Abstract
Nephrolithiasis has a high recurrence rate and imposes a heavy burden on patients and society. Therefore, there is an urgent need to explore new effective targets for nephrolithiasis prevention and treatment. Protein quantitative trait loci (pQTL) data of plasma proteins and plasma protein–protein ratios were used as plasma protein related targets (PPRTs), and genome-wide association studies (GWASs) of nephrolithiasis were used as outcomes. Bidirectional Mendelian randomization analyses were then conducted. The mediating factors between PPRTs and nephrolithiasis were identified. After colocalization analyses, the safety of targeted PPRTs for nephrolithiasis were evaluated. Subsequently, multi-omics data were used to further validate the effects of candidate PPRTs on nephrolithiasis. Finally, molecular docking operations were performed. Six PPRTs were confirmed to be causally associated with nephrolithiasis. The glomerular filtration rate (GFR) was identified as a mediating factor between five PPRTs and nephrolithiasis. There were strong evidences of colocalization of three PPRTs. After safety assessments and multi-omics analyses, BTN3A2 was considered as a potential therapeutic target. Molecular docking analyses indicated that digitoxigenin had the strongest binding ability with BTN3A2. These comprehensive analyses suggested causal relationships between PPRTs and nephrolithiasis, and BTN3A2 might be an advantageous drug target for nephrolithiasis.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-025-17664-0.
Keywords: Nephrolithiasis, Plasma proteins, Mendelian randomization, Target
Subject terms: Urology, Data mining
Introduction
Nephrolithiasis is a common disease in urology, and its overall prevalence is approximately 14.8%. In recent years, the incidence of nephrolithiasis has increased significantly, and 25% of nephrolithiasis patients require hospitalization. Moreover, the recurrence rate within 10 years is 50%1,2. Urinary tract obstruction caused by nephrolithiasis can cause urinary tract or kidney infection, and in severe cases, septic shock can occur. Urinary tract obstruction can also cause hydronephrosis. Long-term chronic hydronephrosis can damage kidney function and even lead to renal failure. Additionally, nephrolithiasis can cause intense low back pain (renal colic) when kidney stones enter the ureter. The high recurrence rate of nephrolithiasis places a heavy burden on patients and has become a major public health problem3. Age, sex, obesity, hypertension, and diabetes are considered important risk factors for nephrolithiasis, but the current understanding of the pathophysiological processes of nephrolithiasis is incomplete, which hinders effective prevention4–7. Therefore, the identification of risk factors related to nephrolithiasis is urgently needed and can provide valuable references for disease prevention and treatment strategies, helping to reduce medical and economic burdens.
Some sites and regions of proteins can specifically bind small-molecule drugs, allowing the development of more accurate drugs8. The development of proteomics technology enables the detection of single-nucleotide polymorphisms (SNPs) in genome-wide association studies (GWASs), also known as protein quantitative trait loci (pQTL), defined as genetic variation sites associated with protein expression levels. These variations affect the content of specific proteins in plasma or tissues by regulating processes such as gene transcription, translation or protein degradation. These SNPs can be used as instrumental variables (IVs) in Mendelian randomization (MR) analyses9,10. MR is a tool that uses SNPs as IVs to assess causal relationships between exposures and outcomes. It can effectively avoid the influence of potential confounding factors and can be used as a new method to identify potential drug targets11. Although studies have confirmed the causal relationships between nephrolithiasis and obesity, diabetes, and alcohol intake through MR analyses12–14, MR analyses of plasma protein data are lacking. MR analyses can provide important evidence for potential plasma protein-related targets (PPRTs) for the prevention and treatment of nephrolithiasis. In our study, PPRTs were defined as targets obtained in plasma proteins that were significantly associated with nephrolithiasis. In subsequent analyses, other multi-omics data of PPRTs were also included for verification.
We conducted a protein-wide MR study supplemented by a variety of comprehensive methods to explore the correlations between PPRTs and the pathogenesis of nephrolithiasis (Fig. 1).
Fig. 1.
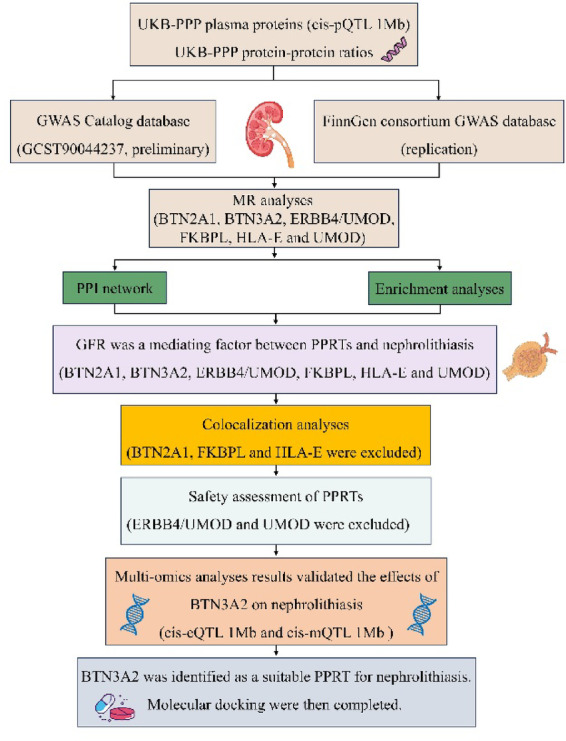
Overview of the study design. UKB-PPP UK Biobank Pharma Proteomics Project, MR Mendelian randomization, GWAS genome-wide association analysis, PPI protein–protein interaction, GFR glomerular filtration rate, BTN2A1 butyrophilin subfamily 2 member A1, BTN3A2 butyrophilin subfamily 3 member A2, ERBB4 receptor tyrosine-protein kinase erbB-4, FKBPL FK506-binding protein-like, HLA-E HLA class I histocompatibility antigen, alpha chain E, UMOD uromodulin, PPRTs plasma protein-related targets, pQTLs protein quantitative trait loci, cis-eQTLs cis expression quantitative trait loci, cis-mQTLs cis-DNA methylation quantitative trait loci.
Materials and methods
Identification of instrumental variables
The current study was performed following the STROBE-MR checklist, and the checklist has been attached as supplemental material. The exposure data for the MR analyses were obtained from the UK Biobank Pharmaceutical Proteomics Project (UKB-PPP), which included 34,557 participants of European ancestry with 2940 plasma proteins15. The initial screening of IVs is based on the following criteria. (1) IVs were cis-pQTLs, defined as SNPs with genome-wide significance (P value < 5.00e-08) within 1 Mb of the measured transcription start site of the protein-coding gene. (2) The clump algorithm was applied to remove linkage disequilibrium (LD), and we appropriately relaxed the clump threshold (kb = 500, r2 = 0.01) to obtain as many SNPs as possible for MR. (3) R2 and F values were calculated, and SNPs with F < 10 were identified as undesirable genetic variants and removed16. (4) After performing the above operations, each protein needed to contain at least three SNPs. Retaining at least three independent SNPs was to ensure the robustness of the statistical results. Multiple SNPs can be weighted or aggregated to improve the accuracy of effect estimates. Sensitivity analyses to detect MR pleiotropy required at least three SNPs to complete. Finally, 1376 proteins were included in the subsequent analyses.
Additionally, other exposure data containing SNPs called ratio QTLs (rQTLs) (a total of 2821 protein‒protein ratios, calculated from UKB-PPP data, for 43,509 participants of European ancestry), defines as SNPs representing protein–protein ratios, were also included17. The introduction of rQTLs could identify pQTLs that were not found by standard GWAS protein levels from the UKB PPP and reveal more than two or more protein‒protein associations in diseases. A P value < 5.00e-08, suitable clump threshold (kb = 10,000, r2 = 0.001) and F > 10 were the preliminary screening standards for IVs. Finally, all 2821 rQTLs met the criteria.
When evaluating the causal relationships between 4197 QTLs (including 1376 pQTLs and 2821 rQTLs) and nephrolithiasis, both Bonferroni and FDR corrections were used. The P value threshold for Bonferroni correction was 1.19e-05 (0.05/4197), and the P value threshold for FDR correction was 0.05.
In the replication analyses, since there were 8 plasma proteins included in the analyses, the P value threshold for Bonferroni correction was 6.25e-03 (0.05/8), and the P value threshold for FDR correction was 0.05.
The outcome data for nephrolithiasis were obtained from the GWAS catalog database (GCST90044237) and the FinnGen consortium database (R10). Three complications of nephrolithiasis (including low back pain, hydronephrosis, and renal failure) were derived from the FinnGen consortium database, and one complication, urinary tract infection (UTI), was from the GWAS catalog database (GCST90044231). The GWAS data for the glomerular filtration rate (GFR) were obtained from a catalog GWAS database (GCST008059). The specific information of the datasets included in this study can be found in Table 1.
Two-sample Mendelian randomization and sensitivity analyses
The application of MR methods should satisfy three assumptions: relevance (there should be a significant association between included IVs and exposure), independence (there should be no common causes between included IVs and the outcome) and exclusion restrictions (IVs were not directly related to the outcome). Two-sample MR was used to evaluate the causal relationships between candidate PPRTs and nephrolithiasis. The GCST90044237 dataset was used for preliminary analyses, and the FinnGen consortium dataset was used for replication analyses. The inverse variance weighted (IVW) method was used for MR analyses. In addition, reverse MR analyses were also performed during screening for significant PPRTs and subsequent MR analyses of the mediating factor.
Some of the datasets included in this study have the risk of sample overlap, but the potential maximum overlap rate of the dataset pairs used for MR analyses was less than 10%, which could provide effective MR analyses with a validity of more than 90%18. To truly evaluate the sample overlaps between each dataset and eliminate the impacts of sample overlaps, the linkage disequilibrium score regression (LDSC) function in the “MRlap” R package was used to calculate the intercepts between dataset pairs with potential sample overlaps (P > 0.05 indicated that there were no sample overlaps)19. If the results indicated that there were sample overlaps, the “MRlap” package could correct the MR results. Datasets with no clear sample overlap were not evaluated for LDSC analyses.
To assess the reliability of the MR results, statistically significant MR results should be followed up with sensitivity analyses. The Q test was performed to assess heterogeneity (P > 0.05 indicated no heterogeneity), and the MR–Egger intercept test was performed to assess pleiotropy (P > 0.05 indicated no pleiotropy). The Steiger test was used to assess whether there was a potential reverse causal association (P < 0.05 indicated no reverse causal association). The Steiger test was also used to verify the reverse MR. Steiger filtering was performed for all MR analyses.
Enrichment analyses
A PPI network of candidate PPRTs was constructed via the GeneMANIA website (https://genemania.org/). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses for the PPRTs were performed using the “Ontologies” and “Pathways” modules of the Enrichr website (https://maayanlab.cloud/Enrichr/), respectively.
Identification of mediating factors
The glomerular filtration rate (GFR) is an important indicator of kidney function. Therefore, we wanted to confirm whether the GFR was a mediating factor between candidate PPRTs and nephrolithiasis. The nephrolithiasis dataset used in the mediation analyses was the FinnGen dataset. We conducted MR analyses between candidate PPRTs and the GFR (effect value was beta1) and MR analyses between the GFR and nephrolithiasis (effect value was beta2). The effect value (beta3) between PPRTs and nephrolithiasis was obtained via previous batch MR analyses. All MR analyses should not have reverse causal relationships. The calculation of the mediated proportion was defined as beta1 × beta2/beta3.
Colocalization analyses
Colocalization analyses (coloc.abf) were conducted to assess the probability that nephrolithiasis risk loci and QTL data of PPRTs share the same variant rather than variants shared coincidentally due to LD association. The probability that a given SNP was associated with nephrolithiasis was denoted P1, the probability that a given SNP is a significant QTL was denoted P2, and the probability that a given SNP was an outcome of both nephrolithiasis and QTLs was denoted P12. The colocalization priors were set as P1 = 1 × 10−4, P2 = 1 × 10−4, and P12 = 1 × 10−5. The genomic window was set as 500 KB. In colocalization analyses, five different posterior probabilities (PP) were reported, corresponding to the following hypotheses: (1) not related to nephrolithiasis and PPRTs (PP.H0), (2) related only to nephrolithiasis and not related to PPRTs (PP.H1), (3) related to PPRTs and not related to nephrolithiasis (PP.H2), (4) related to both nephrolithiasis and PPRTs but also related to two other traits (PP.H3), and (5) related to both nephrolithiasis and PPRTs and shared the same causal variation (PP.H4). Strong evidence of colocalization was considered present when PP.H4 > 0.8. Subsequently, the PPRTs that passed the colocalization analyses (coloc.abf) were revalidated using SuSiE (coloc.susie). The sensitivity of both the prior probabilities and posterior probabilities in all colocalization analyses was assessed.
Safety of drug targets
To evaluate the safety of these PPRTs significantly by colocalization analyses (PP.H4 > 0.8), we performed MR analyses between PPRTs and common complications of nephrolithiasis, including low back pain, hydronephrosis, renal failure, and urinary tract infection (UTI). Although we acknowledge that the patients in the datasets with these complications might not have had these symptoms due to nephrolithiasis, these data still have important implications for the safety of targeted PPRTs in patients with nephrolithiasis. If candidate PPRTs caused these symptoms, these PPRTs could have significant impacts on the aggravation or alleviation of nephrolithiasis.
After confirming that PPRTs did not worsen nephrolithiasis by affecting complications, the potential adverse drug reactions and pleiotropic effects of PPRTs were investigated by phenome-wide association studies (PheWASs). In PheWAS, exposures were final PPRTs, IVs were screened as previously described, the results included 2405 phenotypes from the FinnGen database (R10), and the IVW method was used. The reason for evaluating the causal effect of PPRTs on nephrolithiasis complications before PheWAS was that the large-scale false discovery rate (FDR) correction of the PheWAS results would mask the significance of the effects of PPRTs on nephrolithiasis complications, which should be a priority in the treatment of nephrolithiasis. Both Bonferroni and FDR corrections were used. The P value threshold for Bonferroni correction was 2.08e-05 (0.05/2405). The P value threshold for FDR correction was 0.05.
However, we acknowledged that we did not assess phenotypes outside the FinnGen consortium, which might miss rare side events or effects in non-European populations. Therefore, a wider phenotypic database should be included in future PheWAS analyses. And in the future, longitudinal studies or pharmacovigilance should be conducted after clinical trials to monitor long-term effects.
Analyses of combined multiomics data
The cis DNA methylation QTL (cis-mQTL, 1 Mb) data of the selected PPRTs, defined as SNPs located near DNA methylation sites (within 1 Mb) that can significantly affect the methylation level of local CpG sites, were obtained from the GoDMC database (http://mqtldb.godmc.org.uk/index). In addition, the cis expression QTL (cis-eQTL, 1 Mb) data of the selected PPRTs, defined as SNPs located near the transcription start site of a gene in the genome (within 1 Mb) that could directly regulate gene expression, were downloaded from the eQTLGen consortium (https://www.eqtlgen.org/cis-eqtls.html). A P value < 5.00e-08 and F > 10 were the preliminary screening standards for IVs. The clump threshold of the cis-eQTLs was kb = 10,000 and r2 = 0.001. Owing to the relatively insufficient number of total SNPs, the clump thresholds of the cis-mQTLs were kb = 500 and r2 = 0.01. MR analyses of multiomics data (cis-eQTLs and cis-mQTLs) and nephrolithiasis data were performed. The regulation of gene expression by locus methylation and the regulation of protein abundance by gene expression were also confirmed by MR analyses. Finally, colocalization analyses of multiomics data and nephrolithiasis data were performed.
Additional validation
The nephrolithiasis dataset GSE73680 from the GEO database was used for further additional validation. This dataset included 29 Randall’s plaque tissues and 33 normal tissues. GSE73860 was used to evaluate the expression differences of PPRTs that met all the above criteria in Randall’s plaque tissues and normal tissues. Then we further explored the immune landscape of nephrolithiasis patients via CIBERSORT algorithm20.
Molecular docking
The protein-targeted drugs used were identified through the "Diseases/drugs" module of the Enrichr website. The 3D structures of the PPRTs and candidate drugs can be downloaded from the UniProt database (https://www.uniprot.org/) and the PubChem database (https://pubchem.ncbi.nlm.nih.gov/), respectively. Molecular docking was performed using AutoDock Vina software version 1.5.7. The formation of stable hydrogen bonds between the PPRTs and the drug was considered to indicate effective docking. The free energy values were subsequently calculated, and the results of molecular docking were visualized via PyMOL software (3D) and Ligplus software (2D).
Statistical analyses
All the above operations were implemented using R software version 4.3.2. The R packages used were “TwoSampleMR” (version 0.5.8), “coloc” (version 5.2.3) and “MendelianRandomization” (version 0.9.0). Except for the preliminary and replication MR analyses for screening PPRTs, the other MR analyses only applied the more flexible FDR correction, with a P value threshold of 0.05.
Results
Causal associations between PPRTs and nephrolithiasis
The LDSC analysis results revealed that there were no significant sample overlaps between the dataset pairs that might have potential sample overlaps in this study (Table S1). The results of preliminary analyses suggested that eight PPRTs were significantly causally associated with nephrolithiasis. After the MR analyses of the eight PPRTs were replicated via the nephrolithiasis data of FinnGen, the results of the six PPRTs remained significant (Fig. 2). Among them, butyrophilin subfamily 2 member A1 (BTN2A1), receptor tyrosine-protein kinase erbB-4 (ERBB4)/uromodulin (UMOD), HLA class I histocompatibility antigen, alpha chain E (HLA-E) and UMOD were found to be inhibitory factors for nephrolithiasis [odds ratio (OR) < 1], and butyrophilin subfamily 3 member A2 (BTN3A2) and FK506-binding protein-like (FKBPL) were found to be promoting factors for nephrolithiasis (OR > 1). Reverse MR analyses confirmed that nephrolithiasis did not affect the levels of the six PPRTs (Fig. S1). Sensitivity analyses revealed no heterogeneities except for ERBB4/UMOD (P = 3.77e-03), no pleiotropies and no associations of reverse causation between these six PPRTs and nephrolithiasis (Table S2).
Fig. 2.
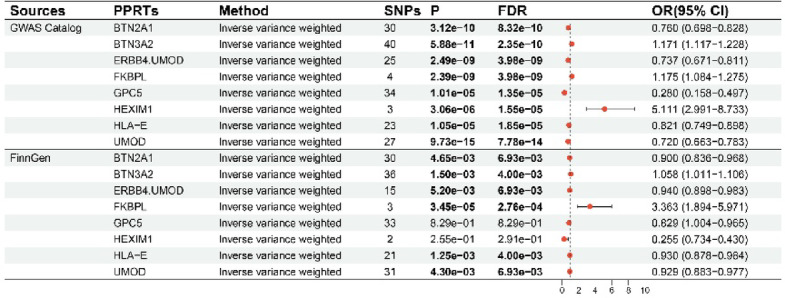
Results of preliminary and replication MR analyses. MR Mendelian randomization, FDR false discovery rate, BTN2A1 butyrophilin subfamily 2 member A1, BTN3A2 butyrophilin subfamily 3 member A2, ERBB4 receptor tyrosine-protein kinase erbB-4, FKBPL FK506-binding protein-like, GPC5 glypican 5, HEXIM1 HEXIM P-TEFb complex subunit 1, HLA-E HLA class I histocompatibility antigen, alpha chain E, UMOD uromodulin, PPRTs plasma protein-related targets, SNPs single-nucleotide polymorphisms. In the primary analyses, the P value threshold for Bonferroni correction was 1.19e-05 (0.05/4197), and the P value threshold for FDR correction was 0.05. In the replication analyses, since there were 8 plasma proteins included in the analyses, the P value threshold for Bonferroni correction was 6.25e-03 (0.05/8), and the P value threshold for FDR correction was 0.05.
Enrichment analyses
A PPI network of PPRTs and related proteins was constructed (Fig. S2). The results of the GO and KEGG enrichment analyses suggested that the PPRTs were closely related to immune-related functions, especially the regulation of NK cells and T cells (Table S3 and S4).
GFR-mediated candidate targets and nephrolithiasis
MR analyses revealed that five PPRTs were significantly related to the GFR (BTN2A1, BTN3A2, ERBB4/UMOD, HLA-E and UMOD), and there was a positive correlation between the GFR and nephrolithiasis (Table S5 and S6). The reverse MR analyses also proved again that there were no reverse causalities in the entire mediation analysis (Fig. S3). These findings indicate that the GFR is a mediating factor between proteins and nephrolithiasis. The percentages of each protein were subsequently calculated and were 5.549% for BTN2A1, 21.495% for BTN3A2, 7.695% for ERBB4/UMOD, 10.050% for HLA-E and 22.350% for UMOD (Table 2).
Colocalization analyses
According to the colocalization analyses (coloc.abf and coloc.susie) of PPRTs, BTN3A2, ERBB4/UMOD and UMOD strongly colocalized (Table S7 and S8). Therefore, these three PPRTs could be used for subsequent safety evaluation, and BTN2A1, FKBPL, and HLA-E were excluded. Sensitivity analyses results suggested that all colocalization analysis results were robust (Fig. S4A-F and S5A-F).
Safety assessment of PPRTs
MR analyses of three PPRTs and nephrolithiasis complications revealed that. ERBB4/UMOD and UMOD were positively associated with renal failure (ERBB4/UMOD, OR = 1.121; UMOD, OR = 1.190). BTN3A2 was positively associated with UTI (OR = 1.052) (Fig. 3, Table S9). As protective factors for nephrolithiasis, increased levels of UMOD and ERBB4/UMOD could reduce the incidence of nephrolithiasis but promote the occurrence of renal failure, which is not conducive to the relief of nephrolithiasis. However, lowering the level of BTN3A2 could reduce the incidence of nephrolithiasis and the occurrence of UTI, which is helpful for the treatment of nephrolithiasis.
Fig. 3.
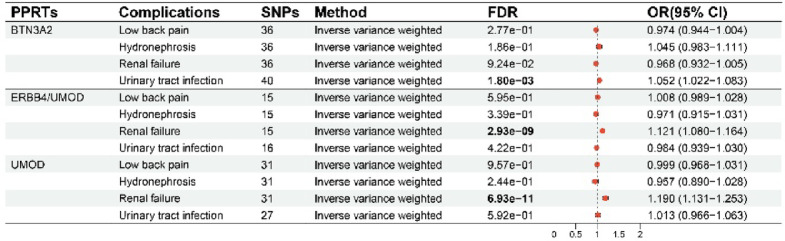
Results of MR analyses assessing the causal association between PPRTs and nephrolithiasis complications. MR Mendelian randomization, FDR false discovery rate, BTN3A2 butyrophilin subfamily 3 member A2, ERBB4 receptor tyrosine-protein kinase erbB-4, UMOD uromodulin, PPRTs plasma protein-related targets, SNPs single-nucleotide polymorphisms. The P value threshold for FDR correction was 0.05.
After confirming that BTN3A2 did not aggravate the complications of nephrolithiasis, PheWAS was used. The results revealed no significant relationships between BTN3A2 and phenotypes in FinnGen (FDR > 0.05), indicating that the risk of potential adverse drug reactions and pleiotropic effects in targeting BTN3A2 was relatively low (Fig. 4A, B).
Fig. 4.

PheWAS analyses of the associations between BTN3A2 and other diseases. P values were corrected for Bonferroni (A) and FDR (B), respectively. PheWAS phenome-wide association study, BTN3A2 butyrophilin subfamily 3 member A2, FDR false discovery rate. The P value threshold for Bonferroni correction was 2.08e-05 (0.05/2405). The P value threshold for FDR correction was 0.05.
Analyses of multiomics data
Three methylation sites of BTN3A2 were retrieved from the GoDMC database (cg02045355, cg14345882 and cg23465465). After primary and replication MR analyses, the results indicated that BTN3A2 (cis-eQTLs) and BTN3A2 (cis-mQTLs of cg14345882 and cg23465465) were causally associated with nephrolithiasis (Fig. S6, Table S10). The methylation sites (cg14345882 and cg23465465) could negatively regulate the gene expression of BTN3A2, and the gene expression of BTN3A2 was significantly positively correlated with the protein abundance of BTN3A2. There was significant evidence of colocalization between BTN3A2 (cis-eQTL and cis-mQTL of cg14345882 and cg23465465) and nephrolithiasis (Table S11 and S12). Sensitivity analyses results suggested that all colocalization analysis results were robust (Fig. S7A-F and SS88A-F).
Further validation
By mining the GSE78680 dataset in the GEO database, we found that the expression of BTN3A2 in Randall’s plaque tissue was significantly higher than that in normal tissue (Fig. S9A). The infiltration levels of T cells CD4 memory activated, NK cells activated and Macrophages M1 in Randall’s plaque tissue were significantly higher than those in normal tissue (Fig. S9B). In addition, BTN3A2 was significantly positively correlated with Dendritic cells activated, NK cells activated, Plasma cells and Macrophages M1 (Fig. S9C-F).
Molecular docking
Drugs targeting BTN3A2 were significantly differentially expressed (P < 0.05) (Table S13). These drugs were molecularly docked, and the free energy of each drug was calculated. The top five drugs in terms of free energy are displayed in Table 3 and include digitoxigenin (Fig. 5A), lycorine (Fig. 5B), piperlongumine (Fig. 5C), cycloheximide (Fig. 5D), and minoxidil (Fig. 5E).
Fig. 5.
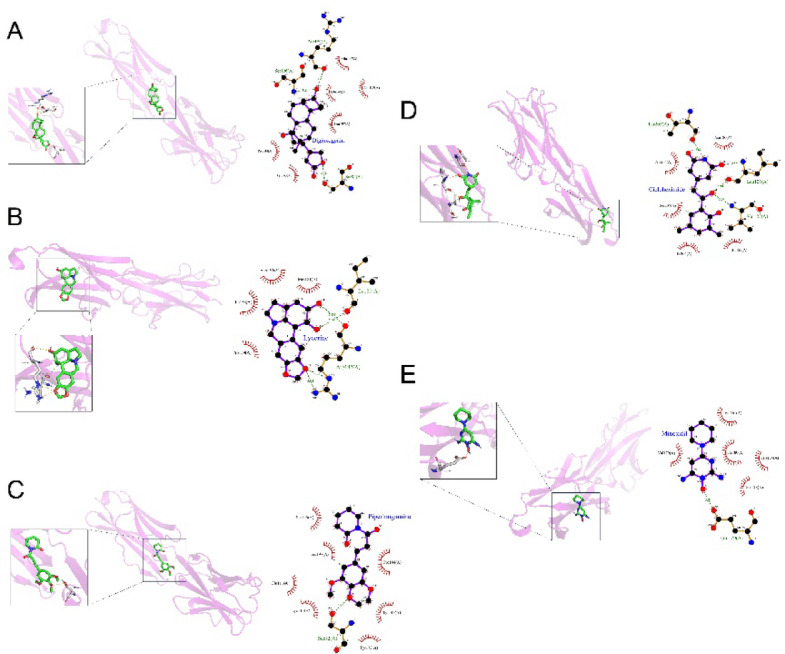
Visualization of the molecular docking results for candidate PPRTs. Molecular docking results of candidate PPRTs and drugs [including digitoxigenin (A), lycorine (B), piperlongumine (C), cycloheximide (D), and minoxidil (E)]. PPRTs plasma protein-related targets.
Discussion
In this study, MR analyses and other methods were used to study the causal relationships between potential PPRTs and nephrolithiasis. The results show that BTN3A2 is an advantageous drug target for nephrolithiasis. The binding efficacy of BTN3A2 to drugs was further evaluated through molecular docking.
On the basis of the preliminary and replication MR analyses, we confirmed causal associations between six PPRTs and nephrolithiasis (BTN2A1, BTN3A2, ERBB4/UMOD, FKBPL, HLA-E and UMOD). Among these six proteins, the role of urinary UMOD in nephrolithiasis has been the most comprehensively studied, but there are no relevant studies on UMOD in plasma. UMOD is a protein produced only by the kidneys, and a reduced level of UMOD in urine is associated with an increased risk of nephrolithiasis via activation of TRPV5/6 channels21–23. Although none of the remaining proteins have been reported to be related to nephrolithiasis, they are involved in the regulation of immunity and metabolism. Abnormalities in immunity and metabolism are also key factors in the occurrence and development of nephrolithiasis24,25. BTN2A1 and BTN3A2 are involved in the regulation of Vγ9Vδ2 T cells, and the functions of these T cells include eliminating various pathogens, controlling inflammation and maintaining immune tolerance26,27. BTN2A1 can trigger Vγ9Vδ2 T-cell activation, but elevated expression of BTN3A2 may mediate immune evasion and inhibit Vγ9Vδ2 T cells under certain circumstances28–30. ERBB4 limits the secretion of excessive inflammatory factors through feedback inhibition, thereby inhibiting the activity of macrophages and producing anti-inflammatory effects31. FKBPL is an angiogenesis-related protein that regulates the migration of endothelial cells and the activity of glucocorticoid receptors32–34. It is involved in the pathogenesis of metabolic diseases (diabetes and cardiovascular disease) and inflammation35,36. HLA-E can increase its expression during infection and regulate the proliferation and function of NK cells by binding to different receptors37,38. The results of reverse MR analyses indicated that nephrolithiasis could not cause changes in the expression of these five proteins, indicating that there was no bias in the previous MR analysis results due to potential reverse causality.
The results of the functional enrichment analyses indicated that five proteins were related mainly to the regulation of NK cells and T cells. There are currently no direct reports linking NK cells to nephrolithiasis. We found that NK cell activation and inhibition occur simultaneously in biological functions. Among the five proteins, HLA-E was the main protein involved in the regulation of NK cells and was negatively correlated with the occurrence of nephrolithiasis. We assumed that increased NK cell activity was a potential risk factor for nephrolithiasis. The results of the T-cell enrichment analyses were significant because the interaction between BTN3A2 and BTN2A1 had a certain effect on Vγ9Vδ2 T cells.
We subsequently identified the GFR as a mediating factor between five PPRTs and nephrolithiasis (BTN2A1, BTN3A2, ERBB4/UMOD, HLA-E and UMOD). A high GFR is a risk factor for nephrolithiasis. Although somewhat counterintuitive, some studies have reached similar conclusions, that low levels of GRF could inhibit the occurrence of nephrolithiasis and decreased GRF level would lead to decreased urinary calcium excretion39–42. But this did not mean that preventing nephrolithiasis by affecting GFR could cause damage to renal function. The results of the mediation analyses (especially the β value) showed that when the level of BTN3A2 in plasma was reduced, the GFR would be slightly downregulated, and only a mild reduction in GFR could significantly reduce the risk of nephrolithiasis. Subsequent results also suggested that changes in BTN3A2 levels did not cause renal failure. However, we should acknowledge that this result needs to be further verified.
Among the five PPRTs, only UMOD had direct evidence that it could affect the GFR, but the results were controversial. Previous observational studies revealed a positive correlation between plasma UMOD and the GFR43,44, and a series of MR studies confirmed that higher plasma UMOD concentrations lead to a lower GFR45,46. One speculated that observational studies confounded populations with rare UMOD variants that could reduce both the plasma UMOD and the GFR47. Our results were consistent with those of previous MR studies. The relationships between other PPRTs and the GFR are unclear and need to be explored in the future.
Furthermore, we determined the probability that a specific genetic variant was associated with PPRT concentration and nephrolithiasis through colocalization analyses. If the MR results suggested a causal association between a protein and nephrolithiasis and were also supported by evidence of colocalization, then a specific genetic variant was considered associated with protein concentration and nephrolithiasis. If the MR results suggest a causal association between a protein and nephrolithiasis but are not supported by evidence of colocalization, then the specific genetic variant might be independently associated with nephrolithiasis through a mechanism other than affecting the concentration of the protein47. The colocalization results revealed that BTN3A2, ERBB4/UMOD and UMOD were strongly colocalized, which meant that the concentrations of these three PPRTs in plasma could directly affect the occurrence of nephrolithiasis.
Three PPRTs were evaluated for safety as drug targets. Increasing the expression levels of the two protective factors against nephrolithiasis in plasma (UMOD and ERBB4/UMOD) could increase the occurrence of renal failure, which was related to the fact that both PPRTs could significantly negatively regulate the GFR. Moreover, reducing the plasma levels of the risk factor for nephrolithiasis could reduce the incidence of UTI (BTN3A2), which is significantly beneficial for the remission of nephrolithiasis. The main reason might be that the immune evasion effect mediated by BTN3A2 causes exhaustion of anti-inflammatory Vγ9Vδ2 T cells, which promotes inflammation. The PheWAS suggested that the risk of potential adverse drug reactions and pleiotropic effects of BTN3A2 was relatively low, which strengthened the evidence supporting the druggable potential of BTN3A2.
Multiomics analyses revealed that both the BTN3A2 methylation level and the gene expression level were causally related to nephrolithiasis. Methylation levels can affect gene expression and thus regulate protein abundance. All three sites (methylation of the BTN3A2 site, gene level and protein level) were significantly colocalized with nephrolithiasis. These findings indicate that epigenetic regulatory mechanisms play important roles in nephrolithiasis and that epigenetic intervention may be beneficial for the treatment of nephrolithiasis, suggesting its potential in therapeutic research.
In subsequent database mining, we found that higher expression of BTN3A2 in Randall’s plaque tissue was significantly positively correlated with activated Dendritic cells, activated NK cells, plasma cells and M1 macrophages. M1 macrophages and NK cells played a pro-inflammatory role in nephrolithiasis formation, inducing renal epithelial cell damage and promoting CaOx crystal deposition48,49. Activated Dendritic cells were important antigen presenting cells, which could mature and migrate to secondary lymphoid organs to activate T cells and promote immune response after capturing antigens50.
Drugs with statistically significant differences were found to target BTN3A2. After molecular docking, five drugs met the requirements. Since the database included only drugs that target BTN3A2, whether they are effective in patients with nephrolithiasis remains unknown. As a key drug for the treatment of congestive heart failure, the positive inotropic effect of digitoxigenin on the heart is controversial for the safety of normal people in the treatment of nephrolithiasis, so digitoxigenin may not be a suitable drug. Other drugs are not directly related to nephrolithiasis, but some potential indirect evidence deserves attention. Lycorine is the main active ingredient of Lycoris and has a strong anti-inflammatory effect51. Piperlongumine attenuates high calcium/phosphate-induced vascular calcification by increasing P53/PTEN signaling, and both vascular calcification and nephrolithiasis are related to calcium receptor disorders52,53. Although the binding ability suggested by the docking analyses suggested the potential of these drugs in treating nephrolithiasis, these were only preliminary results with certain uncertainties. The efficacy and safety of related drugs required rigorous clinical trial evaluation.
Our study focused on plasma biomarkers for nephrolithiasis. Plasma biomarkers are easy to collect, direct in action, cause minimal trauma, and are conducive to dynamic follow-up, making them suitable for assessing the risk of nephrolithiasis and developing preventive measures and treatments. Previous studies have explored therapeutic targets for nephrolithiasis, but screening of the corresponding targets has been performed using stones, kidney tissue or urine, which lack the advantages of therapeutic targets in plasma, and no genetic associations between targets and nephrolithiasis or safety assessments have been performed54–57. Therefore, the use of BTN3A2, which we identified through multidimensional evaluation, could provide an important reference for the treatment of nephrolithiasis.
This study has several advantages. Numerous GWASs were included, comprehensive analyses were performed on these datasets, and the results of each analysis were mutually supportive. The limitations of this study deserve to be discussed. First, because the data included in our study were from individuals of European ethnicity, the generalizability of our findings to other ethnic groups was limited. Future studies should include multi-ethnic cohorts (e.g., African, Asian, and Hispanic populations) to validate the role of BTN3A2. And collaboration with international consortia should be undertaken to expand the diversity of GWAS data. Second, there was still potential residual pleiotropy during MR analyses. Third, the GWAS datasets included in the study lack detailed information on stone composition (oxalate, urate, phosphate, cystine, etc.), which may affect the correlation between PPRTs (especially BTN3A2) and nephrolithiasis. In subsequent studies, datasets containing detailed stone type information should be included for subgroup analyses, and validation for specific stone types is required. Additionally, molecular docking only showed the binding potential of plasma proteins with candidate drugs. We lacked in vitro and in vivo experimental confirmation of the effects of BTN3A2 on nephrolithiasis and its role as a candidate drug target for the treatment of nephrolithiasis. And further clinical trials were necessary.
In summary, by applying a series of comprehensive analytical methods, we found that BTN3A2 has advantages over other PPRTs and could be used as a potential target for the early prevention and treatment of nephrolithiasis. However, in the future, the intrinsic mechanism of the target should be explored through detailed basic experiments, and the effects of targeted drugs should also be confirmed through larger-scale, multicenter and multiethnic clinical trials.
Conclusion
After a series of comprehensive analyses, the results suggested that BTN3A2 could be a suitable therapeutic target for nephrolithiasis. Our findings provide more accurate targets for the treatment of nephrolithiasis, which could help clinicians develop personalized treatment plans and reduce the burden on patients.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We acknowledge all the authors who participated in this study. This study was funded by the National Nature Science Foundation of China (Grant No. 82172568). We would like to thank the UK Biobank, FinnGen consortium, and GWAS catalog. We would like to thank the anonymous reviewers for their constructive comments. We are very grateful to the image website Vecteezy (https://www.vecteezy.com/) for providing the free materials for Fig. 1.
Author contributions
Yang Fu, Yutao Wang and jianbing Bi participated in the design of this study. Yang Fu and Shanshan Sun collected and analyzed all data. Yang Fu and Shanshan Sun drafted the manuscript draft. Yang Fu, Yutao Wang and jianbing Bi conducted the Validation test. Yang Fu, Shanshan Sun, Qingzhuo Dong and jianbing Bi revised the manuscript. Yang Fu, and Shanshan Sun supervised the study. All authors contributed to the manuscript and approved the submitted version.
Data availability
The datasets generated and/or analyzed during the current study are available in the UK Biobank, https://www.synapse.org/Synapse:syn51365303, FinnGen consortium, https://r10.finngen.fi/, and catalog repository, https://www.ebi.ac.uk/gwas/. The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request. In this study, external replication was necessary, and therefore more GWAS databases with detailed information needed to be included in the future to complete replication analyses and further subgroup analyses.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Romero, V., Akpinar, H. & Assimos, D. G. Kidney stones: a global picture of prevalence, incidence, and associated risk factors. Rev. Urol.12, e86-96 (2010). [PMC free article] [PubMed] [Google Scholar]
- 2.Pearle, M. S. et al. Medical management of kidney stones: AUA guideline. J. Urol.192, 316–324 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Johnson, R. J. et al. Climate change and the kidney. Ann. Nutr. Metab.74, 38–44 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Antonelli, J. A., Maalouf, N. M., Pearle, M. S. & Lotan, Y. Use of the national health and nutrition examination survey to calculate the impact of obesity and diabetes on cost and prevalence of urolithiasis in 2030. Eur. Urol.66, 724–729. 10.1016/j.eururo.2014.06.036 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khan, S. R. et al. Kidney stones. Nat. Rev. Dis Primers2, 16008. 10.1038/nrdp.2016.8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moe, O. W. Kidney stones: pathophysiology and medical management. The lancet367, 333–344 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Howles, S. A. et al. Genetic variants of calcium and vitamin D metabolism in kidney stone disease. Nat. Commun.10, 5175 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng, J. et al. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat Genet52, 1122–1131. 10.1038/s41588-020-0682-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Folkersen, L. et al. Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nat Metab2, 1135–1148. 10.1038/s42255-020-00287-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reay, W. R. & Cairns, M. J. Advancing the use of genome-wide association studies for drug repurposing. Nat. Rev. Genet.22, 658–671. 10.1038/s41576-021-00387-z (2021). [DOI] [PubMed] [Google Scholar]
- 11.Wu, Y. et al. Exploration of potential novel drug targets and biomarkers for small cell lung cancer by plasma proteome screening. Front Pharmacol14, 1266782. 10.3389/fphar.2023.1266782 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan, S. & Larsson, S. C. Assessing causal associations of obesity and diabetes with kidney stones using Mendelian randomization analysis. Mol Genet Metab134, 212–215. 10.1016/j.ymgme.2021.08.010 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Yang, S. et al. Association between alcohol and urolithiasis: a mendelian randomization study. Urolithiasis51, 103. 10.1007/s00240-023-01472-0 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lovegrove, C. E. et al. Central adiposity increases risk of kidney stone disease through effects on serum calcium concentrations. J Am Soc Nephrol34, 1991–2011. 10.1681/asn.0000000000000238 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun, B. B. et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature622, 329–338. 10.1038/s41586-023-06592-6 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yun, Z. et al. Genetically predicted 486 blood metabolites in relation to risk of colorectal cancer: A Mendelian randomization study. Cancer Med12, 13784–13799. 10.1002/cam4.6022 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suhre, K. Genetic associations with ratios between protein levels detect new pQTLs and reveal protein-protein interactions. Cell Genom4, 100506. 10.1016/j.xgen.2024.100506 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han, Q. J. et al. PTGES2 and RNASET2 identified as novel potential biomarkers and therapeutic targets for basal cell carcinoma: insights from proteome-wide mendelian randomization, colocalization, and MR-PheWAS analyses. Front Pharmacol15, 1418560. 10.3389/fphar.2024.1418560 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mounier, N. & Kutalik, Z. Bias correction for inverse variance weighting Mendelian randomization. Genet Epidemiol47, 314–331. 10.1002/gepi.22522 (2023). [DOI] [PubMed] [Google Scholar]
- 20.Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods12, 453–457. 10.1038/nmeth.3337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romero, M. C., Nocera, S. & Nesse, A. B. Decreased Tamm-Horsfall protein in lithiasic patients. Clin. Biochem.30, 63–67 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Glauser, A., Hochreiter, W., Jaeger, P. & Hess, B. Determinants of urinary excretion of Tamm-Horsfall protein in non-selected kidney stone formers and healthy subjects. Nephrol. Dial. Transplant.15, 1580–1587 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Lau, W.-H., Leong, W.-S., Ismail, Z. & Gam, L.-H. Qualification and application of an ELISA for the determination of Tamm Horsfall protein (THP) in human urine and its use for screening of kidney stone disease. Int. J. Biol. Sci.4, 215 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian, L. et al. Lactiplantibacillus plantarum J-15 reduced calcium oxalate kidney stones by regulating intestinal microbiota, metabolism, and inflammation in rats. FASEB J.36, e22340 (2022). [DOI] [PubMed] [Google Scholar]
- 25.Capolongo, G., Ferraro, P. M. & Unwin, R. Inflammation and kidney stones: cause and effect?. Curr. Opin. Urol.33, 129–135 (2023). [DOI] [PubMed] [Google Scholar]
- 26.Davis, M. M. & Bjorkman, P. J. T-cell antigen receptor genes and T-cell recognition. Nature334, 395–402. 10.1038/334395a0 (1988). [DOI] [PubMed] [Google Scholar]
- 27.Künkele, K. P. et al. Vγ9Vδ2 T cells: can we re-purpose a potent anti-infection mechanism for cancer therapy?. Cells10.3390/cells9040829 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cano, C. E. et al. BTN2A1, an immune checkpoint targeting Vγ9Vδ2 T cell cytotoxicity against malignant cells. Cell Rep36, 109359. 10.1016/j.celrep.2021.109359 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Messal, N. et al. Differential role for CD277 as a co-regulator of the immune signal in T and NK cells. Eur. J. Immunol.41, 3443–3454 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Benyamine, A. et al. BTN3A is a prognosis marker and a promising target for Vγ9Vδ2 T cells based-immunotherapy in pancreatic ductal adenocarcinoma (PDAC). Oncoimmunology7, e1372080. 10.1080/2162402x.2017.1372080 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schumacher, M. A. et al. NRG4-ErbB4 signaling represses proinflammatory macrophage activity. Am J Physiol Gastrointest Liver Physiol320, G990-g1001. 10.1152/ajpgi.00296.2020 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valentine, A. et al. FKBPL and peptide derivatives: novel biological agents that inhibit angiogenesis by a CD44-dependent mechanism. Clin Cancer Res17, 1044–1056. 10.1158/1078-0432.Ccr-10-2241 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yakkundi, A. et al. The anti-migratory effects of FKBPL and its peptide derivative, AD-01: regulation of CD44 and the cytoskeletal pathway. PLoS ONE8, e55075. 10.1371/journal.pone.0055075 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKeen, H. D. et al. A novel FK506-like binding protein interacts with the glucocorticoid receptor and regulates steroid receptor signaling. Endocrinology149, 5724–5734. 10.1210/en.2008-0168 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Januszewski, A. S. et al. FKBPL is associated with metabolic parameters and is a novel determinant of cardiovascular disease. Sci Rep10, 21655. 10.1038/s41598-020-78676-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Annett, S. et al. FKBPL-based peptide, ALM201, targets angiogenesis and cancer stem cells in ovarian cancer. Br J Cancer122, 361–371. 10.1038/s41416-019-0649-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caccamo, N. et al. Human CD8 T lymphocytes recognize Mycobacterium tuberculosis antigens presented by HLA-E during active tuberculosis and express type 2 cytokines. Eur J Immunol45, 1069–1081. 10.1002/eji.201445193 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Rölle, A., Meyer, M., Calderazzo, S., Jäger, D. & Momburg, F. Distinct HLA-E peptide complexes modify antibody-driven effector functions of adaptive NK cells. Cell Rep24, 1967-1976.e1964. 10.1016/j.celrep.2018.07.069 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Liu, M. et al. Lifestyle factors, serum parameters, metabolic comorbidities, and the risk of kidney stones: a Mendelian randomization study. Front Endocrinol (Lausanne)14, 1240171. 10.3389/fendo.2023.1240171 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jian, Z. et al. Kidney function may partially mediated the protective effect of urinary uromodulin on kidney stone. Urolithiasis51, 65. 10.1007/s00240-023-01441-7 (2023). [DOI] [PubMed] [Google Scholar]
- 41.Coyne, D. et al. Paricalcitol capsule for the treatment of secondary hyperparathyroidism in stages 3 and 4 CKD. Am J Kidney Dis47, 263–276. 10.1053/j.ajkd.2005.10.007 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Liu, J. et al. Determinants and outcomes associated with urinary calcium excretion in chronic kidney disease. J Clin Endocrinol Metab107, e281–e292. 10.1210/clinem/dgab574 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melchinger, H. et al. Urine uromodulin as a biomarker of kidney tubulointerstitial fibrosis. Clin. J. Am. Soc. Nephrol.17, 1284–1292 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steubl, D. et al. Association of serum uromodulin with ESKD and kidney function decline in the elderly: the cardiovascular health study. Am. J. Kidney Dis.74, 501–509 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olinger, E. et al. An intermediate-effect size variant in UMOD confers risk for chronic kidney disease. Proc. Natl. Acad. Sci.119, e2114734119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sjaarda, J. et al. Blood HER2 and uromodulin as causal mediators of CKD. J Am Soc Nephrol29, 1326 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lanktree, M. B. et al. A novel multi-ancestry proteome-wide Mendelian randomization study implicates extracellular proteins, tubular cells, and fibroblasts in estimated glomerular filtration rate regulation. Kidney Int104, 1170–1184. 10.1016/j.kint.2023.08.025 (2023). [DOI] [PubMed] [Google Scholar]
- 48.Taguchi, K., Okada, A., Unno, R., Hamamoto, S. & Yasui, T. Macrophage function in calcium oxalate kidney stone formation: a systematic review of literature. Front Immunol12, 673690. 10.3389/fimmu.2021.673690 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kucuksezer, U. C. et al. The role of natural killer cells in autoimmune diseases. Front Immunol12, 622306. 10.3389/fimmu.2021.622306 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu, C. J. et al. Renal ischemia/reperfusion injury inhibits differentiation of dendritic cells derived from bone marrow monocytes in rats. Life Sci78, 1121–1128. 10.1016/j.lfs.2005.06.043 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Cao, Z., Yang, P. & Zhou, Q. Multiple biological functions and pharmacological effects of lycorine. Sci China Chem56, 1382–1391. 10.1007/s11426-013-4967-9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu, C. J., Cheng, C. W., Tsai, Y. S. & Huang, H. S. Crosstalk between renal and vascular calcium signaling: The link between nephrolithiasis and vascular calcification. Int J Mol Sci10.3390/ijms22073590 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi, W. et al. Piperlongumine attenuates high calcium/phosphate-induced arterial calcification by preserving P53/PTEN signaling. Front Cardiovasc Med7, 625215. 10.3389/fcvm.2020.625215 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kovacevic, L., Kovacevic, N. & Lakshmanan, Y. Proteomic analysis of inhibitory protein profiles in the urine of children with nephrolithiasis: Implication for disease prevention. Int Urol Nephrol54, 2783–2788. 10.1007/s11255-022-03310-5 (2022). [DOI] [PubMed] [Google Scholar]
- 55.Zhu, W. et al. Proteomics and transcriptomics profiling reveals distinct aspects of kidney stone related genes in calculi rats. BMC Genomics24, 127. 10.1186/s12864-023-09222-7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khusid, J. A. et al. Comparative proteomic profiling of uric acid, ammonium acid urate, and calcium-based kidney stones. Am J Clin Exp Urol11, 265–274 (2023). [PMC free article] [PubMed] [Google Scholar]
- 57.Yang, Y. et al. Proteomic analysis reveals some common proteins in the kidney stone matrix. PeerJ9, e11872. 10.7717/peerj.11872 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the UK Biobank, https://www.synapse.org/Synapse:syn51365303, FinnGen consortium, https://r10.finngen.fi/, and catalog repository, https://www.ebi.ac.uk/gwas/. The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request. In this study, external replication was necessary, and therefore more GWAS databases with detailed information needed to be included in the future to complete replication analyses and further subgroup analyses.


