity for DNA amplification from formalin-fixed fecal samples was 200 spores for either the Chelex method or the immunomagnetic bead separation method. Thus, we developed two methods for rapid, inexpensive detection of microsporidial spores in formalin-fixed fecal specimens.
The term microsporidia is a nontaxonomic name used to describe protozoan parasites belonging to the phylum Microspora. Members of several genera of microsporidia (Encephalitozoon cuniculi, Enterocytozoon bieneusi, Encephalitozoon intestinalis, and Encephalitozoon hellem) have been reported to be agents of disease in humans, especially immunocompromised individuals. Recent studies have shown that more than 40% of AIDS patients with associated diarrhea shed microsporidia (10). Enterocytozoon bieneusi and Encephalitozoon intestinalis in particular have been shown to be responsible for intestinal disease manifestations in AIDS patients. Until recently, however, inefficient detection methods have prevented adequate documentation of the occurrence of microsporidia, especially in immunocompetent individuals (7).
Enterocytozoon bieneusi and Encephalitozoon intestinalis cells are approximately 0.5 by 1.5 and 0.5 by 3 to 4 μm, respectively, and are difficult to detect and differentiate in fecal samples by using common protozoan staining methods, such as trichrome staining (3). In addition, transmission electron microscopy is needed to confirm the presence of presumptively identified microsporidia and determine to what species they belong (7). Due to these difficulties there is a need for specific and sensitive molecular methods to detect and identify the etiological agents of microsporidial infections.
The PCR has proven to be useful for detection and identification of microsporidia (2, 7, 9, 11). However, it is very difficult to amplify low-copy-number DNA, such as that in microsporidial spores, especially from formalin-fixed fecal specimens. This is because of the interfering nature of the fixative, which perfuses the microsporidial spores and reacts with the DNA. Currently used methods for the extraction and purification of target DNA from both formalin-fixed and non-formalin-fixed fecal specimens are efficient but extremely time-consuming. These methods usually involve the use of chemical treatment (9) and mechanical disruption, which often requires specialized equipment, such as bead beaters (2) or sonication devices (11). In some cases up to 4 days is required for DNA isolation procedures (8).
This study describes the use of a PCR in conjunction with two simple and inexpensive DNA extraction methods for PCR confirmation and subsequent identification of the organisms in presumptive microsporidial infections to the species level. The first extraction method involved the use of immunomagnetic beads for selective extraction from fecal specimens of microsporidial spores of organisms belonging to certain genera, followed by DNA extraction and PCR detection. The second method involved mass extraction of community DNA from fecal specimens, followed by Chelex purification and PCR analysis.
A total of 41 fecal specimens (27 animal specimens and 14 human specimens) were obtained as previously described (5). The samples contained Encephalitozoon sp. spores presumptively identified by a monoclonal antibody (MAb)-based indirect fluorescent-antibody assay (IFA) as previously reported (6). Three fecal samples were also obtained which were negative for microsporidial spores as determined by the MAb-based IFA, and these samples were used as negative controls and in the detection sensitivity studies (2). Fecal specimens were fixed with 10% formalin in phosphate-buffered saline (PBS) (10 mM sodium phosphate, 0.1 M NaCl; pH 7.2) and then purified by ethyl acetate sedimentation and stored at 4°C (5).
Isolates of Encephalitozoon intestinalis, Encephalitozoon cuniculi, and Encephalitozoon hellem were obtained from the American Type Culture Collection, Rockville, Md. (strains ATCC 50506, ATCC 50602, and ATCC 50604, respectively). Isolates of Vairimorpha ephestiae and Nosema apis were originally obtained from Oleg Ditrich and Tomas Tonka at the Institute of Entomology of the Czech Academy of Sciences, Czech Republic. Enterocytozoon bieneusi spores were obtained from Charles P. Gerba, University of Arizona.
Encephalitozoon intestinalis spore stock preparations were used in all positive-control spore and DNA extractions and all detection sensitivity and positive-control reactions. Dilutions made directly from American Type Culture Collection stock preparations were formalin fixed and used in determining detection sensitivities. These stock dilutions were also used in all of the positive-control PCRs which were performed. The spores in Encephalitozoon intestinalis spore stock preparations were initially enumerated in triplicate by using a hemocytometer at a magnification of ×400. The spores in dilutions were enumerated in duplicate by fluorescent staining with BacLight fluorogenic stain (Molecular Probes, Eugene, Oreg.). After staining, spores were transferred onto 13-mm-diameter, 0.2-μm-pore-size polycarbonate membrane filters (Nuclepore, Pleasanton, Calif.), followed by fluorescence microscopy. Each limit-of-detection sensitivity study was performed in triplicate with each of the three control fecal samples which were used. All of the concentrations given below are the means from these determinations and are reported as the nearest whole numbers (e.g., a dilution which provided a mean of 2.05 × 102 spores per volume used for inoculation of controls is given below as 200 spores).
Sheep anti-mouse immunoglobulin G-conjugated magnetic immunobeads (diameter, 1 μm) in sterile PBS were obtained from Immunotech, Marseille, France. Beads were prepared for immunoseparation as follows. After gentle vortexing, the beads were diluted 1:20 in PBS–0.4% bovine serum albumin to a final volume of 1 ml. The beads were then separated from the buffer solution by using a microcentrifuge tube magnetic separation device (Dynal A.S., Oslo, Norway). The supernatant was removed, and the beads again were suspended in the initial volume of buffer. The murine anti-Encephalitozoon sp. MAb immunoglobulin G2b designated 3B6 (5) was then added to the solution at a concentration of 6 μg of antibody per mg of beads. The mixture was mixed gently and incubated for 30 min. After this conjugation step, the beads were washed twice as described above and subsequently resuspended in the original volume of PBS containing 1.0% bovine serum albumin. This mixture was used as the working stock.
Immunomagnetic separation of spores from fecal samples was accomplished as follows. Fifty microliters (pelleted volume) of fecal material was suspended in 1 ml of 1× PCR buffer B (10 mM Tris-HCl [pH 8.3], 1.5 mM MgCl2, 50 mM KCl). Magnetic beads (50 μl of the working stock) were then added, and the preparation was mixed gently on a shaking incubator for 20 min. By using the microcentrifuge magnetic separator, the beads were separated from the suspension, and the supernatant was carefully removed. The immunobeads with the attached spores were then resuspended in 60 μl of 1× PCR buffer B and heated at 98°C for 10 min. The suspension was subsequently centrifuged at 14,000 rpm (16,000 relative centrifugal force) for 5 min in an Eppendorf model 6415 C microcentrifuge (Brinkmann Instruments, Westbury, N.Y.). Fifty microliters of the supernatant was ultimately used in the PCR, taking into account the resultant buffer concentrations.
A Chelex-based method for DNA extraction was also evaluated. The procedure was performed as follows. Fifty microliters of a pelleted fecal specimen was resuspended in 100 μl of PCR buffer B and heated in a water bath at 98°C for 10 min. The sample was then centrifuged for 10 min, and the supernatant was removed and placed in a sterile microcentrifuge tube. Chelex 100 (100 μl of a 50% suspension in PCR buffer B) was added to the lysate. This suspension was then vortexed and centrifuged, and the supernatant was subsequently used for a PCR. A DNA concentration step was also evaluated by using Microcon 50 microconcentrators (Amicon, Inc., Beverly, Mass.). This concentration step was performed by using the manufacturer’s protocols.
The PCR primers used in this analysis have been described previously (8). The forward primer (5′-CAC CAG GTT GAT TCT GCC TGA C-3′) and the reverse primer (5′-CCT CTC CGG AAC CAA ACC CTG-3′) amplify the small-subunit ribosomal DNAs, which results in amplicon sizes of 250 bp for Enterocytozoon bieneusi, 268 bp for Encephalitozoon cuniculi, 270 bp for Encephalitozoon intestinalis, and 279 bp for Encephalitozoon hellem (Fig. 1). These primers have also been used successfully to amplify and differentiate Encephalitozoon intestinalis and Enterocytozoon bieneusi DNAs in stools taken from infected patients. However, the methods described previously required up to 4 days before results were obtained (8). In addition, these primers were determined to have a high secondary structure by using computer software and, as determined under nonstringent conditions, to form high concentrations of primer dimers. Therefore, PCR conditions were optimized for use with fecal samples by using a Stratagene Opti-prime PCR optimization kit and a Stratagene Robocycler with a gradient temperature annealing block (Stratagene, La Jolla, Calif.). The optimized conditions were as follows: PCR buffer A containing 10 mM Tris-HCl (pH 8.8), 1.5 mM MgCl2, and 7.5 mM KCl and Taq Gold (Perkin-Elmer Corp., Norwalk, Conn.)-induced hot-start cycling conditions consisting of 5 min of denaturation at 98°C, followed by 40 cycles of denaturation at 94°C for 45 s, annealing at 58°C for 20 s, and extension at 72°C for 40 s. A final extension step consisting of 5 min at 72°C was also included. Other than these parameters, the conditions described by Fedorko et al. (8) were used. This optimization procedure resulted in a stock dilution detection sensitivity of two spores (Encephalitozoon intestinalis) based on the median of three separate direct microscopic enumerations. The optimization also considerably decreased the formation of primer dimers.
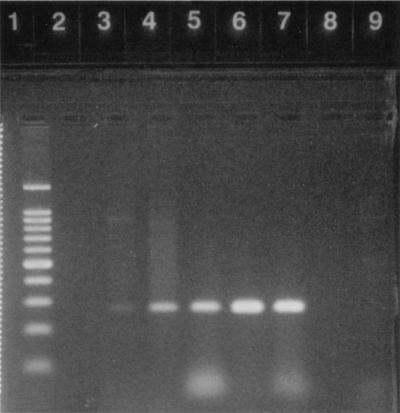
FIG. 1.
Results of the magnetic bead and Chelex methods for extraction of microsporidial spores and DNA, respectively, followed by PCR amplification. Lanes 1 through 3, preparations spiked with 200 spores (Encephalitozoon intestinalis), using the magnetic bead method; lanes 4 through 6, preparations spiked with 2,000 spores (Encephalitozoon intestinalis), using the Chelex method without concentration; lane 7, 20 spores (Encephalitozoon intestinalis) after two rounds of PCR, using Chelex with Centricon 50 concentration; lane 8, 200 spores (Encephalitozoon intestinalis), using Chelex with Centricon 50 concentration; lane 9, 2,000 spores (Encephalitozoon intestinalis), using Chelex with Centricon 50 concentration; lane 10, 100-bp ladder; lane 11, Enterocytozoon bieneusi; lane 12, Encephalitozoon cuniculi; lane 13, Encephalitozoon hellem; lane 14, Encephalitozoon intestinalis.
Two methods were used for species identification. The first method involved the use of PCR restriction fragment length polymorphism analysis, while the second involved the use of PCR sequencing, followed by a computer database BLAST A comparison. Restriction analysis was used to differentiate Encephalitozoon intestinalis from Enterocytozoon bieneusi, while sequencing was used to confirm these results and to differentiate Encephalitozoon intestinalis from other known microsporidia which have similar restriction patterns.
Restriction digestion was performed with all PCR-positive samples. Restriction analysis of the PCR products was performed by using restriction enzyme PstI and 10× universal buffer (Stratagene). The conditions used were as follows. Eighteen microliters of PCR products was added to a sterile 0.2-μl thin-wall PCR tube (Perkin-Elmer), and 2 μl of 10× universal restriction buffer was added, followed by 2 μl (40 U) of restriction enzyme. The restriction reaction was carried out at 37°C in a model 9600 thermocycler (Perkin-Elmer) for 1 h, after which 5 μl of 0.5 M EDTA was added to chelate the Mg2+ and halt the reaction. The products were electrophoresed on a 4% agarose gel (Metaphor; FMC, Rockland, Maine). PstI restriction of the amplicon easily differentiated Enterocytozoon bieneusi from Encephalitozoon intestinalis, Encephalitozoon hellem, and Encephalitozoon cuniculi (8).
For sequence analysis, five of the human fecal samples which were positive for microsporidia as determined by the PCR were randomly chosen for sequencing along with controls for Encephalitozoon intestinalis, Enterocytozoon bieneusi, Encephalitozoon hellem, and Encephalitozoon cuniculi. The PCR products were purified by using a QIAquick PCR purification kit (Qiagen, Santa Clarita, Calif.) and were resuspended in sterile H2O. The forward PCR primer was used for the sequencing, which was performed in the University of Arizona Laboratory of Molecular Systematics and Evolution DNA Sequencing Facility. Sequences obtained from the samples were then entered into the National Center for Biotechnology Information’s World Wide Web site, and an advanced BLAST A search was done to identify homologous sequences. BLAST performs fast database searching combined with rigorous statistics for judging the significance of matches. The BLAST algorithm has been described by Altschul et al. (1). Results are provided in order of highest homology with a score given based on the statistical homology analysis. Essentially, the higher the score, the higher the homology. Statistical P values are also provided as an indication of the probability of error in the comparison.
Results from our study indicate that the extraction methods used allowed subsequent PCR detection of microsporidia in formalin-fixed fecal specimens. In initial pure-culture studies, amplification of microsporidial spore DNA from in vitro cultures was accomplished following a 94°C heat treatment for 10 min. Dilutions (10-fold) containing mean spore numbers ranging from 2 to 20,000 were added directly to the PCR mixtures and were amplified without any additional DNA extraction procedures (Fig. 2). This indicates that no harsh chemical treatment was necessary for the release of microsporidial DNA. The detection sensitivity was shown to be two spores based on the median of three separate direct microscopic enumerations.
FIG. 2.
Limit-of-detection sensitivity for pure-culture PCR amplification of microsporidial (Encephalitozoon intestinalis) spores. Lane 1, 100-bp DNA ladder; lane 2, empty lane; lane 3, 2 spores; lane 4, 20 spores; lane 5, 200 spores; lane 6, 2,000 spores; lane 7, 20,000 spores; lane 8, empty lane; lane 9, negative control.
Subsequent studies were performed by using representatives of two nonhuman (insect) microsporidial genera (the genera Vairimorpha and Nosema). Members of these two genera cross-react with the MAb (MAb 3B6) (6) which was used for magnetic bead spore selection in this study. Thus, if members of these genera of microsporidia were present in a sample, the immunomagnetic beads could potentially extract them. This proved to be problematic if the DNAs were amplified by the PCR. However, the DNAs of the isolates used were not amplified by the PCR primers (data not shown). This indicates that the magnetic bead method described here theoretically extracts these isolates but the PCR does not amplify their DNAs. Thus, the PCR magnetic bead method permits highly specific extraction of the spores of members of certain genera of microsporidia, followed by enzymatic amplification of only the human target microsporidial DNA.
Forty-one human and animal stool samples which had previously been presumptively identified by the MAb-based IFA as positive for Encephalitozoon spp. were used to assess the efficiency of the extraction methods. Each of the fecal samples was split into two equal portions. The first portion was used for DNA or spore extraction with each of the two methods (Chelex and magnetic beads), followed by PCR amplification. The second portion was spiked with spores and used as a positive control to ensure that false negatives did not occur during processing. The results showed that more than 50% of the IFA-positive clinical samples were positive as determined by the PCR. This illustrates the ability of the two methods to detect spores in formalinized fecal material. In addition, since the MAb binds fresh and formalin-fixed spores, both fresh and fixed spores may be used with this method. Thus, immunomagnetic separation and PCR can be used to detect naturally occurring organisms in formalinized samples.
Fecal samples not containing microsporidial spores were also used to assess the overall sensitivity of detection of both DNA extraction methods. These samples were prepared as described above, split into 50-μl (pelleted volume) portions, and spiked with various concentrations of formalin-fixed Encephalitozoon intestinalis spores. The mean counts in 50 μl of pelleted fecal sample necessary for 100% positive PCR results were determined to be 2,000 spores for the Chelex method and 200 spores for the magnetic bead method (Fig. 1). We estimate that the spore burden of an infected individual is between 103 and 107 spores per 100 μl of feces based on IFA analysis, and thus this method is a sensitive method for detection. Unfortunately, because of the recent association of microsporidia with disease in humans, little definitive information concerning the actual spore loads in feces is available.
The magnetic bead method was also used for Enterocytozoon bieneusi, Encephalitozoon cuniculi, Encephalitozoon hellem, and Encephalitozoon intestinalis by adding spores of each species to negative fecal samples. This experiment showed that the four human-infecting species could be efficiently extracted and amplified from fecal material (Fig. 1). This suggests that the magnetic immunobead method has potential for application to a wide range of bodily fluids and in particular should aid in identification of disseminated infections with Encephalitozoon cuniculi and Encephalitozoon hellem.
In species determinations in which restriction digestion was used we were able to differentiate between Encephalitozoon intestinalis and Enterocytozoon bieneusi, but we were not able to differentiate Encephalitozoon intestinalis from Encephalitozoon hellem and Encephalitozoon cuniculi without electrophoresing products on dense agarose (5%) gels or polyacrylamide (12%) gels. Sequencing and BLAST analysis provided positive identification of unknown and control samples. The cost per sample ($16.00) for sequencing was comparable to the cost of high-percentage gel electrophoresis. For all of the positive controls, the sequencing and database comparisons identified the correct species. In the five fecal samples presumptively identified by the IFA, sequencing and BLAST analysis identified all of the microsporidia as Encephalitozoon (Septata) intestinalis (the name of Septata intestinalis was recently changed to Encephalitozoon intestinalis) with an extremely low probability of error (Table 1).
TABLE 1.
PCR product sequence-BLAST A results for unknown presumptive human fecal and control microsporidia
| Sample | Presence of microsporidia as determined by:
|
BLAST A resultsa
|
Probability of errorb | ||
|---|---|---|---|---|---|
| IFA | PCR | Species | Nucleotide sequence accession no. | ||
| Unknown 42 | + | + | Septata intestinalis | U39297 | 1.2 × 10−71 |
| Unknown 60 | + | + | Septata intestinalis | U39297 | 3.8 × 10−72 |
| Unknown 64 | + | + | Septata intestinalis | U39297 | 1.8 × 10−71 |
| Unknown 70 | + | + | Septata intestinalis | U39297 | 3.9 × 10−75 |
| Unknown 208 | + | + | Septata intestinalis | U39297 | 1.8 × 10−73 |
| Enterocytozoon bieneusi positive control | NDc | + | Enterocytozoon bieneusi | L07123 | 1.3 × 10−73 |
| Encephalitozoon intestinalis positive control | + | + | Septata intestinalis | U39297 | 3.4 × 10−73 |
| Encephalitozoon cuniculi positive control | + | + | Encephalitozoon cuniculi | L39107 | 1.3 × 10−69 |
| Encephalitozoon hellem positive control | + | + | Encephalitozoon hellem | L19070 | 1.6 × 10−77 |
The organism whose sequence was identified by the BLAST A method as having the highest homology and the nucleotide sequence accession number of this sequence.
The probability (on a scale of 0 to 1) that the sequence identified occurred by chance.
ND, not determined.
The cost per sample and the time involved for each method are outlined below. For the magnetic bead method, the cost per sample, considering the prices of commercially available antibody for Giardia spp. (as the antibody for microsporidia was not yet on the market), the sheep anti-mouse antibody-conjugated magnetic beads, and the PCR, was $3.50 (when the cost of the PCR was estimated to be $2.00). The time required, including PCR amplification (with the Perkin-Elmer model 9600 thermocycler) as described above, was around 3 h. For the Chelex extraction method, the cost per sample, considering the Chelex and PCR costs, was around $2.00 and the time required was 2.5 h. With subsequent concentration by using Centricon 50, the cost was just under $4.50 and the time required was 3 h. However, with this concentration method the sensitivity of the Chelex method was improved to a mean of 200 spores (Fig. 1).
Overall, the magnetic bead method provided an inexpensive, reproducible, specific, and sensitive method for the preparation of formalin-fixed fecal samples for PCR. The Chelex method combined with subsequent Centricon 50 concentration had similar attributes. However, without DNA concentration the extraction sensitivity was reduced 10-fold. PCR sequencing followed by a computer BLAST analysis showed that it was possible to identify the species of microsporidia. In addition, the magnetic bead extraction procedure has also been shown to have potential applications for isolation of microsporidia from environmental water (4), as well as detection of low spore numbers in cell cultures (data not shown). Both of these applications are currently being evaluated in our labs.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.da Silva A J, Schwartz D A, Visvesvara G S, de Moura H, Slemenda S B, Pieniazek N J. Sensitive PCR diagnosis of infections by Enterocytozoon bieneusi (microsporidia) using primers based on the region coding for small-subunit rRNA. J Clin Microbiol. 1996;34:986–987. doi: 10.1128/jcm.34.4.986-987.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Didier E S, Rogers L B, Brush A D, Wong S, Traina-Dorge V, Bertucci D. Diagnosis of disseminated microsporidian Encephalitozoon hellem infection by PCR-Southern analysis and successful treatment with albendazole and fumagillin. J Clin Microbiol. 1996;34:947–952. doi: 10.1128/jcm.34.4.947-952.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dowd, S. E. 1997. Personal communication.
- 5.Enriquez, F. J., D. Taren, M. Muramoto, J. D. Palting, and A. Cruz-Lopez. Prevalence of fecal microsporidial Encephalitozoon spp. spores in children and adults and potential risk factors involved. Submitted for publication.
- 6.Enriquez F J, Ditrich O, Palting J D, Smith K. Simple diagnosis of Encephalitozoon sp. microsporidial infections by using a panspecific antiexospore monoclonal antibody. J Clin Microbiol. 1997;35:724–729. doi: 10.1128/jcm.35.3.724-729.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fedorko D P, Hijazi Y M. Application of molecular techniques to the diagnosis of microsporidial infection. Emerg Infect Dis. 1996;2:183–191. doi: 10.3201/eid0203.960304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fedorko D P, Nelson N A, Cartwright C P. Identification of microsporidia in stool specimens by using PCR and restriction endonucleases. J Clin Microbiol. 1995;33:1739–1741. doi: 10.1128/jcm.33.7.1739-1741.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katzwinkel-Wladarsch S, Lieb M, Heise W, Löscher T, Rinder H. Direct amplification and species determination of microsporidian DNA from stool specimens. Trop Med Int Health. 1996;1:373–378. doi: 10.1046/j.1365-3156.1996.d01-51.x. [DOI] [PubMed] [Google Scholar]
- 10.Kotler D P. Gastrointestinal manifestations of immunodeficiency infection. Adv Intern Med. 1995;40:197–241. [PubMed] [Google Scholar]
- 11.Ombrouck C, Ciceron L, Biligue S, Brown S, Marechal P, vanGool T, Datry A, Danis M, Desportes-Livage I. Specific PCR assay for direct detection of intestinal microsporidia Enterocytozoon bieneusi and Encephalitozoon intestinalis in fecal specimens from human immunodeficiency virus-infected patients. J Clin Microbiol. 1997;35:652–655. doi: 10.1128/jcm.35.3.652-655.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]