Abstract
The COVID-19 pandemic has been the most catastrophic global health emergency of the  century, resulting in hundreds of millions of reported cases and five million deaths. Chest X-ray (CXR) images are highly valuable for early detection of lung diseases in monitoring and investigating pulmonary disorders such as COVID-19, pneumonia, and tuberculosis. These CXR images offer crucial features about the lung’s health condition and can assist in making accurate diagnoses. Manual interpretation of CXR images is challenging even for expert radiologists due to the overlapping radiological features. Therefore, Artificial Intelligence (AI) based image processing took over the charge in healthcare. But still it is uncertain to trust the prediction results by an AI model. However, this can be resolved by implementing explainable artificial intelligence (XAI) tools that transform a black-box AI into a glass-box model. In this research article, we have proposed a novel XAI-TRANS model with inception based transfer learning addressing the challenge of overlapping features in multiclass classification of CXR images. Also, we proposed an improved U-Net Lung segmentation dedicated to obtaining the radiological features for classification. The proposed approach achieved a maximum precision of 98% and accuracy of 97% in multiclass lung disease classification. By leveraging XAI techniques with the evident improvement of 4.75%, specifically LIME and Grad-CAM, to provide detailed and accurate explanations for the model’s prediction.
century, resulting in hundreds of millions of reported cases and five million deaths. Chest X-ray (CXR) images are highly valuable for early detection of lung diseases in monitoring and investigating pulmonary disorders such as COVID-19, pneumonia, and tuberculosis. These CXR images offer crucial features about the lung’s health condition and can assist in making accurate diagnoses. Manual interpretation of CXR images is challenging even for expert radiologists due to the overlapping radiological features. Therefore, Artificial Intelligence (AI) based image processing took over the charge in healthcare. But still it is uncertain to trust the prediction results by an AI model. However, this can be resolved by implementing explainable artificial intelligence (XAI) tools that transform a black-box AI into a glass-box model. In this research article, we have proposed a novel XAI-TRANS model with inception based transfer learning addressing the challenge of overlapping features in multiclass classification of CXR images. Also, we proposed an improved U-Net Lung segmentation dedicated to obtaining the radiological features for classification. The proposed approach achieved a maximum precision of 98% and accuracy of 97% in multiclass lung disease classification. By leveraging XAI techniques with the evident improvement of 4.75%, specifically LIME and Grad-CAM, to provide detailed and accurate explanations for the model’s prediction.
Keywords: Early disease detection, Explainable artificial intelligence, Transfer learning, Segmentation, Lung disease detection, LIME and Grad-CAM
Subject terms: Data processing, Machine learning, Computational biology and bioinformatics, Health care, Mathematics and computing
Introduction
Each year, pulmonary illnesses, including COVID-19, Bacterial Pneumonia, Viral Pneumonia, and Tuberculosis (TB), claim the lives of millions of people1. The ratio is intended to increase annually. The global healthcare system is under immense strain due to the severe COVID-19 pandemic and the exponential rise in COVID-19 patients. Likewise, tuberculosis and pneumonia can be fatal illnesses. DL-based techniques have demonstrated high accuracy in CXR image classification2. Though more effective at generating sharp images, computerized tomography (CT) and magnetic resonance imaging (MRI) are radiation-intensive and highly costing procedures. Therefore, the best option for identifying pulmonary illnesses is to use CXR images3. The lung condition has comparable symptoms, such as fever, coughing, and breathing difficulties. Conversely, TB has a delayed onset of illness and a more extended incubation period. Therefore, prompt and accurate identification of such disorders is essential to provide the right care and save lives.
Deep learning algorithms can accurately detect and categorize various diseases without requiring human intervention4–6. Deep learning is more successful than machine learning as the network grows because more profound data representation is possible with more extensive networks. It handles and distinguishes the morphological structures and identifies the classes7. As a result, the model automatically gathers attributes and results in more precise outcomes. The main goal of deep learning is extracting and classifying information from images. It exhibits promising revolution in several sectors, including healthcare.
Furthermore, deep learning can be applied to creating models that use CXR images to forecast and diagnose diseases precisely. Conversely, doctors can employ contemporary technologies like AI and imaging inputs like heatmaps and comparable data as decision-support tools to reduce human error and boost diagnosis accuracy8. Techniques for deep learning differ significantly from those for conventional machine learning. They can construct the input model as the network’s size grows. Consequently, the model automatically collects data and produces more accurate results. Unlike machine learning methods, deep learning models describe features through a series of non-linear functions combined to maximize the model’s accuracy9.
Most researchers and professionals use traditional AI models as “black-box” models for various activities, including medical diagnostics. These conventional AI techniques lack the information and justifications needed to support doctors in reaching improved conclusions. This prospect is made possible by XAI, which converts AI-based black-box models to more explainable and explicit gray-box representations10.
The primary drawbacks of the inability to distinguish different lung conditions from one another are solved with our proposed work with the following significant contributions:
Designed an improved U-Net segmentation specifically for multi-class lung disease classification on CXR images.
Developed a specialized transfer learning framework using four pre-trained models, enabling precise classification accuracy in CXR images.
Proposed an XAI-TRANS, an automated interpretation framework on CXR image classification decisions using a LIME-based heatmap, and Grad-CAM has been implemented.
Evaluated results against traditional methods and examined each component’s impact, ensuring the proposed approach’s quality with accuracy, precision, recall and F1-Score metrics.
Related works
Several experiments have recently been conducted to address the COVID-19 outbreak using machine learning approaches. For the early detection of infected cases, researchers suggested a multi-level threshold with a Support Vector Machine (SVM) classifier11. In the beginning, a multi-level threshold technique was used to extract features. Next, an SVM classifier was used to analyze the 40 contrast-enhanced CXR images’ characteristics that were retrieved, and 97% classification accuracy was attained. The authors used an enhanced SVM classifier in a different study to identify COVID-19 instances12. Apostolopoulos et al. have developed a DL-based machine-learning technique to identify COVID-19 instances13. The technique was utilized for binary and multiclass analysis, and the dataset included 500 no-finding images, 700 bacterial pneumonia cases, and 224 COVID-19 X-rays. Concerning binary (COVID-19 vs. No-findings) and multi-class (COVID-19 vs. No-findings vs. pneumonia), the suggested model yields high accuracy results of 98.78% and 93.48%, respectively. Hemdan et al.14 also developed a DL-based COVID-19 case detection approach utilizing X-ray images. The suggested method was contrasted with seven existing DL-based COVID-19 case detection methods. The technique was solely used to classify binary classes, and an accuracy rate of 74.29% was calculated. Three distinct automated lung disease detection techniques have been established based on three distinct DL models-ResNet50, InceptionV3, and InceptionResNetv215. To automatically detect infected regions, Bandyopadhyay et al.16 created a hybrid model based on two distinct ML techniques: Long Short-Term Memory (LSTM) and Gated Recurrent Unit (GRU). About the confirmed COVID-19 cases, the suggested technique achieved 87% accuracy. To automatically classify from CXR, the ADL approach was provided in17. The suggested model yielded an accuracy of 89.5%, precision of 97%, and recall of 100%.
A multi-dilation deep learning method (CovXNet) was presented to detect COVID-19 and other pneumonia cases from CXR images automatically18. Two distinct datasets were used for experiments to assess CovXNet’s performance. 5,856 X-ray images comprised the first dataset, while 305 X-ray images of various COVID-19 patients comprised the second dataset. According to the findings, the CovXNet approach successfully classified classes in 90.2% of cases, detecting COVID or Normal with an accuracy of 97.4%, binary classes with 96.9%, and multiclass classes with 90.2% .
Several pre-trained models, including VGG16, VGG19, ResNet50, DenseNet121, Xception, and capsule networks, have served as the foundation for designing and developing additional deep learning techniques19–25. Using a Generative Adversarial Network (GAN) and deep transfer learning, Loey et al.26 could diagnose COVID-19 from X-ray images. Three distinct transfer learning pre-trained models-AlexNet, GoogleNet, and RestNet18-were used in the suggested methodology. A set of datasets comprising 69 COVID-19, 79 pneumonia viruses, 79 pneumonia bacteria, and 79 normal patients were used to test the approach. The experimental results showed that the maximum accuracy rate (99.9%) for binary class classification issues is achieved when pre-trained GoogleNet is combined with GAN.Grad-CAM was used to show the extracted region on CXR images using the attention model and VGG1627,28. The goal of the Ashan et al.29 study was to identify COVID-19 cases based on CXR and CT images and to use six deep CNN learning models, such as VGG16,400 CXR and 400 CT images, to train MobileNetV2, InceptionResNetV2, ResNet50, ResNet101, and VGG19. MobileNetV2 outperformed NasNetMobile, with an average accuracy of 82.94% on a dataset with CT images and 93.94% on a dataset with CXR images. In addition to CXR image analysis, DL models have recently been used to study CT image-based detection of TB, COVID-19, and pneumonia. For example, Li et al.30 employed a pre-trained ResNet50 model to identify pneumonia from COVID-19 in CT scans. To increase the explainability of the model, they used GradCAM to display important regions in the CT images after training the DL model using 4356 CT data, resulting in a 95% accuracy rate. However, there aren’t enough heat maps to pinpoint the algorithm’s distinct characteristics to make predictions.
Ukwuoma et al. proposed a vision transformer-based framework for automated lung-related pneumonia and COVID-19 detection, demonstrating the growing interest in leveraging transformer architectures for medical imaging tasks31. Also, Addo et al. developed EVAE-Net, an ensemble variational autoencoder model that demonstrated strong classification performance for COVID-19 diagnosis using CXR data32. Lastly, Monday et al. handled the issue of low-quality images by introducing WMR-DepthwiseNet, a wavelet multi-resolution convolutional neural network designed specifically for COVID-19 diagnosis33. This underlines the importance of integrating explainable AI and segmentation techniques, as pursued in the proposed XAI-TRANS framework, to enhance both transparency and diagnostic reliability in lung disease classification.
Explainable Artificial Intelligence (XAI) is a set of techniques and practices developed to make AI models more transparent, interpretable, and understandable. AI offers previously unseen benefits for various everyday tasks, including manufacturing, finance, and entertainment, which include increased efficiency and broader data analysis34,35. However, high-risk systems, especially the healthcare industry are lagging in the application of AI36. The problem with many lung disease classification AI algorithms is that they are often a black box that humans cannot easily comprehend. XAI aims to address this issue by helping healthcare professionals better understand the decisions made by these models. By improving transparency and interpretability in AI models, XAI plays a crucial role in enabling medical professionals to trust and interpret the decisions made by these models37. This understanding leads to better patient care and treatment planning while ensuring adherence to regulatory requirements38.
Proposed methodology
This research presents an XAI approach for detecting multiple lung diseases and assessing healthy lung conditions using transfer learning with CXR images. Improved U-Net lung segmentation is performed before the classification. Employed the transfer learning techniques with VGG16, VGG19, InceptionV3, and ResNet50 models. Our approach involves inception-based transfer learning leveraging XAI techniques, specifically LIME and Grad-CAM, to provide detailed and accurate explanations for predictions related to COVID-19 and other lung diseases. Basic comparisons for all models experimented, and the inception is integrated with the proposed XAI-TRANS model as it outperformed the other models.
Data description
The Dataset used in this research is the Lung Disease Dataset39. This dataset comprises a diverse collection of 7560 Chest X-ray images, consisting of five classes: Bacterial Pneumonia, Viral Pneumonia, COVID-19, Tuberculosis and Normal as depicted in Fig.1. This dataset contains 1550 Bacterial Pneumonia samples, 1490 Viral Pneumonia samples, 1500 COVID-19 samples, 1460 Tuberculosis samples and 1560 Normal samples.
Normal: These images represent healthy lung conditions and serve as a reference point for comparison.
COVID-19: This category includes X-ray images from individuals with confirmed or suspected COVID-19 cases, helping in the early detection and tracking of the disease.
Viral Pneumonia: Images in this group are linked to cases of viral pneumonia caused by flu, RSV, SARS-Cov-2, etc., adding to our knowledge and ability to identify this particular lung infection.
Tuberculosis: These images feature tuberculosis cases, aiding in recognizing and understanding this infectious lung disease.
Bacterial Pneumonia: This category contains X-ray images associated with bacterial pneumonia cases commonly caused by Streptococcus, assisting in identifying and comprehending this type of lung infection.
Fig. 1.
Sample chest X-rays from each category of Lung disease.
Data preprocessing
Image preprocessing is carried out, namely image resizing and normalization. The dataset is assessed and splitting were carried out for the model training and testing as depicted in Table 1.
Table 1.
Dataset Split for training, validation and testing of the proposed XAI-TRANS model.
| Data Split/Class | Bacterial Pneumonia | Viral Pneumonia | COVID-19 | TB | Normal | Total |
|---|---|---|---|---|---|---|
| Training | 900 | 875 | 885 | 830 | 930 | 4420 |
| Validation | 330 | 305 | 315 | 300 | 310 | 1560 |
| Testing | 320 | 310 | 300 | 330 | 320 | 1580 |
Resizing the image: As every image in the collected dataset are of varied sizes, all the images are resized to the fixed resolution of 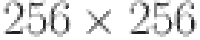 pixels to maintain uniform dimensions in the dataset. This resizing operation was an essential step to ensure consistency and compatibility with our chosen model.
pixels to maintain uniform dimensions in the dataset. This resizing operation was an essential step to ensure consistency and compatibility with our chosen model.
Normalization: Every pixel value in the images ranges between 0 and 255. Normalizing the pixel values of images by dividing by 255 scales the pixel values to a range of [0,1]. This normalization facilitates faster convergence during model training and helps stabilize and improve the neural network’s performance.
Image augmentation techniques
Image augmentation is carried out by applying transformations and modifications like rotations, flips, zooms, shifts, changes in brightness or contrast, etc., to the existing images; it generates new variations of images while preserving the underlying content. By presenting the model with a broader range of variations of the same image, it learns to better recognize objects despite changes in lighting conditions, viewpoints, orientations and other factors. This ultimately leads to more accurate and reliable performance when the model encounters new, unseen data. It can also be used to address class imbalance problems in datasets. The parameters used were a zoom range of 0.2, a shear range of 0.2 and horizontal flipping.
Zooming: This method randomly zooms the image in or out of the images. Thus, it amplifies an image to highlight specific components, enabling the model to capture important details.
Shearing: Modifying an image along a specific axis creates a different perception angle and improves the model’s understanding of diverse viewpoints.
Random flipping: Horizontal flipping is applied in random images to augment dataset diversity further.
This image augmentation process unfolds dynamically during the model training phase, occurring on-the-fly as the model is constructed. By incorporating these pre-processing and augmentation techniques, we equip our model to adeptly accommodate variations in image quality, size, and perspective, culminating in a notable enhancement in the accuracy of our lung disease detection capabilities. We assigned equal weightage to our image augmentation process’s flip, shearing, and zooming techniques. Our approach aimed to ensure a balanced application of these augmentation operations, intending to introduce diversity into the training dataset while avoiding overemphasizing any particular operation. By giving equal weightage to these techniques, we sought to maintain a well-rounded and representative dataset that captures various aspects of variation and distortion that the model might encounter in real-world scenarios. This balanced approach helps enhance the model’s robustness and generalization capabilities, making it better equipped to handle various input variations40.
Improved U-Net lung segmentation
Lung anomalies reveal details about a wide range of illnesses which has more overlapping radiological features, such as ground glass opacities (GGO), paving patterns, reverse halo signs etc. To ascertain what type of lung condition the patient is affected with, our study looks at the CXR with a radical approach. An extra lung segmentation component is added to the pipeline to improve the performance of the proposed model’s detection and explanation components. Manual segmentation is not feasible for CXR images since it has air bronchograms and cavitary lesions. Furthermore, errors and contradictions can arise from human annotations. Thus, the proposed Lung segmentation forces the segmentation of multiclass images and its explanation networks to only detect and explain in the lung sector of the images by feeding them veiled lung images. The explanation section’s output is displayed in the entire CXR for a more accurate understanding.
The U-Net model is a well-known deep-learning architecture designed for image segmentation tasks, such as lung segmentation. The proposed model is shown in Fig.2. Using chest X-rays with dimensions of 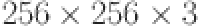 as input images produces binary segmentation masks representing the lungs. The U-Net model was trained on a publicly available dataset containing chest X-ray images and their corresponding masks41,42. The U-Net model uses encoder and decoder structures to process the input images and generate a binary mask representing the lungs. The encoder captures high-level abstractions through convolutional and pooling layers, gradually reducing the spatial dimension and increasing the number of filters to extract hierarchical features. On the other hand, the decoder pathway utilizes skip connections that concatenate feature maps from corresponding layers in the encoder, allowing the recovery of the original image resolution while retaining crucial contextual information. Deconvolution layers (transposed convolutions) have been included to enhance the feature maps. The model outputs a single pixel-wise probability map with a sigmoid activation function. The final output is a binary segmentation mask of the lungs. This improved U-Net effectively analyze X-ray images of the lung by segmenting and interpreting all parts, including the infected and uninfected regions in the lung. We found that including the perimeter outside the lung does not significantly affect the analysis. Still, it is crucial to include the entirety for early diagnosis. Segmentation of the lung is carried out before the classification and explanations.
as input images produces binary segmentation masks representing the lungs. The U-Net model was trained on a publicly available dataset containing chest X-ray images and their corresponding masks41,42. The U-Net model uses encoder and decoder structures to process the input images and generate a binary mask representing the lungs. The encoder captures high-level abstractions through convolutional and pooling layers, gradually reducing the spatial dimension and increasing the number of filters to extract hierarchical features. On the other hand, the decoder pathway utilizes skip connections that concatenate feature maps from corresponding layers in the encoder, allowing the recovery of the original image resolution while retaining crucial contextual information. Deconvolution layers (transposed convolutions) have been included to enhance the feature maps. The model outputs a single pixel-wise probability map with a sigmoid activation function. The final output is a binary segmentation mask of the lungs. This improved U-Net effectively analyze X-ray images of the lung by segmenting and interpreting all parts, including the infected and uninfected regions in the lung. We found that including the perimeter outside the lung does not significantly affect the analysis. Still, it is crucial to include the entirety for early diagnosis. Segmentation of the lung is carried out before the classification and explanations.
Fig. 2.
Improved U-Net architecture for lung segmentation.
Proposed XAI-TRANS workflow architecture
Transfer Learning has found significant success with the rise of deep learning and enables the rapid development of high-performing models and many applications in Natural Language Processing (NLP), image classification and reinforcement learning. It can lead to faster convergence and better model performance, mainly when the new task has limited labelled data. Using the learned characteristics from the first task as a starting point allows the model to understand the second task more quickly and effectively. This can prevent overfitting by enabling the model to learn general features relevant to the second task. Further, it decreases the time-consuming training process and allows the creation of a model with high classification performance43. Ultimately, transfer learning not only enhances accuracy in COVID-19 detection by leveraging pre-learned image features but also plays a crucial role in addressing the global impact of the pandemic, potentially saving lives through swift and precise identification of affected individuals44.
The proposed XAI-TRANS model used the pre-trained weights instead of training a model from scratch for a new task since it adapts the knowledge by fine-tuning it on the new dataset. This process involves updating the model’s parameters to fit the new task better. Lung disease detection is integrated with the improved U-Net model, classification and the explainability as shown in Fig.3. Here, pre-trained convolutional neural networks such as VGG’s, ResNet50, and Inception etc have learned a rich hierarchy of features from vast and diverse image classes. Transfer learning empowers the reuse of these learned features, allowing the development of highly accurate models with substantially reduced data requirements45.VGG16 was selected for its simplicity and strong performance in transfer learning tasks, particularly with limited datasets. ResNet50 was chosen for its deep architecture and skip connections, which help mitigate vanishing gradients and capture complex features effectively in medical images.
Fig. 3.
Proposed XAI-TRANS architecture for lung disease classification on CXR images.
VGG16
In VGG16, VGG refers to the Visual Geometry Group of the University of Oxford, while the number 16 refers to its layers. It has 13 convolutional layers and 3 fully connected layers. VGG16 network is a deep Convolutional Neural Network with 1000 outcomes and capable of handling 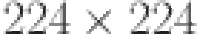 images. Instead of having many hyper-parameters, VGG16 uses a
images. Instead of having many hyper-parameters, VGG16 uses a  filter in convolutional layers and a stride of 1 in the same padding across the entire network. It uses a max pool layer of
filter in convolutional layers and a stride of 1 in the same padding across the entire network. It uses a max pool layer of  of stride 2. It follows convolution and maxpool layers arrangement consistently throughout the architecture. VGG16 is widely used for image classification tasks. A pre-trained version of the VGG16 network is trained on over one million images from the ImageNet database. We can use the pre-trained version of VGG16 as the base model and remove its fully connected layer. Here, the CNN is used as a fixed feature extractor and allows additional layers and fine-tuning based on our classification.
of stride 2. It follows convolution and maxpool layers arrangement consistently throughout the architecture. VGG16 is widely used for image classification tasks. A pre-trained version of the VGG16 network is trained on over one million images from the ImageNet database. We can use the pre-trained version of VGG16 as the base model and remove its fully connected layer. Here, the CNN is used as a fixed feature extractor and allows additional layers and fine-tuning based on our classification.
VGG19
To put it simply, VGG19 is a state-of-the-art convolutional neural network. It includes 16 convolutional layers, 3 fully connected layers, 5 max pool layers, and 1 softmax layer; the VGG19 model is rather complex. Any convolution filter’s reception field is just  in stride 1. Row and column padding are used following convolution to preserve spatial resolution. The largest pooling window is
in stride 1. Row and column padding are used following convolution to preserve spatial resolution. The largest pooling window is  strides46. VGG19 borrows its model architecture from its predecessor, VGG16. When pitted against VGG16, VGG19 performs somewhat better. A pre-trained version of VGG19 is also available, which can be loaded and used. VGG16 is one of the popular networks, trained with more than a million images from the ImageNet dataset with 1,000 different classes. Therefore, the model can be applied as a helpful tool for the feature extractor of new images47.
strides46. VGG19 borrows its model architecture from its predecessor, VGG16. When pitted against VGG16, VGG19 performs somewhat better. A pre-trained version of VGG19 is also available, which can be loaded and used. VGG16 is one of the popular networks, trained with more than a million images from the ImageNet dataset with 1,000 different classes. Therefore, the model can be applied as a helpful tool for the feature extractor of new images47.
ResNet50
ResNet (Residual Network) is a type of deep convolutional neural network (CNN) proposed in 2015 by Kaiming He et al.48at Microsoft Research Asia, has 50 layers deep with 48 convolution layers, 1 Max pool layer, and 1 Average pool layer, was pre-trained on the ImageNet dataset, which has 1000 different classes at resolution 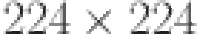 . This model has more than 23 million trainable parameters. The ResNet50 architecture consists of four main parts: convolutional layers, identity block, convolutional block, and fully connected layers. The key element of the ResNet50 architecture is skip connections, sometimes referred to as residual connections. They enable the network to learn deeper architectures without encountering the issue known as vanishing gradients, which arises when the deeper layer’s parameter gradients get too small for the network to learn from. The identity and convolutional blocks in ResNet50 make use of skip connect. After the input has passed through multiple convolutional layers, the identity block adds it back to the output. On the other hand, the convolutional block reduces the number of filters before the
. This model has more than 23 million trainable parameters. The ResNet50 architecture consists of four main parts: convolutional layers, identity block, convolutional block, and fully connected layers. The key element of the ResNet50 architecture is skip connections, sometimes referred to as residual connections. They enable the network to learn deeper architectures without encountering the issue known as vanishing gradients, which arises when the deeper layer’s parameter gradients get too small for the network to learn from. The identity and convolutional blocks in ResNet50 make use of skip connect. After the input has passed through multiple convolutional layers, the identity block adds it back to the output. On the other hand, the convolutional block reduces the number of filters before the  convolutional layer by using a
convolutional layer by using a  convolutional layer before adding the input back to the output. ResNet50 uses skip connections to improve training efficiency and avoid vanishing gradients, allowing the network to learn deeper architectures.
convolutional layer before adding the input back to the output. ResNet50 uses skip connections to improve training efficiency and avoid vanishing gradients, allowing the network to learn deeper architectures.
InceptionV3
The InceptionV3 is a Convolutional Neural network-based deep learning model used for image classification proposed by Szegedy et al. in the paper titled “Rethinking the Inception Architecture for Computer Vision” in 201549. It is an improved version of the Inception V1 model, introduced by Google Net in 2014. The inceptionV3 model is 48 layers deep with under 25 million parameters. It is an improved version of the Inception V1 model, introduced as GoogLeNet in 2014.InceptionV3 is a modified version of the previous Inception architectures focusing on reducing computational power. It introduces inception modules that use multiple parallel filter sizes ( ) to capture features at different scales. This design improves parameter efficiency by using
) to capture features at different scales. This design improves parameter efficiency by using  convolutions for dimensionality reduction before more extensive convolutions. Additionally, it includes auxiliary classifiers as regularised during training to encourage the network to learn more discriminative features at intermediate layers.
convolutions for dimensionality reduction before more extensive convolutions. Additionally, it includes auxiliary classifiers as regularised during training to encourage the network to learn more discriminative features at intermediate layers.
Explainable artificial intelligence
The study uses XAI techniques such as LIME and Grad-CAM to identify significant features and regions in chest X-ray images with interpretability of the transfer learning model results. Thereby improving the accuracy of diagnoses and facilitating effective communication between medical experts and patients.
LIME
In our research, we utilized LIME to clarify the predictions of the proposed DL model. LIME, which stands for Local Interpretable Model-agnostic Explanations, is a well-known XAI technique intended to offer local interpretability for individual predictions made by any black-box model, regardless of its underlying algorithm. The primary objective of LIME is to approximate the complex model locally with a simpler, interpretable surrogate model, providing insight into the decision-making process in the vicinity of the original input. LIME functions by slightly altering the input data and measuring the impact of these alterations on the original model’s predictions. It then fits a locally interpretable surrogate model (such as logistic regression) to explain the prediction differences. The central idea behind LIME is that if the input data were perturbed just enough, the simpler surrogate model should exhibit similar behaviour to the black-box model in the local area around the original input. By creating heatmaps, LIME identified the essential areas of input images that influenced the model’s decision-making process. These heatmaps assigned relative importance scores to different pixels or regions of the images based on their contribution to the model’s predictions50,51.
GRAD-Cam
Gradient-weighted Class Activation Mapping (Grad-CAM) is an XAI technique used primarily for interpreting and visualizing the internal workings of convolutional neural networks. It grants users insight into which parts of an input image bear the greatest weight in the model’s decision-making process, contributing to enhanced trust, transparency, and clarity in AI systems52.Grad-CAM operates based on the output’s gradients for the convolutional layers’ activations. It calculates a gradient of the output class score concerning the feature maps produced by the final convolutional layer before the fully connected layers53. The intermediate feature maps then weigh the gradient to create a class activation heatmap. Regions with higher activation scores in the CAM correspond to areas in the input image that have significantly contributed to the model’s prediction and the results are shown in Fig.4. Grad-CAM is a useful technique for visually interpreting a CNN’s decision-making process54. Applications of Grad-CAM include image classification, object detection, semantic segmentation, medical imaging, and autonomous vehicles. Each application leverages Grad-CAM’s ability to identify the most influential regions in input images for the model’s prediction, helping users refine models, address biases, and improve overall performance.
Fig. 4.
Grad-CAM explanation on chest X-ray images.
Results and discussions
This study presents the results for binary classification into COVID-19 and normal and multiclass classification into normal and other diseased lung conditions such as bacterial pneumonia, viral pneumonia, COVID-19, and tuberculosis. Dense objects, like metal blocks and bones, appear white under X-rays since they absorb the radiation. The least dense regions, like the lungs, will be black, and less dense areas will appear in grey tones. The CXR assessment can identify lung complications like any other relevant researches55 implemented.
Experimental setup
The experiments were conducted on a system with the following specifications: an Intel Core i5 processor, 8 GB RAM, with NVIDIA Quadro K600 GPU and a 64-bit Windows Operating System. The experiments were conducted using the Python programming language, and libraries such as pandas, numpy, matplotlib,keras and tensorflow were utilized.
Evaluation metrics
Assessment of Lung Disease detection model performance relies heavily on the confusion matrix. This offers a summary of predicted and actual class labels, facilitating the evaluation of true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN). We can get insights into the model’s accuracy, precision, recall and F1-score, which are crucial for evaluating the efficacy across different classes.
| 1 |
| 2 |
| 3 |
| 4 |
Tunning the hyper parameters
Learning Rate: The learning rate is a hyperparameter that determines the size of the optimisation algorithm’s steps while searching for the minimum loss function. If the learning rate is too small, learning will be slow. Learning may be unstable and fail to converge if it’s too large. We used 0.001 as the learning rate after considering both convergence speed and stability. This choice facilitates gradual parameter adjustments, reducing the risk of overshooting during optimization. Specifically tailored for our COVID-19 detection task, it ensures consistent progress during training, mitigating erratic behaviour.The hyperparameters used in the proposed XAI-TRANS is given in Table 2.
Table 2.
Hyperparameters used in the implementation of proposed XAI-TRANS model.
| Hyperparameters | Specifications |
|---|---|
| Learning Rate | 0.001 |
| Cross validation | 5 fold |
| Optimizer | Adam |
| Activation Function | ReLu and Softmax |
| Batch size | 32 |
| Epochs(Binary classification) | 5 |
| Epochs(Multiclass classification) | 10 |
| Dropout | 0.2 |
Optimizer: It is an algorithm used to find the optimal set parameters of a neural network during training that results in the best performance and minimizes the error. Optimizers work by iteratively updating the parameters of the neural network based on the gradient of the loss function concerning those parameters. We used Adam as an optimizer since it combines the advantages of two other optimization algorithms, RMSprop and momentum. Adam maintains two moving averages of gradients: the first moment (the mean) and the second moment (the uncentered variance). These moving averages adaptively adjust the learning rate for each parameter, leading to faster convergence and improved performance.
Loss function: A loss function measures the difference between a neural network’s predicted output and target values during training. The purpose of training a neural network is to reduce the loss function, which improves the model’s ability to predict accurately. This study uses a loss function called “categorical cross-entropy loss,” which is used for a multi-class classification model with more than one output label and is one-hot encoded.
Activation function: Here, the Rectified Linear Unit (ReLU) is the activation function in our densely connected layers. ReLU has become widely adopted in deep learning because it addresses the vanishing gradient problem and expedites convergence. ReLU enables the model to grasp intricate features within the data by introducing non-linearity. This activation function is well-suited for our task as it enhances the network’s capacity to discern and encode complex patterns in radiological images, ultimately enhancing diagnostic accuracy.
Analysis on binary and multi-class classification model
Table 3 presents the results for binary classification into COVID-19 and normal. For multiclass classification, the classes are normal, bacterial pneumonia, viral pneumonia, COVID-19, and tuberculosis. The CXR assessment can identify lung complications based on the image appearance. Dense objects, like metal blocks and bones, appear white under X-rays since they absorb the radiation. The least dense regions, like the lungs, will be black, and less dense areas will appear grey. Based on these tones, our classifier distinguishes the different lung conditions.
Table 3.
Results obtained for the binary and multiclass classification on CXR images.
| Methods/Metrics | Multiclass Classifications | Binary Classification | ||||||
|---|---|---|---|---|---|---|---|---|
| ACC | PRE | REC | F1 | ACC | PRE | REC | F1 | |
| InceptionV3 | 0.9506 | 0.9691 | 0.9495 | 0.9592 | 0.9753 | 0.9895 | 0.9701 | 0.9797 |
| ResNet50 | 0.9460 | 0.9651 | 0.9457 | 0.9453 | 0.9704 | 0.9853 | 0.9661 | 0.9756 |
| VGG19 | 0.9266 | 0.9485 | 0.9298 | 0.9391 | 0.9501 | 0.9681 | 0.9495 | 0.9587 |
| VGG16 | 0.9117 | 0.9270 | 0.9091 | 0.9180 | 0.9239 | 0.9457 | 0.9279 | 0.9367 |
The models were developed using transfer learning, with segmented CXR images. Weighted averages were used in the calculation process to account for sample size and yield a more precise representation of the averages. Segmentation of lungs is carried out to further enhance the performance of DL models56. On investigating the pre-trained models with segmented CXR images as inputs, the InceptionV3 model stands out in detecting COVID-19 cases with 98% accuracy. This remarkable accuracy, with an F1-score of 0.98, shows the model’s exceptional proficiency in accurately classifying COVID-19 positive and negative cases. The ResNet50, VGG16 and VGG19 models also indicate a similar classification performance with an accuracy of 97% and with F1-score of 0.97. Together, these accuracy scores demonstrate the models’ ability to produce precise predictions, pointing to their potential for efficient COVID-19 detection.
As illustrated in Fig.5 for the InceptionV3 model, the matrix illustrates 127 true positive predictions and 126 true negative predictions, with 3 single false positive predictions and only 4 false negative predictions. The performance of the InceptionV3 model is further improved with the proposed improved U-Net segmented CXR images as input, depicted in Fig.6 with 130 true positive predictions and 129 true negative predictions. On evaluating precision, which measures the models’ ability to accurately predict the positive cases among the predicted positives, we observed a perfect precision of 0.98 for InceptionV3. Similarly, recall, which indicates the models’ ability to identify actual positives among all positive cases, of 0.98, reflecting the models’ effectiveness in both true positive prediction and positive class identification.
Fig. 5.
Confusion matrix of Binary Classification without segmentation.
Fig. 6.
Confusion matrix of Binary Classification with proposed improved UNet segmentation.
Table 4 compares four DL models for multiclass classification. The evaluation of fine-tuned deep learning models to detect various lung diseases reveals insightful findings, which are summarized. It shows various performance metrics illuminating each model’s effectiveness in accurately identifying lung disease from medical images. Among the various sets of models, the proposed XAI-TRANS stands out as the exemplar with a perfect accuracy rate of 97%, with an F1-score of 0.97, precision of 0.98 and recall of 0.97. The other models also significantly performed well, whereas the ResNet50 with a good accuracy of 93% . VGG19 achieved an accuracy rate of 90%, and VGG16 achieved the lowest accuracy rate of 83%. Unlike another model, the proposed model once again proved the efficacy of the explainable AI and the influence of the improved U-Net segmentation with evident improvements in metrics.
Table 4.
Comparative analysis of the metrics with state-of-the-art methods.
| Model | ACC | PRE | REC | F1 |
|---|---|---|---|---|
| VGG16 | 0.8741 | 0.9109 | 0.8952 | 0.9030 |
| VGG19 | 0.9081 | 0.9316 | 0.9153 | 0.9234 |
| ResNet50 | 0.9166 | 0.9476 | 0.9307 | 0.9391 |
| InceptionV3 | 0.9311 | 0.9514 | 0.9344 | 0.9428 |
| Proposed XAI-TRANS | 0.9753 | 0.9895 | 0.9701 | 0.9797 |
| Improvements in % | +4.75 | +4.00 | +3.82 | +3.91 |
For multi-class classification, almost all the five classes are classified correctly by all the models. The inceptionv3 model is classified much better than the remaining models with the least misclassification, which is illustrated in Fig.7.
Fig. 7.
Confusion matrix of multi-class classification of lung diseases.
XAI-TRANS results on CXR images
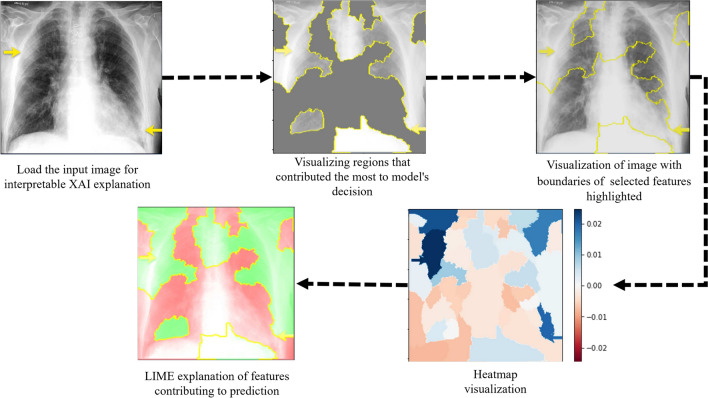
The LIME interpretable model was used for the explainability of lung diseases. The main objective is to provide interpretable explanations for individual predictions of complex deep learning models to gain insights into why the model made its prediction, for instance. This step involves analyzing the visualizations and understanding the contributions of individual features or regions to the model’s decision, thereby increasing trust and transparency in the model’s behaviour. LIME takes the predicted image and provides the step-by-step explanation, shown in Fig.8. The input image shows the region highlighted by a yellow arrow by the doctor, and he describes that these are the main regions that caused COVID-19.
Fig. 8.
Explainability LIME representation on CXR images.
First, Choose the instance for which you want to generate an explanation. Next, it generates boundaries in the input image and finds the distance between the actual and predicted feature maps by creating perturbations. This can be done by making small changes to the instance while keeping its overall characteristics intact. Fit an interpretable model to the perturbed samples. Calculate the explanations for the instance based on the coefficients or feature importance values obtained from the interpretable model. Then, it shows the distance explanation by creating heatmaps.
Further, it creates a visualization highlighting the important features or regions of the instance, represented with blue color in the heatmap and green regions in LIME explanations that influenced the model’s prediction. Grad-CAM is a powerful technique that provides visual insights into why the model makes specific predictions. After the model’s prediction, Grad-CAM can elucidate which regions of the X-ray were instrumental in the model’s decision-making process, as given in Fig 8. Grad-CAM retraces the network’s steps to understand which parts of the image were crucial for the prediction. Next, Grad-CAM computes a weighted sum of these activation maps, where the gradients and the activation values determine the weights. This process generates a heatmap highlighting the regions of the chest X-ray image that significantly contribute to the model’s prediction as resulted in the Fig.9.
Fig. 9.
Proposed XAI-TRANS with Grad-CAM explanation on chest X-Ray images of lung disease.
Significance of the proposed work
The proposed XAI-TRANS integrated with two techniques: LIME and Grad-CAM. The reason for applying both is to evaluate the classification models thoroughly since they work differently are tested with various ablation trials, as depicted in Table 5.Over each epochs the nodes in the networks learns better and the convergence is evident as shown in the Fig 10. They have some significant differences and highlights: (i) LIME is model-agnostic, and Grad-CAM is model-specific; (ii) in LIME, the granularity of important regions is correlated to the granularity of the super-pixel identification algorithm; (iii) Grad-CAM produces a very smoothed output because the dimension of the last convolution layer is much smaller than the dimension of the original input. (iv) The improved U-Net segmentation extracts distinguished features, and transfer learning handles the feature overlapping issues and cuts downs the rate of misclassification. Thus, we can use a more comprehensive approach to increase the model reliability in a real-world context57. Table 4 confirms that the proposed XAI-TRANS results outperform the other state-of-the-art methods in which improvements are recorded and validated against ground truths.
Table 5.
Ablation trail results on different groups of the proposed XAI-TRANS model.
| Model | XAI | Segmentation | Augmentation | Preprocessing | ACC | PRE | REC | F1 |
|---|---|---|---|---|---|---|---|---|
| InceptionV3 | x | x | x | ✓ | 0.9156 | 0.9372 | 0.9225 | 0.9298 |
| x | x | ✓ | x | 0.9195 | 0.9400 | 0.9261 | 0.9330 | |
| x | x | ✓ | ✓ | 0.9217 | 0.9419 | 0.9279 | 0.9349 | |
| x | ✓ | x | x | 0.9250 | 0.9466 | 0.9289 | 0.9377 | |
| x | ✓ | x | ✓ | 0.9283 | 0.9505 | 0.9307 | 0.9405 | |
| x | ✓ | ✓ | x | 0.9311 | 0.9533 | 0.9325 | 0.9428 | |
| x | ✓ | ✓ | ✓ | 0.9311 | 0.9514 | 0.9344 | 0.9428 | |
| ✓ | x | x | x | 0.9339 | 0.9553 | 0.9353 | 0.9452 | |
| ✓ | x | x | ✓ | 0.9391 | 0.9592 | 0.9400 | 0.9495 | |
| ✓ | x | ✓ | x | 0.9448 | 0.9641 | 0.9447 | 0.9543 | |
| ✓ | x | ✓ | ✓ | 0.9506 | 0.9691 | 0.9495 | 0.9592 | |
| ✓ | ✓ | x | x | 0.9565 | 0.9741 | 0.9543 | 0.9641 | |
| ✓ | ✓ | x | ✓ | 0.9625 | 0.9792 | 0.9592 | 0.9691 | |
| ✓ | ✓ | ✓ | x | 0.9686 | 0.9843 | 0.9641 | 0.9741 | |
| ✓ | ✓ | ✓ | ✓ | 0.9753 | 0.9895 | 0.9701 | 0.9797 | |
| ResNet50 | x | x | x | ✓ | 0.9113 | 0.9335 | 0.9189 | 0.9261 |
| x | x | ✓ | x | 0.9151 | 0.9363 | 0.9225 | 0.9293 | |
| x | x | ✓ | ✓ | 0.9173 | 0.9381 | 0.9243 | 0.9312 | |
| x | ✓ | x | x | 0.9205 | 0.9428 | 0.9252 | 0.9339 | |
| x | ✓ | x | ✓ | 0.9249 | 0.9466 | 0.9270 | 0.9367 | |
| x | ✓ | ✓ | x | 0.9266 | 0.9495 | 0.9289 | 0.9391 | |
| x | ✓ | ✓ | ✓ | 0.9166 | 0.9476 | 0.9307 | 0.9391 | |
| ✓ | x | x | x | 0.9294 | 0.9514 | 0.9316 | 0.9414 | |
| ✓ | x | x | ✓ | 0.9345 | 0.9553 | 0.9363 | 0.9457 | |
| ✓ | x | ✓ | x | 0.9402 | 0.9602 | 0.9409 | 0.9505 | |
| ✓ | x | ✓ | ✓ | 0.9460 | 0.9651 | 0.9457 | 0.9453 | |
| ✓ | ✓ | x | x | 0.9518 | 0.9701 | 0.9505 | 0.9602 | |
| ✓ | ✓ | x | ✓ | 0.9577 | 0.9751 | 0.9553 | 0.9651 | |
| ✓ | ✓ | ✓ | x | 0.9637 | 0.9802 | 0.9602 | 0.9701 | |
| ✓ | ✓ | ✓ | ✓ | 0.9704 | 0.9853 | 0.9661 | 0.9756 | |
| VGG19 | x | x | x | ✓ | 0.8933 | 0.9180 | 0.9038 | 0.9109 |
| x | x | ✓ | x | 0.8970 | 0.9207 | 0.9073 | 0.9140 | |
| x | x | ✓ | ✓ | 0.8991 | 0.9225 | 0.9091 | 0.9057 | |
| x | ✓ | x | x | 0.9022 | 0.9270 | 0.9100 | 0.9184 | |
| x | ✓ | x | ✓ | 0.9054 | 0.9307 | 0.9117 | 0.9211 | |
| x | ✓ | ✓ | x | 0.9081 | 0.9335 | 0.9135 | 0.9134 | |
| x | ✓ | ✓ | ✓ | 0.9081 | 0.9316 | 0.9153 | 0.9234 | |
| ✓ | x | x | x | 0.9108 | 0.9153 | 0.9162 | 0.9257 | |
| ✓ | x | x | ✓ | 0.9156 | 0.9391 | 0.9207 | 0.9298 | |
| ✓ | x | ✓ | x | 0.9211 | 0.9438 | 0.9252 | 0.9344 | |
| ✓ | x | ✓ | ✓ | 0.9266 | 0.9485 | 0.9298 | 0.9391 | |
| ✓ | ✓ | x | x | 0.9322 | 0.9533 | 0.9344 | 0.9438 | |
| ✓ | ✓ | x | ✓ | 0.9379 | 0.9582 | 0.9391 | 0.9485 | |
| ✓ | ✓ | ✓ | x | 0.9437 | 0.9631 | 0.9438 | 0.9533 | |
| ✓ | ✓ | ✓ | ✓ | 0.9501 | 0.9681 | 0.9495 | 0.9587 | |
| VGG16 | x | x | x | ✓ | 0.8401 | 0.8978 | 0.8843 | 0.8910 |
| x | x | ✓ | x | 0.8736 | 0.9004 | 0.8776 | 0.8940 | |
| x | x | ✓ | ✓ | 0.8756 | 0.9021 | 0.8793 | 0.8957 | |
| x | ✓ | x | x | 0.8786 | 0.9065 | 0.8902 | 0.8982 | |
| x | ✓ | x | ✓ | 0.8616 | 0.9100 | 0.8818 | 0.9008 | |
| x | ✓ | ✓ | x | 0.8841 | 0.9026 | 0.8835 | 0.9030 | |
| x | ✓ | ✓ | ✓ | 0.8741 | 0.9109 | 0.8952 | 0.9030 | |
| ✓ | x | x | x | 0.8867 | 0.9044 | 0.8961 | 0.9052 | |
| ✓ | x | x | ✓ | 0.8913 | 0.9180 | 0.9004 | 0.9091 | |
| ✓ | x | ✓ | x | 0.8965 | 0.9225 | 0.9147 | 0.9135 | |
| ✓ | x | ✓ | ✓ | 0.9117 | 0.9270 | 0.9091 | 0.9180 | |
| ✓ | ✓ | x | x | 0.9070 | 0.9316 | 0.9135 | 0.9225 | |
| ✓ | ✓ | x | ✓ | 0.9124 | 0.9363 | 0.9180 | 0.9270 | |
| ✓ | ✓ | ✓ | x | 0.9178 | 0.9409 | 0.9225 | 0.9316 | |
| ✓ | ✓ | ✓ | ✓ | 0.9239 | 0.9457 | 0.9279 | 0.9367 |
Fig. 10.
Accuracy and loss graph for the Proposed XAI-TRANS.
Conclusion and future works
This work presents an XAI method leveraging transfer learning with CXR images to diagnose various lung diseases, including COVID-19, bacterial pneumonia, viral pneumonia, and tuberculosis. Our proposed model makes accurate predictions and provides explanations, enhancing transparency and trust. Incorporating lung segmentation significantly improves the model’s classification and explanation performance. Heatmaps generated by XAI techniques such as LIME and Grad-CAM offer clear visualizations of the affected regions, aiding non-expert users in understanding the AI’s decisions. While lung segmentation increases processing time and the model’s reliance on a limited CXR dataset poses some challenges, additional data would enhance classification performance. The XAI component, despite its limitations of being a complex model, helps interpret CXR images effectively. When given a healthy CXR image, the model produces heatmaps indicating regions affected by different lung diseases, emphasizing the importance of the classification section. This study highlights the potential of XAI-TRANS approaches in healthcare, particularly for diagnosing lung diseases and providing insights into AI predictions. The noise removal in the CXR images is a another challenge that is to be addressed seriously in the future implementation for early diagnosis in real-time images. The proposed model demonstrates strong potential for real-world applications by aiding healthcare professionals in accurately diagnosing lung diseases, even under non-ideal conditions. The use of transfer learning enables quick and precise analysis, while XAI-TRANS facilitates interpretation of doubtful results, helping identify the correct disease state and severity. This supports the formulation of appropriate treatment plans and drug suggestions, ultimately leading to faster patient recovery with the right medicines in a shorter time.
Supplementary Information
Acknowledgements
Our earnest thanks to SASTRA Deemed University for providing research facilities at the Computer Vision and Soft Computing Laboratory to proceed with the research.
Author contributions
The contributions for this research article are as follows: formal analysis, Conceptualization, methodology, algorithm implementation, draft preparation, visualization, done by N.V., R.S., S.P., and S.S. Validation, formal analysis, supervision, and project administration by N.V. and P.J. All authors have read and agreed to the published version of the manuscript.
Funding
The author(s) received no financial support for this article’s research, authorship, and/or publication.
Data availability
The datasets generated and/or analysed during the current study are available in the Kaggle repository, can be downloaded from https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nirmala Veeramani, Email: nirmalaveeramani@ict.sastra.ac.in.
Premaladha Jayaraman, Email: premaladha@ict.sastra.edu.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-025-07603-4.
References
- 1.Mahbub, M. K., Biswas, M., Gaur, L., Alenezi, F. & Santosh, K. Deep features to detect pulmonary abnormalities in chest x-rays due to infectious diseasex: Covid-19, pneumonia, and tuberculosis. Information Sciences592, 389–401 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhandari, M., Shahi, T. B., Siku, B. & Neupane, A. Explanatory classification of cxr images into covid-19, pneumonia and tuberculosis using deep learning and xai. Computers in Biology and Medicine150, 106156 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubin, G. D. et al. The role of chest imaging in patient management during the covid-19 pandemic: a multinational consensus statement from the fleischner society. Radiology296(1), 172–180 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghadezadeh, M. et al. Deep convolutional neural network-based computer-aided detection system for covid-19 using multiple lung scans: Design and implementation study. Journal of Medical Internet Research23, 10.2196/27468 (2021). [DOI] [PMC free article] [PubMed]
- 5.Ghadezadeh, M., Aria, M. & Asadi, F. X-ray equipped with artificial intelligence: Changing the covid-19 diagnostic paradigm during the pandemic. BioMed Research International2021, 16. 10.1155/2021/9942873 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghadezadeh, M. et al. Efficient framework for detection of covid-19 omicron and delta variants based on two intelligent phases of cnn models. Computational and Mathematical Methods in Medicine2022, 1–10. 10.1155/2022/4838009 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Self, W.H., Courtney, D.M., McNaughton, C.D., Wunderink, R.G. & Kline, J.A. High discordance of chest x-ray and computed tomography for detection of pulmonary opacities in ed patients: implications for diagnosing pneumonia. The American journal of emergency medicine 31(2), 401–405 (2013) [DOI] [PMC free article] [PubMed]
- 8.Badža, M. M. & Barjaktarović, M. Č. Classification of brain tumors from mri images using a convolutional neural network. Applied Sciences10(6), 1999 (2020). [Google Scholar]
- 9.Sarp, S., Zhao, Y., Kuzlu, M.: Artificial intelligence-powered chronic wound management system: Towards human digital twins. PhD thesis, Virginia Commonwealth University (2022)
- 10.Ayebare, R. R., Flick, R., Okware, S., Bodo, B. & Lamorde, M. Adoption of covid-19 triage strategies for low-income settings. The Lancet Respiratory Medicine8(4), 22 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahdy, L.N., Ezzat, K.A., Elmousalami, H.H., Ella, H.A. & Hassanien, A.E. Automatic x-ray covid-19 lung image classification system based on multi-level thresholding and support vector machine. MedRxiv, 2020–03 (2020)
- 12.Moraes Batista, A.F., Miraglia, J.L., Rizzi Donato, T.H. & Porto Chiavegatto Filho, A.D. Covid-19 diagnosis prediction in emergency care patients: a machine learning approach. MedRxiv, 2020–04 (2020)
- 13.Apostolopoulos, I. D. & Mpesiana, T. A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Physical and engineering sciences in medicine43, 635–640 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemdan, E.E.-D., Shouman, M.A., Karar, M.E.: Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv preprint arXiv:2003.11055 (2020)
- 15.Narin, A., Kaya, C. & Pamuk, Z. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. Pattern Analysis and Applications24, 1207–1220 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bandyopadhyay Sr, S., & DUTTA, S. Machine learning approach for confirmation of covid-19 cases: Positive, negative, death and release (preprint). (No Title) (2020)
- 17.Khan, A. I., Shah, J. L. & Bhat, M. M. Coronet: A deep neural network for detection and diagnosis of covid-19 from chest x-ray images. Computer methods and programs in biomedicine196, 105581 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahmud, T., Rahman, M. A. & Fattah, S. A. Covxnet: A multi-dilation convolutional neural network for automatic covid-19 and other pneumonia detection from chest x-ray images with transferable multi-receptive feature optimization. Computers in biology and medicine122, 103869 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, L., Lin, Z. Q. & Wong, A. Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Scientific reports10(1), 19549 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sethy, P.K., Behera, S.K., Ratha, P.K. & Biswas, P. Detection of coronavirus disease (covid-19) based on deep features and support vector machine. International Journal of Mathematical, Engineering and Management Sciences5 (2020).
- 21.Afshar, P. et al. Covid-caps: A capsule network-based framework for identification of covid-19 cases from x-ray images. Pattern Recognition Letters138, 638–643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horry, M. J. et al. X-ray image based covid-19 detection using pre-trained deep learning models (2020).
- 23.Singh, M. et al. Transfer learning-based ensemble support vector machine model for automated covid-19 detection using lung computerized tomography scan data. Medical & biological engineering & computing59, 825–839 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das, N. N., Kumar, N., Kaur, M., Kumar, V. & Singh, D. Automated deep transfer learning-based approach for detection of covid-19 infection in chest x-rays. Irbm43(2), 114–119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heidari, M. et al. Improving the performance of cnn to predict the likelihood of covid-19 using chest x-ray images with preprocessing algorithms. International journal of medical informatics144, 104284 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loey, M., Smarandache, F. & M. Khalifa, N.E. Within the lack of chest covid-19 x-ray dataset: a novel detection model based on gan and deep transfer learning. Symmetry12(4), 651 (2020).
- 27.Nirmala, V., Shashank, H., Manoj, M., Satish, R.G., Premaladha, J.: Skin cancer classification using image processing with machine learning techniques. In: Intelligent Data Analytics, IoT, and Blockchain, pp. 1–15. Auerbach Publications, ? (2023)
- 28.Selvaraju, R. R. et al. Grad-cam: Visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 618–626 (2017)
- 29.Ahsan, M. M. et al. Covid-19 symptoms detection based on nasnetmobile with explainable ai using various imaging modalities. Machine Learning and Knowledge Extraction2(4), 490–504 (2020). [Google Scholar]
- 30.Li, L. et al. Artificial intelligence distinguishes covid-19 from community acquired pneumonia on chest ct. Radiology. 200905 (2020).
- 31.Ukwuoma, C. et al. Automated lung-related pneumonia and covid-19 detection based on novel feature extraction framework and vision transformer approaches using chest x-ray images. Bioengineering. 9, 709 10.3390/bioengineering9110709 (2022). [DOI] [PMC free article] [PubMed]
- 32.Addo, D. et al. Evae-net: An ensemble variational autoencoder deep learning network for covid-19 classification based on chest x-ray images. Diagnostics. 12(11) 10.3390/diagnostics12112569 (2022). [DOI] [PMC free article] [PubMed]
- 33.Monday, H. N. et al. Wmr-depthwisenet: A wavelet multi-resolution depthwise separable convolutional neural network for covid-19 diagnosis. Diagnostics 12(3) (2022). [DOI] [PMC free article] [PubMed]
- 34.Kuzlu, M., Cali, U., Sharma, V. & Güler, Ö. Gaining insight into solar photovoltaic power generation forecasting utilizing explainable artificial intelligence tools. Ieee Access8, 187814–187823 (2020). [Google Scholar]
- 35.Garg, P., Sharma, M. & Kumar, P. Transparency in diagnosis: Unveiling the power of deep learning and explainable ai for medical image interpretation. Arabian Journal for Science and Engineering, 1–17 (2025)
- 36.Veeramani, N. & Jayaraman, P. Yolov7-xai: Multi-class skin lesion diagnosis using explainable ai with fair decision making. International Journal of Imaging Systems and Technology34(6), 23214 (2024). [Google Scholar]
- 37.Das, A. & Rad, P. Opportunities and challenges in explainable artificial intelligence (xai): A survey. arXiv preprint arXiv:2006.11371 (2020).
- 38.Islam, M. K., Rahman, M. M., Ali, M. S., Mahim, S. & Miah, M. S. Enhancing lung abnormalities detection and classification using a deep convolutional neural network and gru with explainable ai: A promising approach for accurate diagnosis. Machine Learning with Applications14, 100492 (2023). [Google Scholar]
- 39.Kermany, D. S. et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell172(5), 1122–1131 (2018). [DOI] [PubMed]
- 40.Veeramani, N., Premaladha, J., Krishankumar, R. & Ravichandran, K. S. Hybrid and automated segmentation algorithm for malignant melanoma using chain codes and active contours. In: Deep Learning in Personalized Healthcare and Decision Support, pp. 119–129. Elsevier, ??? (2023)
- 41.Jaeger, S. et al. Automatic tuberculosis screening using chest radiographs. IEEE transactions on medical imaging33(2), 233–245 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Candemir, S. et al. Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE transactions on medical imaging33(2), 577–590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarp, S. et al. An xai approach for covid-19 detection using transfer learning with x-ray images. Heliyon9(4) (2023) [DOI] [PMC free article] [PubMed]
- 44.Talukder, M. A., Layek, M. A., Kazi, M., Uddin, M. A. & Aryal, S. Empowering covid-19 detection: Optimizing performance through fine-tuned efficientnet deep learning architecture. Computers in Biology and Medicine168, 107789 (2024). [DOI] [PubMed] [Google Scholar]
- 45.Premaladha, J., Surendra Reddy, M., Hemanth Kumar Reddy, T., Sri Sai Charan, Y. & Nirmala, V. Recognition of facial expression using haar cascade classifier and deep learning. In: Inventive Communication and Computational Technologies: Proceedings of ICICCT 2021, pp. 335–351 (2022). Springer
- 46.V, N., K, J.A.S., G, A., N, S.S., S, P.: Automated template matching for external thread surface defects with image processing. In: 2023 Fifth International Conference on Electrical, Computer and Communication Technologies (ICECCT), pp. 1–6 (2023). 10.1109/ICECCT56650.2023.10179653
- 47.Nirmala, V. An automated detection of notable abcd diagnostics of melanoma in dermoscopic images. In: Artificial Intelligence in Telemedicine, pp. 67–82. CRC Press, ??? (2023)
- 48.He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
- 49.Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J. & Wojna, Z. Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2818–2826 (2016)
- 50.Ahsan, M. M. et al. Deep transfer learning approaches for monkeypox disease diagnosis. Expert Systems with Applications216, 119483 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Främling, K., Westberg, M., Jullum, M., Madhikermi, M. & Malhi, A.. Comparison of contextual importance and utility with lime and shapley values. In: International Workshop on Explainable, Transparent Autonomous Agents and Multi-Agent Systems, pp. 39–54 (2021). Springer
- 52.Song, D. et al. A new xai framework with feature explainability for tumors decision-making in ultrasound data: comparing with grad-cam. Computer Methods and Programs in Biomedicine235, 107527 (2023). [DOI] [PubMed] [Google Scholar]
- 53.Zou, L. et al. Ensemble image explainable ai (xai) algorithm for severe community-acquired pneumonia and covid-19 respiratory infections. IEEE Transactions on Artificial Intelligence4(2), 242–254 (2022). [Google Scholar]
- 54.Marmolejo-Saucedo, J. A. & Kose, U. Numerical grad-cam based explainable convolutional neural network for brain tumor diagnosis. Mobile Networks and Applications29(1), 109–118 (2024). [Google Scholar]
- 55.Shah, P.M., Zeb, A., Shafi, U., Zaidi, S.F.A., Shah, M.A.: Detection of parkinson disease in brain mri using convolutional neural network. In: 2018 24th International Conference on Automation and Computing (ICAC), pp. 1–6 (2018). 10.23919/IConAC.2018.8749023
- 56.Veeramani, N., Jayaraman, P., Krishankumar, R., Ravichandran, K. S. & Gandomi, A. H. Ddcnn-f: double decker convolutional neural network’f’feature fusion as a medical image classification framework. Scientific Reports14(1), 676 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Teixeira, L. O. et al. Impact of lung segmentation on the diagnosis and explanation of covid-19 in chest x-ray images. Sensors21(21), 7116 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the Kaggle repository, can be downloaded from https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia.