Abstract
Although the nephrotoxicity of uranium has been established through numerous animal studies, relatively little is known about the effects of long-term environmental uranium exposure. Using a combination of conventional biochemical studies and serial analysis of gene expression (SAGE), we examined the renal responses to uranyl nitrate (UN) chronic exposure. Renal uranium levels were significantly increased 4 months after ingestion of uranium in drinking water. Creatinine levels in serum were slightly but significantly increased compared with those in controls. Although no further significant differences in other parameters were noted, substantial molecular changes were observed in toxicogenomic profiles. UN induced dramatic alterations in expression levels of more than 200 genes, mainly up-regulated, including oxidative-response–related genes, genes encoding for cellular metabolism, ribosomal proteins, signal transduction, and solute transporters. Seven differentially expressed transcripts were confirmed by real-time quantitative polymerase chain reaction. In addition, significantly increased peroxide levels support the implication of oxidative stress in UN toxicant response. This report highlights the potential of SAGE for the discovery of novel toxicant-induced gene expression alterations. Here, we present, for the first time, a comprehensive view of renal molecular events after uranium long-term exposure.
Keywords: drinking water, gene expression profiles, long-term exposure, mice, SAGE, toxicogenomics, uranyl nitrate
Uranium, the heaviest of the naturally occurring elements, is widely present in the environment as a result of leaching from natural deposits, release in mill tailings, emissions from the nuclear industry, the combustion of coal and other fuels, and the use of phosphate fertilizers and weapons that contain uranium. Thus, uranium is found in various chemical forms and different levels in all soils, rocks, sea, and bedrock (Bosshard et al. 1992; Kurttio et al. 2002; Moss et al. 1983). It is also found in both food and drinking water. The wide range of levels of uranium in drinking water, together with the observation of consistently higher levels in certain community water supplies, has raised concerns regarding the potential hazard of such sources of uranium to human health.
Many isolated studies conducted on the mechanisms for the toxic effects of uranium at moderate to high acute doses on experimental animals have shown that the major health effect of uranium is chemical kidney toxicity rather than a radiation hazard (Lin et al. 1993; Miller et al. 1998, 2002). In addition only a few studies have attempted to characterize the effects of chronic exposure to uranium through drinking water (Gilman et al. 1998a, 1998b, 1998c; Kurttio et al. 2002; McDonald-Taylor et al. 1997; Zamora et al. 1998). Although chronic uranium exposure in humans has been clearly associated with increasing urinary glucose, alkaline phosphatase, and β2-microglobulin supporting proximal tubule alterations, the urinary albumin levels, which are indicators of glomerular function, are conflicting (Kurttio et al. 2002; Zamora et al. 1998). Although both functional and histologic damage to the proximal tubules resulting from acute uranium exposure has been clearly demonstrated (Schramm et al. 2002; Sun et al. 2002), little is known about the effect of long-term environmental uranium exposure in both humans and animals (Gilman et al. 1998a, 1998b, 1998c; Kultima et al. 2002; Mao et al. 1995; McDonald-Taylor et al. 1997; Zamora et al. 1998).
Toxicogenomics is presently used to evaluate risk assessment of environmental toxicants through the identification of gene expression networks, as well as to evaluate toxicant-induced gene expression as a biomarker to assess human exposure. Several researchers are currently combining the identification of gene expression patterns representative of adverse outcomes with traditional biochemical parameter measures to categorize and classify toxic responses through direct comparison in exposed and control samples. The use of oligonucleotide-based or cDNA microarrays for understanding the biochemical processes associated with environmental chemical exposures has proven successful in recent experiments on human health risk assessment for several toxicants (Andrew et al. 2003; Bartosiewicz et al. 2001).
Because the risk assessment and establishment of exposure limits for uranium in drinking water are of considerable importance in various areas, including Finland, we used for the first time the SAGE (serial analysis of gene expression) approach to identify gene expression profiles associated with this hazard exposure. Because toxicogenomics provides increased confidence in extrapolation of hazards observed in animals studies to likely hazards in humans, we examined renal molecular effects of chronic exposure to uranium in mice.
Materials and Methods
Animals
The C57BL/6J mouse was chosen because of the current state of knowledge about this transcriptome and numerous databases such as Mouse SAGE Site (http://mouse.biomed.cas.cz/sage/). This animal model should help improve the overall quality of SAGE gene expression data. Experiments were performed with 16 male C57BL/6J mice, weighing 25–30 g (Harlan, Gannat, France) at the beginning of the study. The mice were randomly divided into three groups: one control group (group 0, six animals) and two uranyl nitrate (UN)-treated mice (groups 1 and 2, six and four animals, respectively). Exposed groups 1 and 2 received UN mineral water at concentrations of 80 or 160 mg UN/L of water, respectively, approximately 3- or 6-fold higher than levels found in bedrock of southern Finland (Juntunen 1991). Uranium in water, given to control mice, was determined to be < 0.002 mg/L uranium. Body weights were measured weekly. Food intake and fluid consumption data were recorded. After 4 months of treatment all animals were euthanized by exsanguination using cardiac puncture. Urine and blood were collected for each group. The kidneys were either embedded in Epon for morphologic examination or snap-frozen in liquid nitrogen and then stored at −70°C until further study.
Assessment of Renal Function Parameters
Uranium contents were determined in samples of kidney using a kinetic phosphorescence analyzer (KPA; Ejnik et al. 2000). Serum creatinine and urea levels and urinary concentrations of glucose and γ-glutamyl transpeptidase (γ-GT) were measured by routine methods.
RNA Isolation
Total RNAs, extracted from renal tissue using the RNA isolation mini kit (Qiagen, Courtaboeuf, France), were pooled for SAGE or used individually for real-time reverse transcriptase polymerase chain reaction (RT-PCR) analyses. The amount of total RNA was determined using a fluorescent nucleic acid stain (RiboGreen RNA Quantitation Kit; Molecular Probes, Montluçon, France). The quality of the RNA was evaluated by measuring the 260:280-nm ratios and confirmed by visualization of intact 18S and 28S RNA bands after agarose gel electrophoresis.
Analysis of Gene Expression
Production of kidney library.
Kidney libraries were generated from 50 μg of total RNA using the I SAGE kit (Invitrogen Corp., Cergy Pontoise, France) following the manufacturer’s instructions (Invitrogen Corporation 2004), adapted from initial description (SAGE 2004; Velculescu et al. 1995). Because of budgetary restrictions, SAGE was performed for only control [UN(−)] and 80 mg UN/L–treated mice [UN(+)], that is, groups 0 and 1, respectively.
Tag quantification.
Concatemer sequences were analyzed by using SAGE software (version 4.0; Invitrogen), which automatically detects and counts tags from sequence files. SAGE software excludes replicate ditags from the tag sequence catalog because the probability of any two tags being coupled in the same ditag is small, even for abundant transcripts. For tag identification, the tag list of each library was matched against a mouse tag database extracted by SAGE software from GenBank (http://www.ncbi.nlm.nih.gov/). Usually, SAGE tag sequences matched more than one transcript. The average p-value computed by the SAGE software, based on a Monte Carlo analysis (Zhang et al. 1997), serves as ranking parameter to produce a list of differentially expressed genes. SAGE data for the libraries described here are available at Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo); accession nos. GSM24256 and GSM24257).
Real-Time RT-PCR
Total RNAs (1 μg) from UN(−) and UN(+) renal tissue (extracted as described above) were used to generate cDNA using Moloney Murine Leukemia Virus Reverse Transcriptase (Invitrogen) according the manufacturer’s conditions. Primers and probes specifically designed for selected cDNA using Primer Express software, version 2.0 (PE; Applied Biosystems, Courtaboeuf, France), are listed in Table 1. The ABI PRISM 7000 Sequence Detection System was used for detected real-time RT-PCR products with the SYBR Green I assay, according to recommendations of the manufacturer (PE; Applied Biosystems). For two cases in which we encountered difficulties with the SYBR Green I assay, we used TaqMan probe assays (Applied Biosystems) (Table 1). Each PCR reaction was optimized to ensure that a single band of the appropriate size was amplified and that no bands corresponding to genomic DNA amplification or primer–dimer pairs were present. The PCR cycling conditions were performed for all samples as follows: 50°C, 2 min for AmpErase UNG (Applied Biosystems) incubation; 95°C, 10 min for AmpliTaq Gold (Applied Biosystems) activation; and 40 cycles for the melting (95°C, 15 sec) and annealing/extension (60°C for 1 min) steps. PCR reactions for each template were done in triplicate in 96-well plates. The comparative threshold cycle (Ct) method using Primer Express software, version 2.0, was used to determine relative quantitation of gene expression for each gene compared with the hypoxanthine guanine phosphoribosyl transferase control (listed in Table 1).
Table 1.
SYBR Green and TaqMan primer sequences used for RT-PCR reactions.
| Gene symbola | Gene namea | Accession no.a | Primer 5′→3′ sequence or assay ID | Amplicon size (bp) |
|---|---|---|---|---|
| Primers using SYBR Green detection | ||||
| Hprt | hypoxanthine phosphoribosyl transferase | NM_013556 | Forward 5′-TTGCTGACCTGCTGGATTAC-3′ b | 112 |
| Reverse 5′-CCCGTTGACTGATCATTACA-3′ | ||||
| Sod1 | superoxide dismutase 1 | XM_128337 | Forward 5′-TGGTGGTCCATGAGAAACAA-3′ | 75 |
| Reverse 5′-TCCCAGCATTTCCAGTCTTT-3′ | ||||
| Odc | ornithine decarboxylase, structural | NM_013614 | Forward 5′-TTGCCACTGATGATTCCAAA-3′ | 129 |
| Reverse 5′-CATGGAAGCTCACACCAATG-3′ | ||||
| Fau | Finkel-Biskis-Reilly murine sarcoma virus | NM_007990 | Forward 5′-GCTGGGAGGTAAAGTTCACG-3′ | 125 |
| Reverse 5′-TGTACTGCATTCGCCTCTTG-3′ | ||||
| Tctp | translationally regulated transcript | NM_009429 | Forward 5′-CCGGGAGATCGCGGAC-3′ | 92 |
| Reverse 5′-TTCCACCGATGAGCGAGTC-3′ | ||||
| Primers using TaqMan technology | ||||
| Hprt | hypoxanthine phosphoribosyl transferase | NM_013556 | Mm00446968m1c | |
| Kap | kidney androgen regulated protein | NM_010594 | Mm00495104m1 | |
| NaPi-II | solute carrier family 34, member 1 | NM_011392 | Mm00441450m1 | |
| Umod | uromodulin | NM_009470 | Mm00447649m1 | |
From Applied Biosystems (http://myscience.appliedbiosystems.com/cdsEntry/Form/gene_expression_keyword.jsp).
Primer 5′→3′ sequence.
Assay ID – Applied Biosystems.
Hydrogen Peroxide Assay
To determine the impact of UN on the oxidative balance status, hydrogen peroxide levels were determined using a PeroxiDetect kit (Sigma, Lyon, France). Briefly, kidney samples from different groups (0, 1, and 2) were homogenized in the indicated phosphate buffer on ice, then centrifuged at 15,000×g for 15 min at 4°C. Supernatant samples (100 μL) were incubated for 30 min with 1 mL of aqueous peroxide color reagent (aqueous solution containing 100 mM sorbitol and 125 μM xylenol orange) and 10 μL of ferrous ammonium sulfate reagent (25 mM ferrous ammonium sulfate in 2.5 M sulfuric acid), and the hydrogen peroxide level was measured by the absorbance at 560 nm.
Results
General Observations
To examine the general parameters, we performed gross end-point analysis such as body and organ weight changes and histologic observations, as well as the dosage of uranium content in renal tissue and biochemical markers. No significant dose-related effects were observed on body weight gain, food intake, or water consumption. Because the concentrations of UN in the drinking water remained constant throughout the study, it is natural to assume that the measurement of UN per kilogram body weight decreased with age.
Gross pathologic examination was performed in all animals, and the histopathologic analysis did not identify any significant differences between control and exposed groups. We observed a significant dose-dependent increase in renal uranium tissue levels in groups 1 and 2 compared with control mice, using KPA. Compared with controls, there were no significant differences in kidney weights in any dose group (Table 2). Serum creatinine levels appeared to increase in dose-independent manner with UN treatment, and groups 1 and 2 showed creatinine levels significantly higher than those of controls.
Table 2.
Physiologic parameters in serum and urine and uranium amount in control group 0 and contaminated groups 1 and 2 after 4 months of daily contamination (mean ± SE).
| Group
|
|||
|---|---|---|---|
| Parameter | 0 | 1 | 2 |
| Exposure (mg UN/L) | 0 | 80 | 160 |
| Kidney | |||
| Weight (g) | 0.47 ± 0.01 | 0.46 ± 0.01 | 0.47 ± 0.02 |
| Uranium amount (μg/g) | 0.16 ± 0.04 | 0.35 ± 0.02* | 1.05 ± 0.21* |
| Serum | |||
| Urea (mg/dL) | 59 ± 5 | 57 ± 5 | 54 ± 7 |
| Creatinine (mg/dL) | 0.12 ± 0.02 | 0.23 ± 0.02* | 0.25 ± 0.02* |
| Urine | |||
| Glucose (g/L) | 0.08 ± 0.03 | 0.08 ± 0.03 | 0.04 ± 0.01 |
| γ -GT (U/L) | 86 ± 44 | 94 ± 42 | 119 ± 66 |
*p < 0.05 versus control; n = 4.
Genes Responding to Toxic UN Exposure
We investigated the transcriptomic response that underlies the induction of the metal-elicited molecular modification in C57/Bl6J mice. SAGE was used to determine the global gene expression profile in UN toxicity. This approach allows an analysis of gene expression by the sequencing of approximately 21,000 transcripts from kidney libraries of the groups 0 and 1, which represent 5,252 and 4,069 unique tags, respectively.
We validated the quality of both libraries by comparing both with previous data on the kidney (Chabardes-Garonne et al. 2003; El-Meanawy et al. 2000; Virlon et al. 1999). For example, known markers for proximal tubules [kidney androgen-regulated protein (kap)] and thick ascending limbs [uromodulin (Umod)] were evidenced in both libraries. As expected, a large fraction of the most abundant tags matched with widely expressed mitochondrial genes or ribosomal proteins such as ribosomal proteins P1 and S26. Because the kidney mass consists predominantly of proximal tubules, a significant fraction of tags are mapped to genes known to be expressed in proximal tubular epithelial cells. Particularly, the most abundant transcripts in normal kidney were kap and glutathione peroxidase 3 (GPx3), in agreement with previous data (El-Meanawy et al. 2000).
Tags that are significantly up- or down-regulated in the UN RNA library are listed in Table 3 with their frequency and their relevant accession number. We considered only the transcripts with a significant expression change (p < 0.05). Considering the large number of sequenced tags, the number of genes expressed in kidney was evaluated by excluding tags matching mitochondrial sequences, tags with multiple matches, and nonreliable matches. Tags were arbitrarily separated in categories according to gene function. As illustrated in Table 3, most of these changes involved up-regulation. SAGE analysis revealed the expression changes of genes related to lipid metabolism [crystalline, zeta (Cryz); phosphatidic acid phosphatase type 2c (PPap2c)], carbohydrate metabolism [phosphoglycerate kinase 1 (Pgk1); sorbitol dehydrogenase 1 (Sdh1)], and amino acid metabolism (glutamate dehydrogenase (Glud); ornithine decarboxylase, structural (Odc). The UN-induced transcripts consisted mainly of genes encoding proteins associated with protein biosynthesis [ribosomal protein S25 (Rps25); S26 (Rps26); large, P1 (Rplp1); L19 (Rpl19)], protein folding [heat-shock 10 kDa protein 1 (chaperonin 1) (Hspe1)], and proteolysis [kallikrein 5 (Klk5), protein C (Proc)]. Many genes involved in signaling were up-regulated, such as hormonal receptors [growth hormone receptor (Ghr), cholecystokinin A receptor (Cckar)]. Chronic exposure to UN also increased the expression of a number of genes related to oxidative process and detoxification. Among these is cytochrome P450 (Cyp4b1), which catalyzes the oxidation of a wide variety of substrates, including endogenous lipids and xenobiotics (Heng et al. 1997). Other relevant enzymes under- or overexpressed include thioredoxin, mitochondrial (Txn2); superoxide dismutase 1, soluble (Sod1); and thioether S-methyltransferase (Temt). We also mainly observed up-regulation of genes related with ion transporters including solute carrier family 34 (sodium phosphate), member 1 (Slc34a1, NaPi-II); and with electron transporters such as ATPase inhibitor, and cytochrome c oxidase, subunit IVa (Cox4a); subunit VIIIa (Cox8a); and subunit XVII assembly protein homolog (Cox 17 ). Finally, expression levels of several genes, in the category related to stress/apoptosis [Bcl2 associated athanogene 1 (Bag1); nerve growth factor receptor (TNFRSF16) associated protein 1 (Ngfrap1)]; immunity (Ia-associated invariant chain (Ii)]; and translationally regulated transcripts (21 kDa) (Trt, Tpt1, Tctp, Umod) were changed.
Table 3.
List of tags with significant variations in expression level induced by UN long-term ingestion (p < 0.05), their frequency, and their relevant accession number.
| Tag sequence | Count
|
Gene namea | Accession no.a | Regulationb | Gene symbola | |
|---|---|---|---|---|---|---|
| UN(−) | UN(+) | |||||
| Apoptosis | ||||||
| GCTGCCAGGG | 11 | 4 | Bcl2-associated athanogene 1 | NM_009736 | − | Bag1 |
| GAAAGCAATG | 0 | 6 | nerve growth factor receptor (TNFRSF16) associated protein 1 | NM_009750 | + | Ngfrap1 |
| TGCCTTACTT | 3 | 8 | programmed cell death 6 | NM_011051 | + | Pdcd6 |
| Amino acid metabolism | ||||||
| CGTATCTGTA | 4 | 10 | D-amino acid oxidase | NM_010018 | + | Dao1 |
| CAGTTACAAA | 1 | 6 | glutamate dehydrogenase | NM_008133 | + | Glud |
| TTTTACCTGC | 0 | 8 | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | NM_025961 | + | Gatn |
| CTACCACTGC | 4 | 12 | fumarylacetoacetate hydrolase | NM_010176 | + | Fah |
| ATACTAACGT | 40 | 24 | ornithine decarboxylase, structural | NM_013614 | − | Odc |
| AACAGAAAGT | 1 | 8 | phenylalanine hydroxylase | NM_008777 | + | Pah |
| Carbohydrate metabolism | ||||||
| GCAAACAAGA | 11 | 18 | isocitrate dehydrogenase 2 (NADP+), mitochondrial | NM_173011 | + | Idh2 |
| GTGCCATATT | 12 | 26 | isocitrate dehydrogenase 2 (NADP+), mitochondrial | NM_173011 | + | Idh2 |
| /CCAAATAAAA | 17 | 31 | lactate dehydrogenase 1, A chain | NM_010699 | + | Ldh1 |
| TGATATGAGC | 33 | 12 | lactate dehydrogenase 2, B chain | NM_008492 | − | Ldh2 |
| TTGTTAGTGC | 70 | 89 | malate dehydrogenase, soluble | NM_008492 | + | Mor2 |
| GCAATCTGAT | 17 | 31 | phosphoglycerate kinase 1 | NM_008828 | + | Pgk1 |
| GCCCAGACCT | 25 | 41 | sorbitol dehydrogenase 1 | NM_146126 | + | Sdh1 |
| GCTTGTGACG | 1 | 8 | transaldolase 1 | NM_011528 | + | Taldo1 |
| Cell adhesion | ||||||
| CTCTGACTTA | 3 | 8 | basigin | NM_009768 | + | Bsg |
| GAGACTAGCA | 4 | 10 | transmembrane 4 superfamily member 8 | NM_019793 | + | Tm4sf8 |
| Immunity and defense | ||||||
| Immunity | ||||||
| GTTCAAGTGA | 4 | 12 | Ia-associated invariant chain | NM_010545 | + | Ii |
| TATCCTGAAT | 14 | 2 | lymphocyte antigen 6 complex, locus A | NM_010738 | − | Ly6a |
| TTTTATGTTT | 12 | 20 | tumor necrosis factor, alpha-induced protein 1 (endothelial) | NM_009395 | + | Tnfaip1 |
| TATACATCCA | 43 | 26 | uromodulin | NM_009470 | − | Umod |
| TGGGTTGTCT | 151 | 174 | translationally regulated transcript (21 kDa) | NM_009429 | + | Trt, Tpt1, Tctp |
| Antioxidant and free radical removal | ||||||
| CTATCCTCTC | 297 | 341 | glutathione peroxidase 3 | NM_008161 | + | Gpx3 |
| CAGCTTCGAA | 12 | 2 | glutathione S-transferase, theta 2 | NM_010361 | − | Gstt2 |
| AGAAACAAGA | 7 | 18 | superoxide dismutase 1, soluble | XM_128337 | + | Sod1 |
| TTGCTTCTAT | 20 | 8 | thioether S-methyltransferase | NM_009349 | − | Temt |
| CATCAGCCTC | 7 | 0 | thioredoxin, mitochondrial | NM_019913 | − | Txn2 |
| Lipid fatty acid and steroid metabolism | ||||||
| TCTCCTTAGC | 0 | 10 | ATP-binding cassette, subfamily D (ALD), member 3 | NM_008991 | + | Abcd3 |
| TTAAGACCTG | 9 | 18 | crystallin, zeta | NM_009968 | + | CryZ |
| TATAATAAAC | 0 | 8 | cytochrome P450, 2d9 | NM_080006 | + | Cyp2d9 |
| TGTGTGGAAT | 14 | 20 | cytochrome P450, subfamily IV B, polypeptide 1 | NM_007823 | + | Cyp4b1 |
| GGAGGGTGTG | 4 | 10 | phosphatidic acid phosphatase type 2c | NM_015817 | + | Ppap2c |
| Protein metabolism and modification | ||||||
| Protein folding | ||||||
| CCTCCCTTTT | 4 | 14 | heat shock 10 kDa protein 1 (chaperonin 10) | NM_008303 | + | Hspe1 |
| Protein biosynthesis | ||||||
| GATGTGGCTG | 7 | 22 | eukaryotic translation elongation factor 1 beta 2 | NM_018796 | + | Esf1b2 |
| TCACCCAATA | 36 | 49 | eukaryotic translation elongation factor 2 | NM_007907 | + | Eef2 |
| CTAATAAAGC | 18 | 43 | Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed (fox derived) | NM_007990 | + | Fau |
| TGTCATCTAG | 7 | 14 | laminin receptor 1 (67 kDa, ribosomal protein SA) | NM_011029 | + | Lamr1 |
| TGCTGGGATG | 6 | 16 | mitochondrial ribosomal protein S12 | NM_011885 | + | Mrps12 |
| AGGTCGGGTG | 7 | 14 | ribosomal protein L13a | + | Rpl13a | |
| TGGATCAGTC | 47 | 66 | ribosomal protein L19 | NM_009078 | + | Rpl19 |
| CCAGAACAGA | 7 | 20 | ribosomal protein L30 | NM_009078 | + | Rpl30 |
| GGCTTCGGTC | 48 | 68 | ribosomal protein, large, P1 | NM_018853 | + | Rplp1 |
| GTGAAACTAA | 36 | 45 | ribosomal protein S4, X-linked | NM_009094 | + | Rps4x |
| CTGGGCGTGT | 3 | 8 | ribosomal protein S15 | NM_009091 | + | Rps15 |
| GTGGGCGTGT | 0 | 6 | ribosomal protein S15 | NM_009091 | + | Rps15 |
| CAGAACCCAC | 0 | 6 | ribosomal protein S18 | NM_138946 | + | Rps18 |
| GCCTTTATGA | 4 | 10 | ribosomal protein S24 | NM_011297 | + | Rps24 |
| AACAGGTTCA | 11 | 18 | ribosomal protein S25 | NM_024266 | + | Rps25 |
| TAAAGAGGCC | 18 | 29 | ribosomal protein S26 | NM_013765 | + | Rps26 |
| Proteolysis | ||||||
| GGTTAAATGT | 1 | 8 | cathepsin L | NM_009984 | + | Ctsl |
| CAGCAAAAAA | 33 | 41 | kallikrein 5 | NM_008456 | + | Klk5 |
| GAGAGTGTGA | 6 | 14 | kidney-derived aspartic protease-like protein | NM_008437 | + | Kdap |
| CAGAATGGAA | 14 | 29 | peptidase 4 | NM_008820 | + | Pep4 |
| AGGCGGGATC | 3 | 8 | proteasome (prosome, macropain) subunit, alpha type 7 | NM_011969 | + | Psma7 |
| CAACAAACAT | 3 | 10 | protein C | NM_008934 | + | Proc |
| GTAAGCAAAA | 22 | 43 | ubiquitin B | NM_011664 | + | Ubb |
| Signal transduction system, receptor | ||||||
| TGGGACTCAC | 4 | 14 | cholecystokinin A receptor | NM_009827 | + | Cckar |
| AGAAAAAAAA | 7 | 14 | ciliary neurotrophic factor receptor | NM_016673 | + | Cntfr |
| TGATTTTTGT | 1 | 10 | disabled homolog 2 (Drosophila) | NM_023118 | + | Dab2 |
| GGGCAAGCCA | 4 | 14 | estrogen-related receptor, alpha | NM_007953 | + | Esrra |
| CATACGCATA | 7 | 16 | growth hormone receptor | NM_010284 | + | Ghr |
| TTAAGAGGGA | 12 | 0 | transducer of ErbB-2.1 | NM_009427 | − | Tob1 |
| Transport | ||||||
| Electron transport | ||||||
| GCTTTGAATG | 20 | 35 | ATPase inhibitor | NM_007512 | + | Atpi |
| CCAGTCCTGG | 12 | 24 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit c (subunit 9), isoform 1 | NM_007506 | + | Atp5g1 |
| GTTCTTTCGT | 3 | 8 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit c (subunit 9), isoform 2 | NM_026468 | + | Atp5g2 |
| GCCGAGCATA | 6 | 16 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit f, isoform 2 | NM_020582 | + | Atp5j2 |
| GATAGATAAT | 3 | 8 | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit, isoform 1 | NM_007505 | + | Atp5a1 |
| CTAATAAAAG | 33 | 45 | cytochrome c oxidase, subunit IVa | NM_009941 | + | Cox4a |
| TATTGGCTCT | 53 | 74 | cytochrome c oxidase, subunit VIIIa | NM_007750 | + | Cox8a |
| AGGGCACTGG | 3 | 8 | cytochrome c oxidase, subunit XVII assembly protein homolog | AV158262 | + | Cox17 |
| CAGAATGTGC | 3 | 8 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 | NM_010885 | + | Ndufa2 |
| TTATGAAATG | 15 | 24 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 | NM_019443 | + | Ndufa1 |
| ACTGCTTTTC | 1 | 10 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7 | NM_023202 | + | Ndufa7 |
| Ion transport | ||||||
| TTCTAGCATA | 28 | 10 | ATPase, Na+/K+ transporting, beta 1 polypeptide | NM_009721 | − | Atp1b1 |
| CTAGGTACTG | 48 | 91 | solute carrier family 34 (sodium phosphate), member 1 | NM_011392 | + | Slc34a1 |
| ACAAATTATG | 1 | 8 | voltage-dependent anion channel 2 | NM_011695 | + | Vdac2 |
| Lipid fatty acid transport | ||||||
| GCTCTGATAC | 0 | 8 | sterol carrier protein 2, liver | NM_138508 | + | Scp2 |
| Others | ||||||
| TGCTTTTACG | 7 | 20 | 6-pyruvoyl-tetrahydropterin synthase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) | NM_025273 | + | Pcbd |
| ATTACGGTGG | 7 | 18 | aldo-keto reductase family 1, member A4 (aldehyde reductase) | NM_021473 | + | Akr1a4 |
| AAGACCTATG | 12 | 2 | diazepam binding inhibitor | NM_007830 | − | Dbi |
| CTCCTGCAGC | 15 | 29 | esterase 10 | NM_016903 | + | Es10 |
| ATCTGACTCC | 3 | 10 | hemoglobin Y, beta-like embryonic chain | NM_008221 | + | Hbb |
| TAAAGCAAAA | 20 | 43 | H2B histone family, member S | NM_023422 | + | Hist1h2bc |
| GACTTCACGC | 155 | 182 | kidney androgen-regulated protein | NM_010594 | + | Kap |
| GCACGAGCGT | 7 | 0 | low density lipoprotein receptor-related protein 2 | XM_130363 | − | Lrp2 |
| TGCTGTGACC | 9 | 16 | membrane-associated protein 17 pending | NM_026018 | + | Map17-p |
| TGTGCTTCCC | 4 | 12 | neural precursor cell expressed, developmentally down-regulated gene 8 | NM_008683 | + | Nedd8 |
| TGAGCGCTGC | 15 | 24 | PDZ domain containing 1 | NM_021517 | + | Pdzk1 |
| GGGGAGGGGG | 7 | 0 | pre B-cell leukemia transcription factor 2 | NM_017463 | − | Pbx2 |
| GGCTGGGGGC | 3 | 10 | profilin 1 | NM_011072 | + | Pfn1 |
| AAGTAAAGCG | 6 | 12 | SEC61, gamma subunit (S. cerevisiae) | NM_011343 | + | Sec61g |
| CAGCCTGAGC | 4 | 10 | selenoprotein R | NM_013759 | + | Sepr |
| TTTCCAGGTG | 1 | 8 | selenoprotein W, muscle 1 | NM_009156 | + | Sepw1 |
From Applied Biosystems (http://myscience.appliedbiosystems.com/cdsEntry/Form/gene_expression_keyword.jsp.).
+, up-regulation; −, down-regulation.
Real-Time Quantitative PCR Analyses
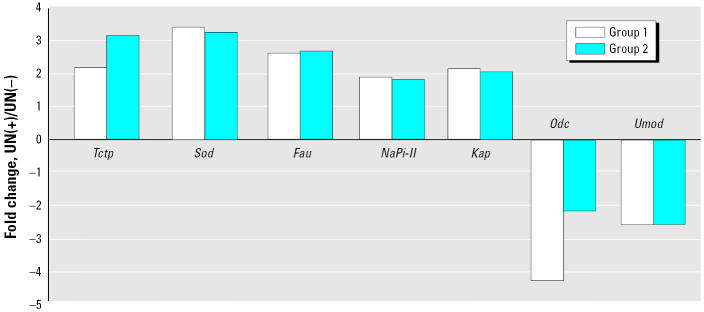
To validate our SAGE data, we conducted real-time quantitative PCR analyses to verify the differential expression of seven selected genes (Figure 1). kap was chosen because of its high abundance level in the normal and contaminated kidney. Solute carrier family 34 [sodium phosphate, NaPi] member 1 (slc34a1, NaPi-II)], Sod1, Finkel-Biskis-Reilly murine sarcoma virus ubiquitously expressed (Fau), and translationally regulated transcript (Trt or Tctp) were chosen because they were increased in our data. Umod and ornithine decarboxylase structural (Odc) were chosen because their expression levels were decreased in the present study as well as in ischemic acute renal failure (ARF) or UN-induced chronic renal failure, respectively (Fleck et al. 2003). Using real-time PCR analyses, Kap, NaPi-II, Sod, Fau, and Tctp were confirmed to be significantly increased whereas Odc and Umod were decreased in chronic exposure to UN. In summary PCR analysis confirmed the accuracy of the differences in expression levels observed in our SAGE analysis for group 1. Moreover, using real-time PCR for group 2, we observed that the expression of the selected transcripts were altered in the same direction compared with group 1, that is, increased or decreased. We noted dose-dependent increases in Tctp mRNA level at the highest concentration, and the observed decrease of Odc mRNA levels was more moderate for group 2.
Figure 1. Confirmation of SAGE data by real-time RT-PCR analysis. The variation of the amplification of the expression in groups 1 and 2 [UN(−)/UN(+)] is plotted. PCR analyses were performed on cDNA from UN(−) or UN(+) tissues.
Peroxide Level Measurement
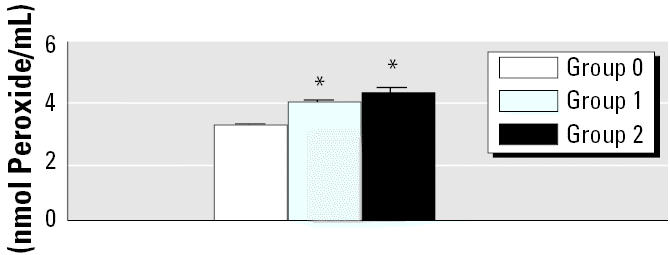
To evaluate whether the variations in both Sod and Gpx transcripts may reflect a potential oxidative stress, we examined the production of H2O2. The concentration of H2O2 in the kidney was found to be significantly higher in groups 1 and 2 compared with the control group (4.06 ± 0.06 and 4.39 ± 0.11 vs. 3.3 ± 0.02 nmol peroxide/mL) (Figure 2). Long-term UN exposure clearly caused the production of H2O2 levels in UN groups 1 and 2, in dose-dependent fashion.
Figure 2. Measurement of hydrogen peroxide already formed in kidney tissue. An increase in H2O2 level was induced by UN in a dose-dependent manner. Data shown represent means ± SE of three independent experiments (n = 4). : *p < 0.05 versus control.
Discussion
Human exposures to metals such as uranium in both occupational and environmental settings are common occurrences. Uranium exposures are a growing concern in our society. Classically, toxicologists assess potential chronic adverse health outcomes resulting from chemical exposures by using gross end points such as body or organ weight changes and histopathologic observations. However, analysis of histologic or biochemical markers often does not provide information about the mechanisms involved in toxicant response. The study of molecular mechanisms of toxicant action might provide information crucial to the understanding of their potential adverse effects on human health. Recent technologies such as SAGE facilitate studies that add insight into the cellular response to chemical exposure. In environmental monitoring, SAGE could not only provide a method for quickly categorizing chemicals and assigning a mode of toxic action but also allow more sensitive end points to address specifically gene expression pattern.
Results reported here identify > 200 genes from approximately 21,000 tags sequenced, for which the expression in kidney changed significantly after UN long-term exposure. Most of these tags represent distinct transcripts; however, some tags, especially those detected only once, may result from PCR or sequencing errors (Velculescu et al. 1997; Zhang et al. 1997). Using classical end-point examination, including histologic appearance of the kidney and clinical and biochemical parameters, we observed that the UN doses used in this study produced only a slight alteration in serum creatinine levels and a significant but nonlinear increase of intrarenal uranium content. The dose-independent induction of the serum creatinine may be attributable, as already reported (Amin et al. 2004), to the fact that this parameter, like serum urea, traditionally used as indices of changes in glomerular filtration rate, is a relatively insensitive marker of glomerular injury. Taken together, these data suggest that the glomerular filtration rate remains relatively normal in mice after UN chronic exposure. Because the degree of renal injury appeared to be minimal in the strain of mouse used in the present study, further work will be needed to correlate the renal toxicity with the chronic uranium treatment, in dose- and time-dependent manner.
At the molecular level we observed that UN induced changes in expression profiles for oxidative response–related genes and genes encoding for ribosomal proteins, cellular metabolism, signal transduction, and solute transporters. Altered expression of these genes likely reflects an altered protein product (not determined in the present study).
Oxidative Stress Response
Reactive oxygen species (ROS) are produced by the metabolism of O2 in all aerobic cells and are essential for normal cellular signaling functions. However, oxidative stress can occur as a result of either increased ROS generation or depressed antioxidant system, or both. Of them, SOD, catalase, and GPx constitute the main components of the antioxidant defense system. These antioxidants protect the cell against cytotoxic ROS such as superoxide anions, hydrogen peroxide, and hydroxyl radicals. The measurement of peroxides in biologic systems is one of the factors allowing the determination of the degree of certain free radicals present in specific tissues. Recently, Jung et al. (2003) suggested that H2O2 produced by arsenite might activate growth factor receptor by increasing its tyrosine phosphorylation. These data indicated that H2O2 might be a pivotal mediator of the tumor-promoting activity of arsenite (Jung 2003). In the present study we observed that UN induces dose-dependent production of H2O2. We also observed an increase in Cu,Zn-SOD mRNA levels in the kidney. SOD is an enzyme responsible for dismutation of highly reactive superoxide radicals to H2O2. Moreover, GPx, which scavenges H2O2 and lipid peroxides, had its gene expression level increased, potentially induced by the high concentrations of H2O2. Induction of oxidative balance perturbation has been previously described in UN-induced ARF (Schramm et al. 2002). In addition, it has also been reported that some toxicants such as cadmium and arsenic are able to induce an increase in H2O2 levels after acute exposure (Ercal et al. 2001). Taken together, these data suggest that UN induces oxidative stress. Exploring this point seems of interest in evaluating the risks of UN long-term exposures.
Involvement of Genes Encoding Ion Transporters
The proximal tubule (especially the S3 segment) and the outer medullary thick ascending limb suffer the most severe injury after toxic and ischemic insult (Kwon et al. 2000; Sun et al. 2000). Although basolateral transport of sodium among the entire nephron and collecting ducts occurs via the active Na-K-ATPase pump, the active absorption is mediated by the Na+-dependent inorganic phosphate co-transporters (NaPi-II). In contrast to a previous study (Park et al. 1997) showing that chronic exposure to cadmium impairs the Pi transport capacity, probably by reducing the effective number of NaPi co-transporter units, we found that UN long-term exposure induces an increase of NaPi-II mRNA levels. As already suggested (Levi et al. 1994; Loghman-Adham 1997), this increase in NaPi-II is probably the result of an increase in Vmax by a transporter-shuttling mechanism, which is sensitive to disruptors of microtubule integrity. In addition, as previously reported (Moz et al. 1999) in hypophosphatemia studies, our observations suggest that UN chronic exposure could enhance the renal translational machinery. Further experiments, for example, examining the in vivo rates of NaPi-II synthesis, should allow clarification of whether UN-like hypophosphatemia affects NaPi-II translation. Moreover, Na-K-ATPase expression levels are down-regulated after UN long-term ingestion. This observation is consistent with previous work, after ischemic injury, that also shows a decreased Na-K-ATPase mRNA transcription (Kwon et al. 2000). The potential significance of this observation is that urine volume might be increased because of decreased Na+ reabsorption. Unfortunately, urine volumes were not recorded throughout the experiments, and the monitoring of the water consumption did not reveal any change in differently treated groups compared with controls. Thus, the role of these proteins in response to UN exposure remains unclear and warrants additional investigation.
Involvement of Protein Biosynthesis–Related Genes
Interestingly, many ribosomal subunits and other factors involved in protein synthesis (elongation factor) were induced upon UN treatment. Ribosomal proteins are major component of ribosomes that catalyze protein biosynthesis in the cytoplasm of cells. Under normal growth conditions, ribosomal proteins are synthesized stoichiometrically, in relation with ribosomal RNA, to produce an equimolar supply of ribosomal components. However, regulation of the transcriptional activity of the genes encoding for ribosomal protein in differentiated human tissues appears to be less concertedly regulated than previously reported (Bortoluzzi et al. 2001). Recent progress in ribosome research provides growing evidence that ribosomal proteins can also have a function during various cellular processes such as replication, transcription, RNA processing, DNA repair, and even inflammation; all these functions are independent of their own involvement in the protein biosynthesis (Wool 1996; Yamamoto 2000). In the present work, up-regulation of transcripts for several ribosomal proteins such as RPL13a, RPL19, RPL30, RPLP1, RPS24, and RPS26 has been observed. This latter has been described as a marker to differentiate either ozone or ultraviolet B radiation environmental stresses in plants (Brosché and Strid 1999). Whereas RPS4, RPL19, and RPS18 have been involved in regulation of the development (Wool 1996), RPL13A, RPS18, and RPS24 have been associated in the maturation of mucosal epithelia (Kasai et al. 2003). Moreover, the latter was markedly decreased in colorectal cancer (Kasai et al. 2003). Taken together, these observations may suggest that UN induce a perturbation in protein synthesis and offer a new putative way of investigation on cellular proliferation study after chronic UN exposure.
Others Genes of Interest
ODC, described as the rate-limiting enzyme of polyamine biosynthesis and a marker of G1 phase, is down-regulated in long-term UN exposure. Recently, Fleck et al. (2003) also observed a decrease in Odc expression levels 10 weeks after a single injection of UN. Kramer et al. (2001) have showed that a depletion of polyamine pool, through inhibition of ODC, causes p21-mediated G1 cell cycle arrest, followed by development of a senescence-like phenotype and loss of cellular proliferative capacity. Thus, the decrease in Odc mRNA levels might be related to an arrest of the cell cycle after UN treatment. However, these data are inconsistent with the observed increase in protein biosynthesis–related genes. It has been previously reported that mammalian ODC protein has a very short half-life; its control is under negative feedback regulation by the polyamines, and its degradation is dependent on 26S proteasome complex (Hascilowicz et al. 2002). Interestingly, we noted an increase in proteasome subunit (Psma7) mRNA expression levels. Nevertheless, further study with added dimensions of time and doses may clarify the observed modest Odc mRNA expression levels for the group 2 and allow a best evaluation of uranium chronic exposure impact on its expression. Of particular interest, Umod (Tamm-Horsfall protein) was decreased in the present study. This protein is one of the most abundant in the renal tubule (Bachmann et al 1990). Moreover, expression levels of UMOD have been previously reported to decrease in ischemic-induced ARF (Yoshida et al. 2002). Unexpectedly, in previous work performed in our laboratory, we showed that its expression level was increased in UN-induced ARF. In addition, an up-regulation of Umod has been observed in the progression of nephrolithiasis (Katsuma et al. 2002). However, the role of this protein remains unclear and requires additional investigation. Finally, and perhaps more interestingly, TCTP, a cytoplasmic protein usually found in both normal and tumor cell lines, is overexpressed after UN long-term ingestion. It was identified as an antiapoptotic protein (Li et al. 2001). TCTP is associated with components of the translational machinery, the elongation factors implicated in tumor formation (Cans et al. 2003). Interestingly, we observed dose-dependent increases in Tctp mRNA levels using RT-PCR analysis. Further investigations are necessary to evaluate the implication of this protein in potentially adverse health effects.
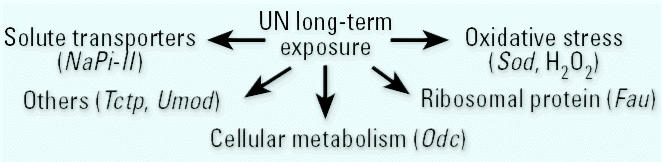
In summary, by using SAGE, we elegantly demonstrated that UN chronic exposure induces changes in expression profiles. The present report provides the first evidence that UN alters the expression of numerous genes including those encoding for oxidative-stress–related proteins, ribosomal proteins, solute transporters, and genes involved in cellular metabolism or signal transduction (Figure 3). Although these molecular changes, resulting from a subclinical toxicity, do not systematically lead to kidney failure or overt illness, our results might constitute a determining step in the identification of sensitive biomarkers to prevent the development of a UN-induced renal injury. Moreover, although studying human biology is ideal, such studies are neither feasible nor ethical. Thus, the vast majority of current biomedical research is conducted using mice and rats. However, we must keep in mind that extrapolation to humans might have some bias because humans can be exposed to many compounds simultaneously, often on a chronic or intermittent basis. Thus, the use of throughput genomic approaches after long-term exposure to mixtures of toxicants might help in the assessment of interactions such as additivity, synergism, or antagonism. The comparison of gene expression profiles could help to identify putative new sensitive biomarkers of chronic nephrotoxicity and then evaluate the impact of environmental toxic contaminants on human health.
Figure 3. Cellular pathways triggered in response to UN long-term exposure. Some genes or molecules, which present an altered expression level after uranium ingestion, emphasize the implication of these cellular processes in UN long-term exposure (in parentheses).
References
- Amin RP, Vickers AE, Sistare F, Thompson KL, Roman RJ, Lawton M, et al. Identification of putative gene-based markers of renal toxicity. Environ Health Perspect. 2004;112(4):465–479. doi: 10.1289/ehp.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew AS, Warren AJ, Barchowsky A, Temple KA, Klei L, Soucy NV, et al. Genomic and proteomic profiling of responses to toxic metals in human lung cells. Environ Health Perspect. 2003;111(6):825–835. doi: 10.1289/ehp.111-1241504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann S, Metzger R, Bunnemann B. Tamm-Horsfall protein-mRNA synthesis is localized to the thick ascending limb of Henle’s loop in rat kidney. Histochemistry. 1990;94(5):517–523. doi: 10.1007/BF00272616. [DOI] [PubMed] [Google Scholar]
- Bartosiewicz MJ, Jenkins D, Penn S, Emery J, Buckpitt A. Unique gene expression patterns in liver and kidney associated with exposure to chemical toxicants. J Pharmacol Exp Ther. 2001;297(3):895–905. [PubMed] [Google Scholar]
- Bortoluzzi S, Alessi F, Romualdi C, Danieli GA. Differential expression of genes coding for ribosomal proteins in different human tissues. Bioinformatics. 2001;17(12):1152–1157. doi: 10.1093/bioinformatics/17.12.1152. [DOI] [PubMed] [Google Scholar]
- Bosshard E, Zimmerli B, Schalatter C. Uranium in diet: risk assessment of its nephro- and radiotoxicity. Chemosphere. 1992;24(3):309–321. [Google Scholar]
- Brosché M, Strid A. The mRNA-binding ribosomal protein S26 as a molecular marker in plants: molecular cloning, sequencing and differential gene expression during environmental stress. Biochim Biophys Acta. 1999;1445(3):342–344. doi: 10.1016/s0167-4781(99)00050-0. [DOI] [PubMed] [Google Scholar]
- Cans C, Passer BJ, Shalak V, Nancy-Portebois V, Crible V, Amzallag N, et al. Translationally controlled tumor protein acts as a guanine nucleotide dissociation inhibitor on the translation elongation factor eEF1A. Proc Natl Acad Sci USA. 2003;100(24):13892–13897. doi: 10.1073/pnas.2335950100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabardes-Garonne D, Mejean A, Aude JC, Cheval L, Di Stefano A, Gaillard MC, et al. A panoramic view of gene expression in the human kidney. Proc Natl Acad Sci USA. 2003;100(23):13710–13715. doi: 10.1073/pnas.2234604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejnik JW, Hamilton MM, Adams PR, Carmichael AJ. Optimal sample preparation conditions for the determination of uranium in biological samples by kinetic phosphorescence analysis (KPA) J Pharm Biomed Anal. 2000;24(2):227–235. doi: 10.1016/s0731-7085(00)00412-x. [DOI] [PubMed] [Google Scholar]
- El-Meanawy MA, Schelling JR, Pozuelo F, Churpek MM, Ficker EK, Iyengar S, et al. Use of serial analysis of gene expression to generate kidney expression libraries. Am J Physiol Renal Physiol. 2000;279(2):F383–F392. doi: 10.1152/ajprenal.2000.279.2.F383. [DOI] [PubMed] [Google Scholar]
- Ercal N, Gurer-Orhan H, Aykin-Burns N. Toxic metals and oxidative stress part I: mechanisms involved in metal-induced oxidative damage. Curr Top Med Chem. 2001;1(16):529–539. doi: 10.2174/1568026013394831. [DOI] [PubMed] [Google Scholar]
- Fleck C, Sutter L, Appenroth D, Koch B, Meinhold T, Pitack M, et al. Use of gene chip technology for the characterisation of the regulation of renal transport processes and of nephrotoxicity in rats. Exp Toxicol Pathol. 2003;54(5–6):401–410. doi: 10.1078/0940-2993-00289. [DOI] [PubMed] [Google Scholar]
- Gilman AP, Moss MA, Villeneuve DC, Secours VE, Yagminas AP, Tracy BL, et al. Uranyl nitrate: 91-day exposure and recovery studies in the male New Zealand white rabbit. Toxicol Sci. 1998a;41(1):138–151. doi: 10.1006/toxs.1997.2369. [DOI] [PubMed] [Google Scholar]
- Gilman AP, Villeneuve DC, Secours VE, Yagminas AP, Tracy BL, Quinn JM, et al. Uranyl nitrate: 28-day and 91-day toxicity studies in the Sprague-Dawley rat. Toxicol Sci. 1998b;41(1):117–128. doi: 10.1006/toxs.1997.2367. [DOI] [PubMed] [Google Scholar]
- Gilman AP, Villeneuve DC, Secours VE, Yagminas AP, Tracy BL, Quinn JM, et al. Uranyl nitrate: 91-day toxicity studies in the New Zealand white rabbit. Toxicol Sci. 1998c;41(1):129–137. doi: 10.1006/toxs.1997.2368. [DOI] [PubMed] [Google Scholar]
- Hascilowicz T, Murai N, Matsufuji S, Murakami Y. Regulation of ornithine decarboxylase by antizymes and antizyme inhibitor in zebrafish (Danio rerio) Biochim Biophys Acta. 2002;1578(1–3):21–28. doi: 10.1016/s0167-4781(02)00476-1. [DOI] [PubMed] [Google Scholar]
- Heng YM, Kuo CS, Jones PS, Savory R, Schulz RM, Tomlinson SR, et al. A novel murine P-450 gene, Cyp4a14, is part of a cluster of Cyp4a and Cyp4b, but not of CYP4F, genes in mouse and humans. Biochem J. 1997;325(pt 3):741–749. doi: 10.1042/bj3250741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Invitrogen Corporation 2004. Guide for Constructing SAGE Libraries, version H. Available: http://www.invitrogen.com/content/sfs/manuals/sage_man.pdf [assessed 7 October 2004].
- Jung DK, Bae GU, Kim YK, Han SH, Choi WS, Kang H, et al. Hydrogen peroxide mediates arsenite activation of p70(s6k) and extracellular signal-regulated kinase. Exp Cell Res. 2003;290(1):144–154. doi: 10.1016/s0014-4827(03)00320-3. [DOI] [PubMed] [Google Scholar]
- Juntunen R. 1991. Uranium and Radon in Wells Drilled into Bedrock in Southern Finland. Report of Investigation, Geological Survey of Finland.
- Kasai H, Nadano D, Hidaka E, Higuchi K, Kawakubo M, Sato TA, et al. Differential expression of ribosomal proteins in human normal and neoplastic colorectum. J Histochem Cytochem. 2003;51(5):567–574. doi: 10.1177/002215540305100502. [DOI] [PubMed] [Google Scholar]
- Katsuma S, Shiojima S, Hirasawa A, Takagaki K, Kaminishi Y, Koba M, et al. Global analysis of differentially expressed genes during progression of calcium oxalate nephrolithiasis. Biochem Biophys Res Commun. 2002;296(3):544–552. doi: 10.1016/s0006-291x(02)00840-9. [DOI] [PubMed] [Google Scholar]
- Kramer DL, Chang BD, Chen Y, Diegelman P, Alm K, Black AR, et al. Polyamine depletion in human melanoma cells leads to G1 arrest associated with induction of p21WAF1/CIP1/SDI1, changes in the expression of p21-regulated genes, and a senescence-like phenotype. Cancer Res. 2001;61(21):7754–7762. [PubMed] [Google Scholar]
- Kurttio P, Auvinen A, Salonen L, Saha H, Pekkanen J, Makelainen I, et al. Renal effects of uranium in drinking water. Environ Health Perspect. 2002;110(4):337–342. doi: 10.1289/ehp.02110337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon TH, Froklaer J, Huan JS, Knepper MA, Nielson S. Decreased abundance of major Na+ transporters in kidneys of rats with ischemia-induced acute renal failure. Am J Physiol Renal Physiol. 2000;278:F925–F939. doi: 10.1152/ajprenal.2000.278.6.F925. [DOI] [PubMed] [Google Scholar]
- Levi M, Lotscher M, Sorribas V, Custer M, Arar M, Kaissling B, et al. Cellular mechanisms of acute and chronic adaptation of rat renal P(i) transporter to alterations in dietary P(i) Am J Physiol. 1994;267:F900–F908. doi: 10.1152/ajprenal.1994.267.5.F900. [DOI] [PubMed] [Google Scholar]
- Li F, Zhang D, Fujise K. Characterization of for-tilin, a novel antiapoptotic protein. J Biol Chem. 2001;276(50):47542–47549. doi: 10.1074/jbc.M108954200. [DOI] [PubMed] [Google Scholar]
- Lin RH, Wu LJ, Lee CH, Lin-Shiau SY. Cytogenetic toxicity of uranyl nitrate in Chinese hamster ovary cells. Mutat Res. 1993;319(3):197–203. doi: 10.1016/0165-1218(93)90079-s. [DOI] [PubMed] [Google Scholar]
- Loghman-Adham M. Adaptation to changes in dietary phosphorus intake in health and in renal failure. J Lab Clin Med. 1997;129:176–188. doi: 10.1016/s0022-2143(97)90137-2. [DOI] [PubMed] [Google Scholar]
- Mao Y, Desmeules M, Schaubel D, Berude D, Dyck R, Brule D, et al. Inorganic components of drinking water and microalbuminuria. Environ Res. 1995;71(2):135–140. doi: 10.1006/enrs.1995.1075. [DOI] [PubMed] [Google Scholar]
- McDonald-Taylor CK, Singh A, Gilman A. Uranyl nitrate-induced proximal tubule alterations in rabbits: a quantitative analysis. Toxicol Pathol. 1997;25(4):381–389. doi: 10.1177/019262339702500406. [DOI] [PubMed] [Google Scholar]
- Miller AC, Fuciarelli AF, Jackson WE, Ejnik EJ, Emond C, Strocko S, et al. Urinary and serum mutagenicity studies with rats implanted with depleted uranium or tantalum pellets. Mutagenesis. 1998;13(6):643–648. doi: 10.1093/mutage/13.6.643. [DOI] [PubMed] [Google Scholar]
- Miller AC, Stewart M, Brooks K, Shi L, Page N. Depleted uranium-catalyzed oxidative DNA damage: absence of significant alpha particle decay. J Inorg Biochem. 2002;91(1):246–252. doi: 10.1016/s0162-0134(02)00391-4. [DOI] [PubMed] [Google Scholar]
- Moss MA, McCurdy RF, Dooley KC, Givner ML, Dymond LC, Slayter JM, et al. 1983. Uranium in drinking water-report on clinical studies in Nova Scotia. In: Chemical Toxicology and Clinical Chemistry of Metals (Brown SS, Savory J, eds). London:Academic Press, 149–152.
- Mouse SAGE site Available: http://mouse.biomed.cas.cz/sage/ [accessed 7 October 2004].
- Moz Y, Silver J, Naveh-Many T. Protein-RNA interactions determine the stability of the renal NaPi-2 cotransporter mRNA and its translation in hypophosphatemic rats. J Biol Chem. 1999;274(36):25266–25272. doi: 10.1074/jbc.274.36.25266. [DOI] [PubMed] [Google Scholar]
- Park K, Kim KR, Kim JY, Park YS. Effect of cadmium on Na-Pi cotransport kinetics in rabbit renal brush-border membrane vesicles. Toxicol Appl Pharmacol. 1997;145(2):255–259. doi: 10.1006/taap.1997.8180. [DOI] [PubMed] [Google Scholar]
- SAGE 2004. Home page. Available: http://www.sagenet.org/ [accessed 7 October 2004].
- Schramm L, La M, Heidbreder E, Hecker M, Beckman JS, Lopau K, et al. L-Arginine deficiency and supplementation in experimental acute renal failure and in human kidney transplantation. Kidney Int. 2002;61(4):1423–1432. doi: 10.1046/j.1523-1755.2002.00268.x. [DOI] [PubMed] [Google Scholar]
- Sun DF, Fujigaki Y, Fujimoto T, Goto T, Yonemura K, Hishida A. Relation of distal nephron changes to proximal tubular damage in uranyl acetate-induced acute renal failure in rats. Am J Nephrol. 2002;22(5–6):405–416. doi: 10.1159/000065265. [DOI] [PubMed] [Google Scholar]
- Sun DF, Fujigaki Y, Fujimoto T, Yonemura K, Hishida A. Possible involvement of myofibroblasts in cellular recovery of uranyl acetate-induced acute renal failure in rats. Am J Pathol. 2000;157(4):1321–1335. doi: 10.1016/S0002-9440(10)64647-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270(5235):484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Zhou W, Vogelstein J, Basrai MA, Basset DE, Jr, et al. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- Virlon B, Cheval L, Buhler JM, Billon E, Doucet A, Elalouf JM. Serial microanalysis of renal transcriptomes. Proc Natl Acad Sci USA. 1999;96(26):15286–15291. doi: 10.1073/pnas.96.26.15286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wool IG. Extraribosomal functions of ribosomal proteins. Trends Biochem Sci. 1996;21(5):164–165. [PubMed] [Google Scholar]
- Yamamoto T. Molecular mechanism of monocyte predominant infiltration in chronic inflammation: mediation by a novel monocyte chemotactic factor, S19 ribosomal protein dimer. Pathol Int. 2000;50(11):863–871. doi: 10.1046/j.1440-1827.2000.01132.x. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Kurella M, Beato F, Min H, Ingelfinger JR, Stears RL, et al. Monitoring changes in gene expression in renal ischemia-reperfusion in the rat. Kidney Int. 2002;61(5):1646–1654. doi: 10.1046/j.1523-1755.2002.00341.x. [DOI] [PubMed] [Google Scholar]
- Zamora ML, Tracy BL, Zielinski JM, Meyerhof DP, Moss MA. Chronic ingestion of uranium in drinking water: a study of kidney bioeffects in humans. Toxicol Sci. 1998;43(1):68–77. doi: 10.1006/toxs.1998.2426. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, et al. Gene expression profiles in normal and cancer cells. Science. 1997;276(5316):1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]