Abstract
Introduction
Embryo selection remains a key challenge in in vitro fertilization (IVF), as many morphologically “normal” embryos fail to implant. Artificial intelligence (AI) offers a promising tool for improving embryo assessment by providing more objective and accurate predictions of pregnancy outcomes. This study aims to systematically review and conduct a diagnostic meta-analysis to evaluate the effectiveness of AI-based tools in embryo selection for predicting pregnancy outcomes in IVF.
Methods
We conducted a systematic review following PRISMA guidelines, searching Web of Science, Scopus, and PubMed. Original research articles evaluating AI’s diagnostic accuracy in embryo selection were included, while duplicates, non-peer-reviewed papers, abstracts, and conference proceedings were excluded. Data on sample sizes, AI tools, and diagnostic metrics were extracted, with quality assessed using the QUADAS-2 tool.
Results
AI-based embryo selection methods showed strong diagnostic performance, with pooled sensitivity of 0.69 and specificity of 0.62 in predicting implantation success. The positive likelihood ratio was 1.84 and the negative likelihood ratio was 0.5. The area under the curve reached 0.7, indicating high overall accuracy. The Life Whisperer AI model achieved 64.3% accuracy in predicting clinical pregnancy, while the FiTTE system, which integrates blastocyst images with clinical data, improved prediction accuracy to 65.2% with an AUC of 0.7.
Conclusion
AI offers a promising advancement in embryo selection for IVF, with the potential to enhance clinical outcomes and improve decision-making. Future studies should focus on refining these models to achieve the ultimate goal of a healthy live birth by developing more sophisticated algorithms and validating them with larger, diverse datasets.
Keywords: In vitro fertilization (IVF), Embryo assessment, Pregnancy, Artificial intelligence (AI), Diagnostic accuracy
Introduction
In vitro fertilization (IVF) stands as a pivotal intervention in the treatment of infertility, involving a complex series of procedures designed to achieve successful pregnancy. This process includes ovarian stimulation to produce multiple oocytes, retrieval of these oocytes, fertilization in vitro, and subsequent embryo culture under meticulously controlled conditions for a period ranging from 1 to 5 days [1]. Despite notable advancements in IVF technology and techniques, the overall success rates remain modest, with average live birth rates hovering around 30% per embryo transfer [2]. Consequently, optimizing the embryo selection process is essential for enhancing the efficacy of IVF treatments and improving success rates.
Traditionally, the selection of embryos for transfer has been based on a range of morphological parameters assessed at specific developmental stages. These include the appearance of pronuclei, early cleavage patterns, multinucleation, and characteristics of the zona pellucida [3]. While these morphological assessments provide valuable insights, they offer only a limited perspective on embryo viability. Recent advancements have introduced time-lapse imaging technology, which facilitates continuous, real-time monitoring of embryo development [4]. This innovation allows for the detection of subtle morphological changes and critical developmental milestones with precise time-stamping, leading to the development of advanced embryo selection models based on morphokinetic parameters [5]. These models have demonstrated promise in predicting key outcomes such as blastocyst formation, genetic status, and live birth rates [6].
Despite the benefits of time-lapse monitoring and morphokinetic models, significant challenges remain in managing embryo selection within the busy environment of a clinical laboratory [7]. To address these issues, automatic scoring systems have been developed to assess embryo morphology and morphokinetics more efficiently. Early systems utilized traditional computer vision techniques, but recent advancements have incorporated sophisticated artificial intelligence (AI) technologies, including artificial neural networks (ANNs), deep learning, and machine learning algorithms [8]. These AI-driven approaches offer advanced decision support by ranking embryos and predicting their suitability for transfer or cryopreservation, thereby reducing the reliance on subjective human judgment [9].
The combination of AI into IVF practices presents the potential to significantly enhance the accuracy and reliability of embryo selection, minimizing subjectivity and providing more objective assessments of embryo viability [10]. However, the rapid evolution of AI technology introduces challenges, including the need for standardized performance metrics and concerns regarding the generalizability and precision of different models [11]. These challenges are partly attributable to the requirements for high-quality data and large sample sizes, which can be addressed by leveraging discarded embryos for model training and validation [12].
The adoption of AI in IVF is rapidly advancing, with numerous studies investigating the effectiveness of various AI systems. For example, models like Tran’s IVY have demonstrated exceptional predictive accuracy, while others, such as Geller’s AI model, have shown varying performance when compared to traditional methods. This variability underscores the necessity for a comprehensive evaluation of AI’s predictive capabilities in IVF [13, 14]. To address this need, our study conducts a systematic review and diagnostic meta-analysis to strictly assess the specificity and sensitivity of AI in identifying embryos with the highest implantation potential. By synthesizing data from multiple sources, our analysis aims to provide a thorough understanding of AI’s strengths and limitations in improving IVF success rates, ultimately guiding future research in this rapidly evolving field.
Compared to previous reviews such as those by Salih et al. (2023) and Sfakianoudis et al. (2022), our study offers a distinct contribution by conducting a diagnostic meta-analysis, which quantitatively synthesizes sensitivity, specificity, likelihood ratios, and AUC [15, 16]. These metrics provide a more clinically relevant understanding of AI’s utility in embryo selection. In contrast, earlier reviews primarily offered narrative assessments or qualitative comparisons between AI tools and traditional embryologist evaluations. For instance, Salih et al. provided a broad overview of AI applications in IVF, but without offering pooled diagnostic metrics. Similarly, Sfakianoudis et al. discussed AI’s potential impact but did not systematically evaluate diagnostic performance [15, 17]. Our study bridges this gap by presenting objective, pooled estimates, thereby enabling direct benchmarking of AI performance and facilitating more informed clinical decision-making.
Methods and materials
Design
This systematic review and diagnostic meta-analysis was conducted in accordance with the PRISMA guidelines for diagnostic test accuracy reviews [18].
Search strategy
To identify all relevant studies evaluating the diagnostic accuracy of AI in IVF, we performed a comprehensive search across four major databases: PubMed, Scopus, Web of Science, and Google Scholar. Our search strategy followed the Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy [19]. The search terms included: Artificial intelligence (AI), Machine learning (ML), Deep learning, Generative artificial intelligence (AI), Language model, Transformer deep neural network, Artificial neural network (ANN), Convolutional neural network (CNN), Computer vision, Supervised learning, Unsupervised learning, Supervised, Unsupervised, Recurrent neural network, Long short-term model, Linear regression, Logistic regression, Decision tree, k-nearest neighbors (k-NN), Support vector machine (SVM), Random forest, k-means, Generative adversarial network (GAN), Large language model (LLM), Proprietary algorithm, Lasso regression Model, light gradient boosting machine, True positive rate (TPR), Positive predicted value (PPV), Embryologists, Female, Fertility, Infertility, Sperm, Oocyte, Cumulus-oocyte complexes (COC), Immature oocyte, Mature oocyte, Germinal vesicle (GV) oocyte, Metaphase II (MII) oocyte, Metaphase I (MI) oocyte, Assisted reproductive biology (ART), In vitro fertilization (IVF), Intracytoplasmic sperm injection (ICSI), Culture, Embryo culture, Fertilization, Zygote, Pronuclear, Pronucleus, Two-pronuclear zygote (2PN), One-pronuclear zygote (2PN), Three-pronuclear zygote (2PN), Multinucleation, Cleavage, Embryo, Grading, Embryo Gading, Day-3 Embryo, Day-5 Embryo, Morphology, Blastomere, Cell symmetry, Symmetry, Vacuoles, Fragmentation, Blastocoel cavity, Blastocoel expansion, Inner cell mass (ICM), Trophectoderm, Morula, Early blastocyst, Blastocyst, Morphological assessment, Cleavage stage evaluation, Blastocyst grading, Hatching, Cytogenetic, Embryo biopsy, Genetic screening, Preimplantation genetic diagnosis (PGD), Preimplantation genetic testing (PGT), Pre-implantation genetic testing for aneuploidy (PGT-A), Ploidy, Aneuploidies, Polyploidy, Metabolomic, Embryo scope, Time-lapse imaging, Time-lapse microscopy, Morphokinetics, Embryo monitoring, Cell cycle timing, Kinetic parameters, Developmental milestones, Non-invasive, Non-invasive embryo selection, Viability, Omics, Embryo selection, Embryo quality, Implantation, Transfer, Embryo transfer, Single embryo transfer, Clinical outcomes, Implantation rate, Pregnancy, Clinical pregnancy, Clinical pregnancy rate, Implantation Failure, Recurrent implantation failure (RIF), Miscarriage, Abortion, Live birth, and Live birth rate.
Study selection
We included original research articles that evaluated the diagnostic accuracy of artificial intelligence (AI) in assessing embryos to predict pregnancy-related outcomes. Studies were eligible if they used image-based or clinical data inputs and reported diagnostic metrics such as sensitivity, specificity, or AUC. We excluded duplicate records, non-peer-reviewed articles, reviews, abstracts, conference proceedings, and book chapters. Eligible AI models included those using methods such as convolutional neural networks (CNNs), support vector machines (SVMs), and ensemble techniques. We considered models trained using supervised learning methods and validated either through internal cross-validation, external datasets, or prospective evaluation.
Data extraction
We extracted all relevant available data consisting of sample size, country, year, true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN), design, used AI tool, outcome, AUC, sensitivity (SEN), specificity (SP), accuracy, precision, PPV, and NPV.
Search strategy syntax
The search strategies and syntax were tailored for each database as follows:
PubMed (Results: 822)
((Artificial intelligence [Title/Abstract] OR Machine learning [Title/Abstract] OR Light gradient boosting machine [Title/Abstract] )) AND ((Blastocyst[Title/Abstract] OR Preimplantation genetic [Title/Abstract] OR Ploidy[Title/Abstract] OR Zygote [Title/Abstract] OR Fertilization[Title/Abstract] OR Embryo [Title/Abstract] OR In vitro fertilization [Title/Abstract] OR Assisted reproductive biology [Title/Abstract] OR Oocyte [Title/Abstract] OR Sperm [Title/Abstract] OR Infertility [Title/Abstract] OR Fertility [Title/Abstract]))
Scopus (Results: 756)
Query string: (TITLE-ABS ( " Artificial intelligence " ) OR TITLE-ABS ( " Machine learning " )) AND ( TITLE-ABS ( " embryo” ) OR TITLE-ABS ( " sperm " ) OR TITLE-ABS ( " Zygote " ) OR TITLE-ABS ( " In vitro fertilization " ) OR TITLE-ABS ( " Assisted reproductive biology " ) OR TITLE-ABS ( " Oocyte " )).
Web of science (Results: 272)
((TI=(Artificial intelligence OR Machine learning )) AND ((TI=( sperm OR embryo OR In vitro fertilization OR Oocyte))))
Risk of bias assessment
We assessed the methodological quality of the selected studies through the revised Quality Assessment of Diagnostic Accuracy Studies (QUADAS- 2) tool, recommended by the Cochrane collaboration for the risk of bias and applicability concerns [20] in which “Patient Selection”, “Details of used index test”, “Details of used standard test”, and “how to follow the samples and perform the procedures” were assessed. Each of the four items was categorized as high, low, or unclear, and disagreements were resolved by a third reviewer (RV).
Statistical issues
We used the MIDAS module in STATA/SE 11.2 to perform all analyses related to diagnostic test accuracy [21]. From each study, we extracted true positive (TP), false positive (FP), false negative (FN), and true negative (TN) values to calculate key diagnostic metrics, including sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), and diagnostic odds ratio (DOR). Pooled estimates and their 95% confidence intervals were calculated using a bivariate random-effects regression model and the hierarchical summary receiver operating characteristic (HSROC) model, which jointly analyze sensitivity and specificity while accounting for between-study heterogeneity [22, 23]. These models were implemented through MIDAS, which allows integrated bivariate and HSROC analyses within STATA.
Results were visualized using coupled forest plots for sensitivity, specificity, and likelihood ratios, as well as an HSROC curve to illustrate overall test performance. Study heterogeneity was evaluated using Cochran’s Q test and the I² statistic. Potential publication bias was assessed through Deek’s funnel plot asymmetry test. Where possible, we extracted details on the development of AI tools from the original studies, including their training-validation processes (e.g., cross-validation, external validation) and the types of algorithms used (e.g., CNNs, SVMs, ensemble methods), to provide further context for interpreting diagnostic performance.
Results
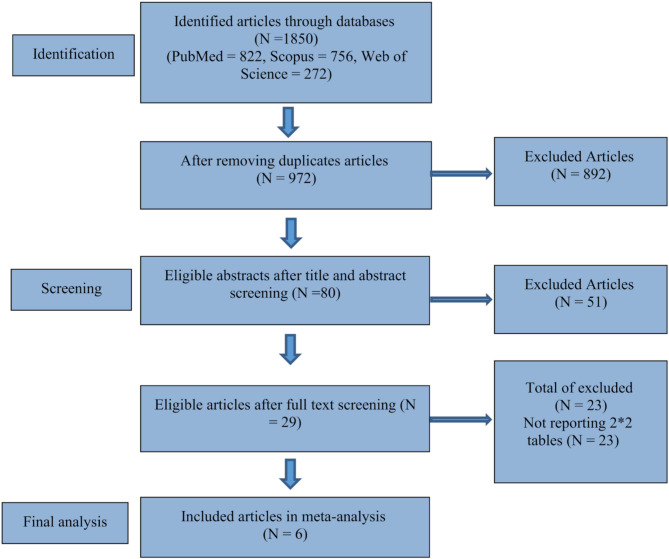
A total of 1,850 records were identified through database searches (PubMed = 822, Scopus = 756, Web of Science = 272). After removing 878 duplicates, 972 articles were screened. Of these, 80 abstracts were reviewed, and 29 full-text articles were assessed for eligibility. Twenty-three were excluded for not reporting diagnostic 2 × 2 data, leaving six studies for final inclusion in the meta-analysis (Table 1). The PRISMA flowchart summarizing the study selection process is shown in Fig. 1.
Table 1.
Summary of main characteristics of studies included in the diagnostic meta-analysis
| Study No (ref) |
Author year | Region | N | AI tool | Outcome | AUC | Sen | SP | TP | FP | FN | TN | Accuracy | Precision | PPV | NPV |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 [24] | Bori et al., 2021 (1) | Spain | 367 | Conventional morphokinetics using artificial neural network | Implantation | 0.64 | 0.88 | 0.46 | 0.71 | |||||||
| 1 [24] | Bori et al., 2021 (2) | Spain | 367 | Novel morphodynamics using artificial neural network | Implantation | 0.73 | 0.86 | 0.58 | 0.75 | |||||||
| 1 [24] | Bori et al., 2021 (3) | Spain | 367 | Conventional morphokinetics þ novel morphodynamics using artificial neural network | Implantation | 0.77 | 0.82 | 0.67 | 0.76 | |||||||
| 1 [24] | Bori et al., 2021 (4) | Spain | 367 | Discriminatory variables from statistical test using artificial neural network | Implantation | 0.68 | 0.85 | 0.57 | 0.74 | |||||||
| 2 [25] | Coticchio et al., 2021 (1) | Italy | 230 | k-Nearest Neighbor/K-NN-2 using artificial neural network | Blastocyst development | 0.76 | 0.74 | 0.72 | 0.74 | |||||||
| 2 [25] | Coticchio et al., 2021 (2) | Italy | 230 | k-Nearest Neighbor/K-NN-1 using artificial neural network | Blastocyst development | 0.82 | 0.60 | 0.71 | 0.66 | |||||||
| 2 [25] | Coticchio et al., 2021 (3) | Italy | 230 | Long-Short Term Memory Neural Network (LSTM-NN-1) | Blastocyst development | 0.76 | 0.65 | 0.71 | 0.68 | |||||||
| 2 [25] | Coticchio et al., 2021 (4) | Italy | 230 | Long-Short Term Memory Neural Network (LSTM-NN-2) | Blastocyst development | 0.61 | 0.80 | 0.71 | 0.75 | |||||||
| 3 [26] | Enatsu et al., 2022 (1) | Japan | 561 | Image only model/FiTTE of blastocyst | Clinical pregnancy | 0.53 | 0.57 | 452 | 345 | 162 | 399 | 0.62 | 0.53 | |||
| 3 [26] | Enatsu et al., 2022 (2) | Japan | 65 | Ensemble model using blastocyst images and clinical data | Clinical pregnancy | 0.66 | 0.64 | 51 | 19 | 28 | 37 | 0.65 | 0.56 | |||
| 3 [26] | Enatsu et al., 2022 (3) | Japan | 214 | Imaging of blastocyst | Live birth | 0.29 | 0.93 | 1030 | 308 | 83 | 131 | 0.75 | 0.30 | |||
| 4 [27] | He et al., 2024 (1) | China | 184 | Timelapse | Blastocyst euploidy | 0.76 | 0.76 | 0.67 | 0.72 | 0.71 | 0.71 | 0.72 | ||||
| 4 [27] | He et al., 2024 (2) | China | 184 | Non-invasive chromosomal screening (NICS) | Blastocyst euploidy | 0.91 | 0.93 | 0.74 | 0.84 | 0.79 | 0.79 | 0.9 | ||||
| 4 [27] | He et al., 2024 (3) | China | 184 | NICS-timelapse | Blastocyst euploidy | 0.94 | 0.86 | 0.87 | 0.86 | 0.87 | 0.87 | 0.86 | ||||
| 5 [28] | Liao et al., 2021 (1) | China | 264 | Pronuclei (PN) estimation model | Blastocyst formation | 0.94 | 0.90 | 0.93 | 0.94 | 0.9 | ||||||
| 5 [28] | Liao et al., 2021 (2) | China | 8346 | Temporal stream network | Blastocyst formation | 0.84 | 0.64 | 0.76 | 0.78 | 0.73 | ||||||
| 5 [28] | Liao et al., 2021 (3) | China | 8346 | Spatial stream network | Blastocyst formation | 0.70 | 0.69 | 0.70 | 0.81 | 0.68 | ||||||
| 5 [28] | Liao et al., 2021 (4) | China | 8346 | Spatial–temporal ensemble model (STEM) | Blastocyst formation | 0.85 | 0.66 | 0.78 | 0.79 | 0.75 | ||||||
| 5 [28] | Liao et al., 2021 (5) | China | 8346 | Temporal stream network | Usable blastocyst formation | 0.74 | 0.70 | 0.71 | 0.57 | 0.83 | ||||||
| 5 [28] | Liao et al., 2021 (6) | China | 8346 | Spatial stream network | Usable blastocyst formation | 0.77 | 0.64 | 0.68 | 0.53 | 0.83 | ||||||
| 5 [28] | Liao et al., 2021 (7) | China | 8346 | Spatial–temporal ensemble model (STEM) | Usable blastocyst formation | 0.75 | 0.70 | 0.71 | 0.57 | 0.84 | ||||||
| 6 [29] | Miyagi et al., 2019a | Japan | 160 | Imaging of blastocyst using machine learning | Live birth without aneuploidy | 0.65 | 0.60 | 0.70 | 0.65 | 0.66 | 0.63 | |||||
| 7 [30] | Miyagi et al., 2019b (1) | Japan | 181 | AI with deep learning in the convolutional neural network | Live birth in < 35 yrs | 0.634 | 0.53 | 0.72 | 0.64 | 0.56 | 0.69 | |||||
| 7 [30] | Miyagi et al., 2019b (2) | Japan | 84 | AI with deep learning in the convolutional neural network | Live birth in 35–37 yrs | 0.688 | 0.65 | 0.68 | 0.67 | 0.51 | 0.79 | |||||
| 7 [30] | Miyagi et al., 2019b (3) | Japan | 33 | AI with deep learning in the convolutional neural network | Live birth in 38–39 yrs | 0.728 | 0.69 | 0.69 | 0.69 | 0.41 | 0.88 | |||||
| 7 [30] | Miyagi et al., 2019b (4) | Japan | 20 | AI with deep learning in the convolutional neural network | Live birth in 40–41 yrs | 0.743 | 0.65 | 0.79 | 0.77 | 0.30 | 0.94 | |||||
| 7 [30] | Miyagi et al., 2019b (5) | Japan | 6 | AI with deep learning in the convolutional neural network | Live birth in ≥ 42 yrs | 0.837 | 0.83 | 0.86 | 0.86 | 0.22 | 0.99 | |||||
| 7 [30] | Miyagi et al., 2019b (6) | Japan | 181 | Combination of AI with conventional embryo evaluation/CEE | < Live birth in 35 yrs | 0.655 | 0.65 | 0.59 | 0.61 | 0.51 | 0.71 | |||||
| 7 [30] | Miyagi et al., 2019b (7) | Japan | 84 | Combination of AI with conventional embryo evaluation/CEE | Live birth in 35–37 yrs | 0.723 | 0.78 | 0.61 | 0.67 | 0.50 | 0.84 | |||||
| 7 [30] | Miyagi et al., 2019b (8) | Japan | 33 | Combination of AI with conventional embryo evaluation/CEE | Live birth in 38–39 yrs | 0.791 | 0.75 | 0.72 | 0.73 | 0.45 | 0.9 | |||||
| 7 [30] | Miyagi et al., 2019b (9) | Japan | 20 | Combination of AI with conventional embryo evaluation/CEE | Live birth in 40–41 yrs | 0.806 | 0.7 | 0.81 | 0.8 | 0.35 | 0.95 | |||||
| 7 [30] | Miyagi et al., 2019b (10) | Japan | 6 | Combination of AI with conventional embryo evaluation/CEE | Live birth in ≥ 42 yrs | 0.888 | 1 | 0.77 | 0.78 | 0.17 | 1 | |||||
| 8 [31] | VerMilyea et al., 2020 | Australia | 262 | AI-based model using static two-dimensional optical light microscope image | Day 5 blastocyst | 0.7 | 0.6 | 100 | 34 | 61 | 67 | 0.64 | ||||
| 9 [32] | Wen et al., 2022 (1) | Taiwan | 1507 | Random forest model/RFM | Clinical pregnancy | 0.76 | 0.7 | 0.71 | 0.71 | 0.62 | 0.78 | |||||
| 9 [32] | Wen et al., 2022 (2) | Taiwan | 1507 | Support vector machine (SVM) | Clinical pregnancy | 0.78 | 0.72 | 0.71 | 0.71 | 0.62 | 0.79 | |||||
| 9 [32] | Wen et al., 2022 (3) | Taiwan | 1507 | Light gradient boosting machine (LightGBM) | Clinical pregnancy | 0.75 | 0.71 | 0.71 | 0.71 | 0.62 | 0.78 | |||||
| 9 [32] | Wen et al., 2022 (4) | Taiwan | 1507 | Multilayer perceptron (MLP) | Clinical pregnancy | 0.76 | 0.72 | 0.63 | 0.67 | 0.57 | 0.77 | |||||
| 9 [32] | Wen et al., 2022 (5) | Taiwan | 1507 | Extreme gradient boosting (XGBoost) | Clinical pregnancy | 0.78 | 0.71 | 0.71 | 0.71 | 0.62 | 0.78 | |||||
| 10 [33] | Yang et al., 2022 | USA | 367 | Random forest learning algorithm | Clinical pregnancy | 0.70 | 0.65 | 0.6 | 0.74 | 0.50 | ||||||
| 11 [34] | Blank et al., 2019 (1) | Belgium | 1052 | Random forest model/RFM | Ongoing pregnancies of 11 weeks | 0.74 | 0.84 | 0.48 | ||||||||
| 11 [34] | Blank et al., 2019 (2) | Belgium | 1052 | Multivariate logistic regression model/MvLRM | Ongoing pregnancies of 11 weeks | 0.66 | 0.66 | 0.58 | ||||||||
| 12 [35] | Amitai et al., 2023 (1) | Israel | 391 | Extreme gradient boosting (XGBoost) | First trimester miscarriage | 0.68 | 14 | 1 | 10 | 5 | ||||||
| 12 [35] | Amitai et al., 2023 (1) | Israel | 391 | Random forest model/RFM | First trimester miscarriage | 0.68 | 9 | 6 | 4 | 11 | ||||||
| 13 [36] | Uyar et al., 2014 | Turkey | 2453 | Machine Learning | Implantation | 0.63 | 24 | 19 | 23 | 107 | 0.80 | |||||
| 14 [37] | Abadjieva et al., 2023 | Bulgaria | 30 | Random forest model/RFM | Sperm quality | 22 | 1 | 1 | 6 | |||||||
| 15 [38] | Campanholi et al., 2023 (1) | Brazil | 290 | Artificial neural network | In vitro embryo production | 78 | 7 | 8 | 69 | |||||||
| 15 [38] | Campanholi et al., 2023 (2) | Brazil | 290 | Artificial neural network | In vitro embryo production | 69 | 25 | 15 | 59 | |||||||
| 15 [38] | Campanholi et al., 2023 (3) | Brazil | 290 | Artificial neural network | In vitro embryo production | 69 | 16 | 17 | 60 |
Fig. 1.
PRISMA flowchart for the meta-analysis
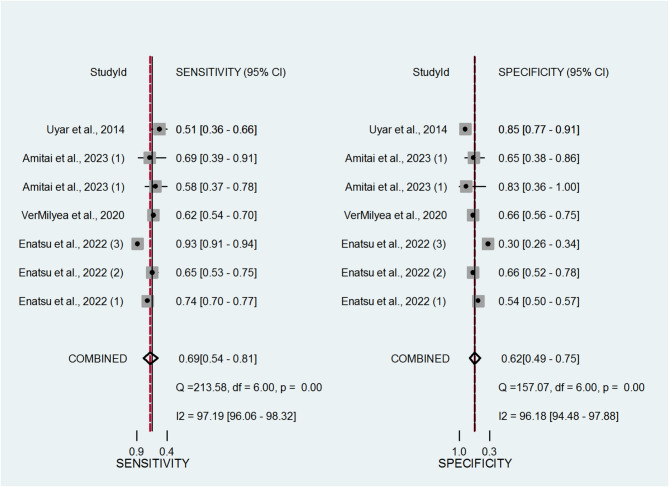
The forest plots in Fig. 2 display the sensitivity and specificity estimates from each study. The pooled sensitivity across studies was 0.69, and the pooled specificity was 0.62. These findings suggest that AI models have good potential to distinguish between embryos likely and unlikely to result in successful implantation. These values correspond to a Diagnostic Odds Ratio (DOR) of 3.69, suggesting moderate-to-strong overall discriminatory power of AI-based assessment tools in IVF.
Fig. 2.
Coupled forest plots displaying the pooled sensitivity and specificity estimates of artificial intelligence (AI) tools in predicting pregnancy-related outcomes in in vitro fertilization (IVF) cycles
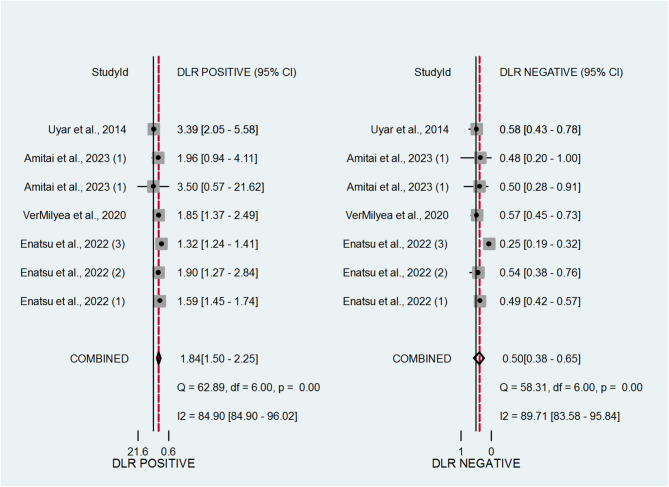
We presented the likelihood ratios (LRs) in Fig. 3, which illustrate the diagnostic value of AI-based embryo selection. The positive likelihood ratio (LR positive) is defined as the probability of a positive test result in patients with the outcome (pregnancy) divided by the probability of the same result in those without the outcome. In our analysis, the LR positive was 1.84, indicating that embryos identified as viable by AI models are approximately 2 times more likely to lead to a pregnancy compared to embryos that are not. The negative likelihood ratio (LR negative) was 0.5, suggesting that a negative result moderately reduces the probability of successful implantation. These findings support the moderate diagnostic utility of AI tools in embryo assessment.
Fig. 3.
Coupled forest plots presenting the pooled estimates of positive and negative likelihood ratios for artificial intelligence (AI) tools in assessing pregnancy-related outcomes in IVF cycles
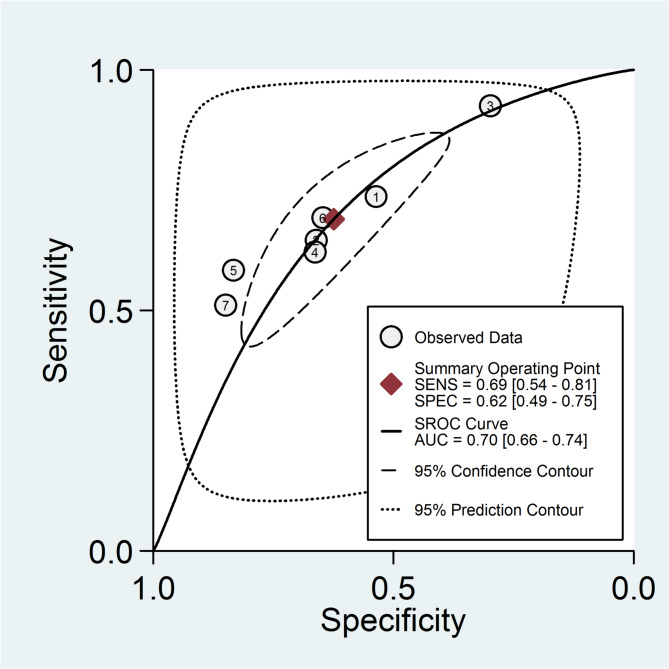
The hierarchical summary receiver operating characteristic (HSROC) curve for the diagnostic accuracy of AI tools is presented in Fig. 4. The area under the curve (AUC) was 0.7 with a 95% confidence interval ranging from 0.66 to 0.74. This value indicates a high level of diagnostic accuracy for AI-based methods in assessing pregnancy-related outcomes in IVF settings.
Fig. 4.
Hierarchical summary receiver operating characteristic (HSROC) curve illustrating the overall diagnostic accuracy of artificial intelligence (AI) tools in predicting pregnancy-related outcomes in IVF cycles
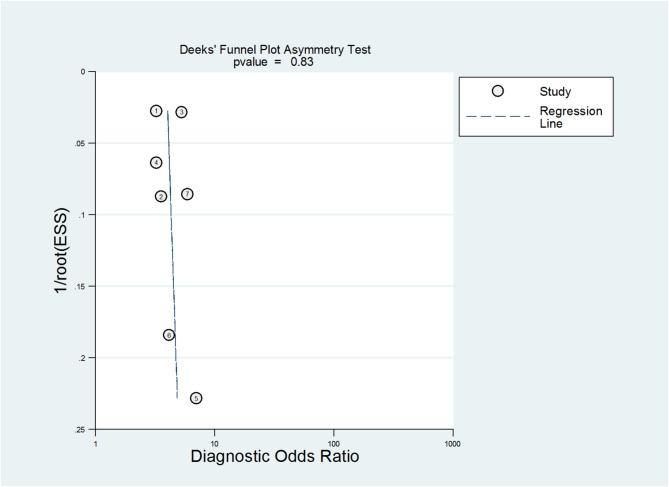
We evaluated potential publication bias using Deek’s funnel plot, as shown in Fig. 5. The Deek asymmetry test produced a p-value greater than 0.1, which indicates that there was no evidence of publication bias in the included studies [39].
Fig. 5.
Deeks’ funnel plot for assessing publication bias among studies evaluating the diagnostic performance of artificial intelligence (AI) tools in predicting pregnancy-related outcomes
Table 2 provides a summary of key pooled diagnostic metrics with interpretation, offering a concise overview of AI tools’ performance in predicting IVF pregnancy outcomes.
Table 2.
Summary of pooled diagnostic performance metrics for AI-based embryo selection tools in IVF
| Metric | Pooled Estimate | 95% Confidence Interval | Acceptable Range | Interpretation |
|---|---|---|---|---|
| Sensitivity | 0.69 | 0.54–0.81 | > 0.70 | Good detection of implantable embryos. |
| Specificity | 0.62 | 0.49–0.75 | > 0.70 | Moderate ability to exclude poor embryos. |
| AUC (Area Under Curve) | 0.70 | 0.66–0.74 | > 0.80 (excellent); 0.70–0.80 (good) | Strong overall diagnostic accuracy. |
| Positive Likelihood Ratio (PLR) | 1.84 | 1.50–2.25 | > 2 (useful); >5 (strong) | Increases confidence in implantation when result is positive |
| Negative Likelihood Ratio (NLR) | 0.50 | 0.38–0.65 | < 0.5 (moderate); <0.2 (strong) | Decreases confidence in implantation when result is negative |
| Diagnostic Odds Ratio (DOR) | 3.69 | 2.72–5.01 | > 5 (moderate); >10 (strong) | Moderate-to-strong predictive power. |
Discussion
In this systematic review and diagnostic meta-analysis, we assessed the effectiveness of AI tools in predicting pregnancy outcomes in in vitro fertilization (IVF). Data from six eligible studies showed that AI-based embryo selection offers promising diagnostic performance, with a pooled sensitivity of 0.69 and specificity of 0.62. These values indicate that AI can reliably distinguish embryos with higher chances of implantation. The positive and negative likelihood ratios were 1.84 and 0.50, respectively, and the pooled Diagnostic Odds Ratio (DOR) was 3.69, reflecting moderate-to-strong discriminatory power. An overall Area Under the Curve (AUC) of 0.7 further supports the diagnostic accuracy of AI systems. These findings suggest that integrating AI into embryo selection may improve IVF success rates by enabling more accurate, consistent, and objective assessments.
Selecting the ideal embryo from a set of viable candidates is a major challenge in IVF [40]. The goal is to achieve the highest live birth rates and minimize the risk of multiple pregnancies by choosing one or two embryos with the greatest implantation potential [41]. This selection process is currently manual, relying on trained embryologists who use optical light microscopes to assess embryo morphology [42]. Traditional methods are subjective and can vary significantly between embryologists. The introduction of AI represents a transformative shift in reproductive medicine. AI’s non-invasive capabilities have sparked significant advancements in IVF, aiming to refine embryo selection by identifying the embryos with the highest likelihood of successful implantation [43]. Furthermore, AI promises to automate various IVF procedures, such as assessing gamete quality, selecting sperm during intracytoplasmic sperm injection (ICSI), and developing enhanced stimulation protocols [16]. This shift from subjective morphological evaluations to more objective, data-driven methods mark a significant advancement in the field.
AI models, such as the Life Whisperer AI evaluated by VerMilyea et al., have demonstrated substantial progress in embryo assessment. The Life Whisperer model, which analyzed data from 8,886 embryos, achieved an overall accuracy rate of 64.3% in predicting embryo viability using images from conventional optical light microscopes. The model showed sensitivity and specificity rates of 70.1% and 60.5%, respectively, indicating robust performance in predicting embryo viability [31]. This model’s ability to provide a reliable confidence score for pregnancy prediction enhances its clinical utility. However, despite encouraging results, the current accuracy still falls short of achieving consistently perfect predictions in embryo selection.
In contrast, recent research has aimed to overcome limitations in current AI algorithms, which often fail to integrate embryonic traits with maternal factors. Enatsu et al.‘s development of the Fertility Image Testing Through Embryo (FiTTE) system represents a notable advancement. FiTTE combines ensemble modeling with a comprehensive dataset of 19,342 static blastocyst images and clinical information, such as age, hormone levels, pregnancy history, and endometrial thickness. This approach achieved a 62.7% improvement in prediction accuracy over traditional methods, with an AUC of 0.71 compared to 0.62 for the Gardner grading system [26]. This improvement underscores the potential of integrating diverse data sources to enhance embryo selection and increase the likelihood of successful pregnancy outcomes.
Further advancements in AI include the development of models that predict miscarriage risk by analyzing time-lapse recordings of embryo development. Amitai et al.’s research utilized a dataset of 314 morphological, morphokinetic, and dynamic parameters, identifying six key features that offer robust predictive power for miscarriage risk [35]. Their model achieved an AUC of 68%, comparable to other established algorithms. Similarly, Uyar et al.’s evaluation of embryo implantation potential through various classifiers demonstrated improved performance, particularly for predicting multiple embryo implantations [36]. These studies illustrate the growing capability of AI to address both implantation potential and miscarriage risk, offering more comprehensive tools for embryo assessment.
Time-lapse monitoring has emerged as a non-invasive method for embryo assessment, offering valuable insights into morphokinetic characteristics associated with implantation, blastocyst formation, and aneuploidy. For instance, the NICS-Timelapse model achieved an AUC of 0.94 and an accuracy of 0.88 in predicting blastocyst euploidy, indicating its effectiveness in predicting clinical pregnancy and live birth rates [27]. This non-invasive approach complements other AI advancements by providing real-time analysis of embryo development, which is crucial for optimizing embryo selection.
The development of semi-automatic grading systems and deep learning models further highlights AI’s role in assessing embryo quality. Filho et al. created a semi-automatic grading system with accuracy rates ranging from 67 to 92% [44]. Similarly, Khosravi et al. and Kragh et al. developed AI models that classify blastocyst images based on Gardner’s scale [43, 45]. Khosravi et al.’s STORK model achieved a remarkable 97.5% accuracy, though this high accuracy may be attributed to its focus on classifying extreme grades of embryos [43]. In contrast, Kragh et al.’s model demonstrated 71.9% accuracy in evaluating the inner cell mass (ICM) and 76.4% accuracy in assessing the trophectoderm, with AUCs of 0.66 and 0.64, respectively [45]. These findings suggest that while some AI models perform well in grading embryos, their predictive power for pregnancy outcomes may still be limited compared to the expertise of human embryologists. However, the development of AI models continues to advance, as evidenced by Tran et al.‘s creation of the deep learning model IVY, which achieved an impressive AUC of 0.93 in predicting the likelihood of detecting a fetal heartbeat from time-lapse recordings [13]. This substantial improvement highlights AI’s growing capability to not only automate and standardize traditional grading procedures but also to enhance predictive accuracy in reproductive medicine.
Building on this trend, recent advances have explored the use of AI in analyzing early-stage cytoplasmic dynamics and proteomic data. Coticchio et al. proposed that AI-driven analysis of early-stage cytoplasmic dynamics could offer a novel approach for predicting embryo viability, achieving an accuracy of 82.6% in forecasting blastocyst development [25]. Moreover, integrating morphological grading with proteomic data has shown promise in estimating live birth probability with an accuracy of 72.7% [24]. These innovative approaches reflect a broader trend towards leveraging AI to enhance embryo selection through a combination of morphological and molecular data. Complementing these efforts, Blank et al. used clinical records to predict ongoing pregnancy after single blastocyst transfer and found that the random forest algorithm outperformed logistic regression based on AUC [34]. Additionally, Wen et al. developed two machine learning models using XGBoost to detect multiple pregnancies, demonstrating superior performance in predicting outcomes and reducing the risk of multiple pregnancies [32]. Collectively, these advancements underscore a growing recognition of AI’s role in refining reproductive technologies and improving patient outcomes through increasingly sophisticated predictive models.
These technological advancements demonstrate strong diagnostic potential, but their true value lies in successful combination into real-world IVF settings. AI-based tools can improve the consistency of embryo selection and reduce reliance on subjective human judgment. In clinical practice, this may lead to more standardized, efficient workflows and better patient outcomes. However, several challenges must be addressed before widespread adoption. These include the need for rigorous clinical validation across diverse patient populations, seamless integration of AI systems into existing laboratory workflows, and training for embryologists and clinicians to understand and trust AI-generated insights. Additionally, cost-effectiveness and regulatory approval are important considerations for broader implementation. Addressing these practical factors is essential to transition AI from research innovation to routine clinical utility.
While our meta-analysis shows encouraging diagnostic performance of AI-based embryo assessment tools, it is important to acknowledge the significant heterogeneity among the included studies. This variability likely stems from differences in patient populations, clinical protocols, imaging platforms, and AI algorithm design and training. Such inconsistencies can affect the comparability of results and may limit the reliability of pooled estimates. Moreover, the relatively small number of studies included restricts the ability to perform subgroup analyses that could further clarify the effectiveness of specific AI models across different settings.
Collectively, to enhance the evidence base and support clinical adoption, future studies should be designed as prospective, multicenter, and blinded investigations. They should use standardized definitions for outcomes and apply consistent data collection methods across different clinical settings. Additionally, external validation of AI tools in diverse patient populations and real-world IVF workflows is crucial to ensure their reliability and generalizability. Adhering to these methodological standards will help future research more effectively demonstrate the clinical value of AI in embryo selection and support its adoption into routine fertility care.
Conclusion
This systematic review and meta-analysis demonstrate that AI holds strong potential to enhance embryo selection in IVF. With a pooled sensitivity of 0.69, specificity of 0.62, AUC of 0.70, and a DOR of 3.69, AI tools show promising diagnostic performance in predicting pregnancy outcomes. However, current models are limited by their narrow data inputs and lack of integration of factors such as genetic, proteomic, and maternal health information. Future research should focus on developing more sophisticated algorithms that incorporate diverse data types, including time-lapse imaging, to better predict embryo viability and long-term clinical outcomes.
Acknowledgements
Not applicable.
Author contributions
MA, MY, TD, and SD contributed to the study concept and design. SD, NAA, and SS performed data screening. RV and HH conducted the data analysis. All authors contributed to drafting the manuscript. MA, SS, and HH reviewed and revised the final version of the manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Ethical guidelines
We complied with all the ethical guidelines.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Samaneh Sheibani and Hossein Hosseinirad contributed equally to this work.
Contributor Information
Samaneh Sheibani, Email: samane.sheibani@yahoo.com.
Hossein Hosseinirad, Email: shfcn@umsystem.edu.
References
- 1.Jain M, Singh M. Assisted reproductive technology (ART) techniques. 2022. [PubMed]
- 2.Kanakasabapathy MK, Thirumalaraju P, Bormann CL, Kandula H, Dimitriadis I, Souter I, et al. Development and evaluation of inexpensive automated deep learning-based imaging systems for embryology. Lab Chip. 2019;19(24):4139–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fishel S, Campbell A, Foad F, Davies L, Best L, Davis N, et al. Evolution of embryo selection for IVF from subjective morphology assessment to objective time-lapse algorithms improves chance of live birth. Reprod Biomed Online. 2020;40(1):61–70. [DOI] [PubMed] [Google Scholar]
- 4.Technology EWGT-L, Apter S, Ebner T, Freour T, Guns Y, Kovacic B, et al. Good practice recommendations for the use of time-lapse technology. Hum Reprod Open. 2020;2020(2):hoaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Storr A, Venetis CA, Cooke S, Susetio D, Kilani S, Ledger W. Morphokinetic parameters using time-lapse technology and day 5 embryo quality: a prospective cohort study. J Assist Reprod Genet. 2015;32:1151–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardner DK, Balaban B. Assessment of human embryo development using morphological criteria in an era of time-lapse, algorithms and ‘OMICS’: is looking good still important? Mol Hum Reprod. 2016;22(10):704–18. [DOI] [PubMed] [Google Scholar]
- 7.Babayev E, Feinberg EC. Embryo through the lens: from time-lapse cinematography to artificial intelligence. Elsevier; 2020. pp. 342–3. [DOI] [PubMed]
- 8.Goyal A, Kuchana M, Ayyagari KPR. Machine learning predicts live-birth occurrence before in-vitro fertilization treatment. Sci Rep. 2020;10(1):20925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curchoe CL, Bormann CL. Artificial intelligence and machine learning for human reproduction and embryology presented at ASRM and ESHRE 2018. J Assist Reprod Genet. 2019;36:591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kragh MF, Karstoft H. Embryo selection with artificial intelligence: how to evaluate and compare methods? J Assist Reprod Genet. 2021;38(7):1675–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curchoe CL, Farias AF-S, Mendizabal-Ruiz G, Chavez-Badiola A. Evaluating predictive models in reproductive medicine. Fertil Steril. 2020;114(5):921–6. [DOI] [PubMed] [Google Scholar]
- 12.Zaninovic N, Rosenwaks Z. Artificial intelligence in human in vitro fertilization and embryology. Fertil Steril. 2020;114(5):914–20. [DOI] [PubMed] [Google Scholar]
- 13.Tran D, Cooke S, Illingworth PJ, Gardner DK. Deep learning as a predictive tool for fetal heart pregnancy following time-lapse incubation and blastocyst transfer. Hum Reprod. 2019;34(6):1011–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geller J, Collazo I, Pai R, Hendon N, Lokeshwar SD, Arora H, et al. Development of an artificial Intelligence-Based assessment model for prediction of pregnancy success using static images captured by optical light microscopy during IVF. Fertil Steril. 2020;114(3):e171. [Google Scholar]
- 15.Salih M, Austin C, Warty R, Tiktin C, Rolnik D, Momeni M, et al. Embryo selection through artificial intelligence versus embryologists: a systematic review. Hum Reprod Open. 2023;2023(3):hoad031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sfakianoudis K, Maziotis E, Grigoriadis S, Pantou A, Kokkini G, Trypidi A, et al. Reporting on the value of artificial intelligence in predicting the optimal embryo for transfer: a systematic review including data synthesis. Biomedicines. 2022;10(3):697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merican ZZ, Yusof UK, Abdullah NL. Review on embryo selection based on morphology using machine learning methods. Int J Adv Soft Comput Appl. 2021;13(2).
- 18.McInnes MDF, Moher D, Thombs BD, McGrath TA, Bossuyt PM, Clifford T, et al. Preferred reporting items for a systematic review and Meta-analysis of diagnostic test accuracy studies: the PRISMA-DTA statement. JAMA. 2018;319(4):388–96. [DOI] [PubMed] [Google Scholar]
- 19.Deeks JJ, Bossuyt PM, Leeflang MM, Takwoingi Y. Cochrane handbook for systematic reviews of diagnostic test accuracy. Wiley; 2023. [DOI] [PMC free article] [PubMed]
- 20.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals Int Med. 2011;155(8):529–36. [DOI] [PubMed] [Google Scholar]
- 21.Harbord RM, Deeks JJ, Egger M, Whiting P, Sterne JA. A unification of models for meta-analysis of diagnostic accuracy studies. Biostatistics. 2007;8(2):239–51. [DOI] [PubMed] [Google Scholar]
- 22.Reitsma JB, Glas AS, Rutjes AW, Scholten RJ, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol. 2005;58(10):982–90. [DOI] [PubMed] [Google Scholar]
- 23.Rutter CM, Gatsonis CA. A hierarchical regression approach to meta-analysis of diagnostic test accuracy evaluations. Stat Med. 2001;20(19):2865–84. [DOI] [PubMed] [Google Scholar]
- 24.Bori L, Dominguez F, Fernandez EI, Del Gallego R, Alegre L, Hickman C, et al. An artificial intelligence model based on the proteomic profile of euploid embryos and blastocyst morphology: a preliminary study. Reprod Biomed Online. 2021;42(2):340–50. [DOI] [PubMed] [Google Scholar]
- 25.Coticchio G, Fiorentino G, Nicora G, Sciajno R, Cavalera F, Bellazzi R, et al. Cytoplasmic movements of the early human embryo: imaging and artificial intelligence to predict blastocyst development. Reprod Biomed Online. 2021;42(3):521–8. [DOI] [PubMed] [Google Scholar]
- 26.Enatsu N, Miyatsuka I, An LM, Inubushi M, Enatsu K, Otsuki J. A novel system based on artificial intelligence for predicting blastocyst viability and visualizing the explanation. Reprod Med Biol. 2022;21(1):e12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He H, Wu L, Chen Y, Li T, Ren X, Hu J, et al. A novel non-invasive embryo evaluation method (NICS-Timelapse) with enhanced predictive precision and clinical impact. Heliyon. 2024;10(9):e30189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao Q, Zhang Q, Feng X, Huang H, Xu H, Tian B, et al. Development of deep learning algorithms for predicting blastocyst formation and quality by time-lapse monitoring. Commun Biol. 2021;4(1):415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyagi Y, Habara T, Hirata R, Hayashi N. Feasibility of predicting live birth by combining conventional embryo evaluation with artificial intelligence applied to a blastocyst image in patients classified by age. Reprod Med Biol. 2019;18(4):344–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyagi Y, Habara T, Hirata R, Hayashi N. Feasibility of artificial intelligence for predicting live birth without aneuploidy from a blastocyst image. Reprod Med Biol. 2019;18(2):204–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.VerMilyea M, Hall JMM, Diakiw SM, Johnston A, Nguyen T, Perugini D, et al. Development of an artificial intelligence-based assessment model for prediction of embryo viability using static images captured by optical light microscopy during IVF. Hum Reprod. 2020;35(4):770–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wen JY, Liu CF, Chung MT, Tsai YC. Artificial intelligence model to predict pregnancy and multiple pregnancy risk following in vitro fertilization-embryo transfer (IVF-ET). Taiwan J Obstet Gynecol. 2022;61(5):837–46. [DOI] [PubMed] [Google Scholar]
- 33.Yang L, Peavey M, Kaskar K, Chappell N, Zhu L, Devlin D, et al. Development of a dynamic machine learning algorithm to predict clinical pregnancy and live birth rate with embryo morphokinetics. F S Rep. 2022;3(2):116–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blank C, Wildeboer RR, DeCroo I, Tilleman K, Weyers B, de Sutter P, et al. Prediction of implantation after blastocyst transfer in in vitro fertilization: a machine-learning perspective. Fertil Steril. 2019;111(2):318–26. [DOI] [PubMed] [Google Scholar]
- 35.Amitai T, Kan-Tor Y, Or Y, Shoham Z, Shofaro Y, Richter D, et al. Embryo classification beyond pregnancy: early prediction of first trimester miscarriage using machine learning. J Assist Reprod Genet. 2023;40(2):309–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uyar A, Bener A, Ciray HN. Predictive modeling of implantation outcome in an in vitro fertilization setting: an application of machine learning methods. Med Decis Mak. 2015;35(6):714–25. [DOI] [PubMed] [Google Scholar]
- 37.Abadjieva D, Georgiev B, Gerzilov V, Tsvetkova I, Taushanova P, Todorova K et al. Machine learning approach for muscovy Duck (Cairina moschata) semen quality assessment. Animals 2023;13(10). [DOI] [PMC free article] [PubMed]
- 38.Campanholi SP, Garcia Neto S, Pinheiro GM, Nogueira MFG, Rocha JC, Losano JDA, et al. Can in vitro embryo production be estimated from semen variables in senepol breed by using artificial intelligence? Front Vet Sci. 2023;10:1254940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005;58(9):882–93. [DOI] [PubMed] [Google Scholar]
- 40.Illingworth PJ, Venetis C, Gardner DK, Nelson SM, Berntsen J, Larman MG et al. Deep learning versus manual morphology-based embryo selection in IVF: a randomized, double-blind noninferiority trial. Nat Med. 2024;1–7. [DOI] [PMC free article] [PubMed]
- 41.Mastenbroek S, Van Der Veen F, Aflatoonian A, Shapiro B, Bossuyt P, Repping S. Embryo selection in IVF. Hum Reprod. 2011;26(5):964–6. [DOI] [PubMed] [Google Scholar]
- 42.Gardner D, Sakkas D. Assessment of embryo viability: the ability to select a single embryo for transfer—a review. Placenta. 2003;24:S5–12. [DOI] [PubMed] [Google Scholar]
- 43.Khosravi P, Kazemi E, Zhan Q, Malmsten JE, Toschi M, Zisimopoulos P, et al. Deep learning enables robust assessment and selection of human blastocysts after in vitro fertilization. NPJ Digit Med. 2019;2(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Filho ES, Noble JA, Poli M, Griffiths T, Emerson G, Wells D. A method for semi-automatic grading of human blastocyst microscope images. Hum Reprod. 2012;27(9):2641–8. [DOI] [PubMed] [Google Scholar]
- 45.Kragh MF, Rimestad J, Berntsen J, Karstoft H. Automatic grading of human blastocysts from time-lapse imaging. Comput Biol Med. 2019;115:103494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.