Abstract
Fecal specimens from patients with acute diarrhea were collected from 10 prefectures in Japan over a 6-month period (November 1992 to April 1993), and the specimens that were negative for human group A rotaviruses were screened for the presence of human group C rotaviruses (CHRVs) by the reverse passive hemagglutination test. Of 784 specimens examined, 53 samples (6.8%) that were collected in 7 of 10 prefectures were positive for CHRV, indicating that CHRVs are widely distributed across Japan. Most of the CHRV isolates were detected in March and April, and CHRVs mainly prevailed in children ages 3 to 8 years. The genome electropherotypes of eight strains isolated in five individual prefectures were surprisingly similar to each other and were different from those of CHRV strains isolated to date. The outer capsid glycoprotein (VP7) gene homologies of the isolates retrieved in 1993 were subsequently analyzed by the dot blot hybridization method. As a result, the VP7 genes of the isolates revealed very high levels of homology not only with each other but also with the VP7 gene of the OK118 strain isolated in 1988. These results suggest that a large-scale outbreak of CHRV occurred during the winter of 1992 and 1993 in Japan.
Rotaviruses are recognized as the major etiologic agents of diarrheal diseases in young children and animals. The genome of rotaviruses consists of 11 segments of double-stranded RNA (dsRNA) enclosed in a double-shelled particle. Rotaviruses are classified into seven groups (groups A to G) on the basis of their dsRNA electropherotypes and a common group antigen on the inner capsid protein (protein VP6) (19).
Group C rotaviruses were first recognized in swine (2, 20) and then were confirmed as human pathogens by Bridger et al. (4). During the last decade, human group C rotaviruses (CHRVs) have been associated with several outbreaks of acute diarrhea in Asia (12, 18), Europe (3, 5), and South America (7). More recently, CHRVs have been detected in patients with sporadic cases of diarrhea in the United States (9). These observations indicate that CHRV is widely distributed and is likely to be an emerging pathogen.
CHRV infections in Japan were first recognized by Oseto et al. (17) in 1985. Ushijima et al. (23) also detected CHRVs from fecal specimens collected in the Tokyo area in 1987. Since then, some researchers have recognized CHRV infections in individual locations (6, 14, 16). However, none of these investigators have carried out an epidemiological study that covers several locations in Japan.
Until recently, the detection of CHRVs in fecal specimens was usually performed by immune electron microscopy with CHRV-specific antisera or polyacrylamide gel electrophoresis (PAGE) of viral dsRNA, because CHRVs were noncultivatable. The difficulty in detection has hindered the epidemiological analysis of CHRV infections. Recently, we have developed a reverse passive hemagglutination (RPHA) test using CHRV-specific monoclonal antibodies (11). This test is very easy to perform and should be a useful tool for the screening of CHRVs on a large scale.
In this study, we collected fecal specimens from patients with diarrhea in 10 prefectures in Japan and examined the specimens for the presence of CHRVs by the RPHA test. As a result, CHRVs were detected in seven prefectures. To define the genetic relationship among the CHRV field isolates, we analyzed the genome electropherotypes and the outer capsid glycoprotein (VP7) gene homologies of the isolates.
MATERIALS AND METHODS
Fecal specimens.
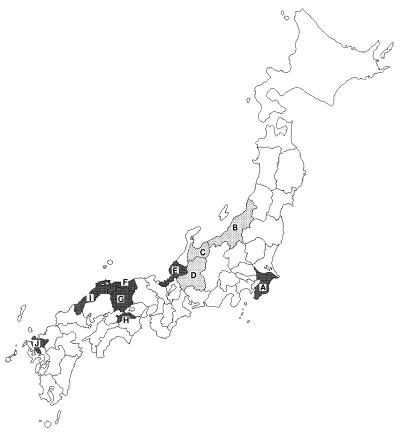
Between November 1992 and April 1993, 1,114 fecal specimens from patients with acute diarrhea were collected at pediatric clinics or outpatient sections of general hospitals in 10 prefectures in Japan. Figure 1 shows the geographic areas in which the specimens were collected.
FIG. 1.
Map of Japan showing the prefectures from which the fecal specimens were collected. Darker shading indicates the prefectures in which CHRVs have been detected. A, Chiba; B, Niigata; C, Toyama; D, Gifu; E, Fukui; F, Tottori; G, Okayama; H, Kagawa; I, Shimane; J, Saga.
All fecal specimens were screened for human group A rotaviruses with an enzyme-linked immunosorbent assay (ELISA) kit (ROTACLONE; Cambridge Biotech, Worcester, Mass.). The specimens that were negative for human group A rotaviruses were further examined for the presence of CHRV by the RPHA test.
RPHA test.
The RPHA test was performed as described previously (11). Briefly, 10% suspensions of the fecal specimens in phosphate-buffered saline (pH 7.2) were centrifuged at 2,000 × g for 10 min and the supernatants were tested. Serial twofold dilutions of the samples were made in duplicate. In one dilution series, a 0.7% suspension of sheep erythrocytes (SRBCs) coated with CHRV-specific monoclonal antibodies was added to each well. In the other series, SRBCs coated with normal mouse immunoglobulin G (control SRBCs) were added. Hemagglutination titers were observed after 1 h. When the RPHA test titer with monoclonal antibody-coated SRBCs was four or more times greater than that obtained with the control SRBCs, the sample was judged to be CHRV positive.
RNA extraction.
Fecal suspensions containing CHRVs were extracted with an equal volume of trichlorotrifluoroethane, and the supernatants were adjusted to contain 10 mM EDTA, 0.6% sodium dodecyl sulfate (SDS), and 300 μg of proteinase K per ml. The suspensions were incubated for 1.5 h at 40°C and then extracted with phenol-chloroform. Viral RNA was further purified with an RNAID kit (Bio 101, Inc., La Jolla, Calif.) according to the manufacturer’s instructions. The purified RNAs were stored at −30°C until use.
Dot blot hybridization.
Hybridization tests were carried out as described previously (10). In brief, the VP7 genes of CHRV isolates were first amplified by the reverse transcription-PCR (RT-PCR) method (10) and were then purified with a Suprec-02 column (Takara Shuzo Co., Ltd., Kyoto, Japan). Equivalent amounts (200 ng) of the genes amplified by RT-PCR were blotted onto nylon membranes and were separately hybridized with K9304, OK118, and OK450 VP7 genes that had been labeled with digoxigenin-11-dUTP (Boehringer, Mannheim, Germany). Hybridization was performed under highly stringent conditions (50% formamide and 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] at 52°C). After hybridization, the membranes were washed twice in 0.1× SSC containing 0.1% SDS at 68°C for 15 min. Colorimetric detection of the hybridized probe was carried out according to the manufacturer’s instructions.
Sequencing of the VP7 genes.
The VP7 gene of a clinical isolate (isolate K9304) was amplified by the RT-PCR method and was then cloned into plasmid pUC18 (10). Five individual recombinants were isolated, and both strands of the cloned DNA were sequenced by the dideoxynucleotide chain-termination method (Applied Biosystems, Foster City, Calif.). Nucleotide sequence data were then analyzed by the GENETYX-MAC, version 6.0, program. The program MAlign, version 1.0, was also used for sequence alignments.
Nucleotide sequence accession number.
The nucleotide sequence data for the VP7 gene from strain K9304 has been submitted to the DDBJ DNA database and has been assigned accession number AB004250.
RESULTS
Detection of CHRVs from fecal specimens by RPHA test.
The results of the detection of rotaviruses are summarized in Table 1. Human group A rotaviruses were detected in 330 of 1,114 specimens. Group A rotavirus-negative samples were further examined for CHRVs by the RPHA test, and 53 were positive. The geographic distribution of the seven prefectures in which CHRVs have been detected is shown in Fig. 1. CHRVs were mainly distributed in the western area of Japan. The rates of positivity for CHRV were considerably lower than those for human group A rotavirus (Table 1). No significant difference was observed between the rates of positivity for males and females (data not shown).
TABLE 1.
Detection of rotaviruses in 10 prefectures
| Prefecture | Detection resulta
|
No. of CHRV isolates detected in at the following times:
|
||||||
|---|---|---|---|---|---|---|---|---|
| Human group A rotavirusesb | CHRVsc | 1992
|
1993
|
|||||
| November | December | January | February | March | April | |||
| Chiba | 24/97 (24.7) | 2/73 (2.7) | 1 | 1 | ||||
| Niigata | 7/55 (12.7) | 0/48 | ||||||
| Toyama | 49/119 (41.2) | 0/70 | ||||||
| Gifu | 20/74 (27) | 0/54 | ||||||
| Fukui | 6/21 (28.6) | 2/15 (13.3) | 2 | |||||
| Tottori | 46/123 (37.4) | 6/77 (7.8) | 6 | |||||
| Okayama | 22/176 (12.5) | 11/154 (7.1) | 2 | 2 | 3 | 4 | ||
| Kagawa | 83/163 (50.9) | 8/80 (10) | 2 | 1 | 1 | 3 | 1 | |
| Shimane | 30/187 (16) | 19/157 (12.1) | 1 | 4 | 6 | 6 | 2 | |
| Saga | 43/99 (43.4) | 5/56 (8.9) | 2 | 2 | 1 | |||
| Total | 330/1,114 (29.6) | 53/784 (6.8) | 1 | 4 | 7 | 11 | 14 | 16 |
Values indicate number of samples that were positive/total number of samples tested. Values in parentheses indicate percentage of tests that were positive.
Human group A rotaviruses were detected with a commercially available ELISA kit.
CHRVs were detected by the RPHA test.
The epidemiological features of CHRV infections were then compared with those of human group A rotavirus infections. CHRVs were mainly detected in March and April (Table 1), whereas most human group A rotavirus infections occurred between January and February (data not shown). The age-specific attack rates for human group C and group A rotaviruses are compared in Table 2. Although CHRVs principally prevailed in children ages 3 to 8 years, the target age groups of human group A rotavirus were below 3 years. The mean age of the patients infected with CHRV (4.36 years) was significantly (P < 0.01) higher than that of patients infected with human group A rotavirus (2.19 years).
TABLE 2.
Comparison of age-specific attack rates for CHRV and human group A rotavirus
| Age group (yr) | Attack rate (%)
|
|
|---|---|---|
| CHRV | Human group A rotavirus | |
| <1 | 4.2 | 38.7 |
| 1–2 | 8.9 | 42.3 |
| 3–4 | 12.8 | 24.2 |
| 5–6 | 6.2 | 14.5 |
| 7–8 | 10.0 | 10.7 |
| 9–10 | 4.2 | 4.0 |
| >10 | 2.6 | 10.0 |
Genome electropherotypes of CHRV field isolates.
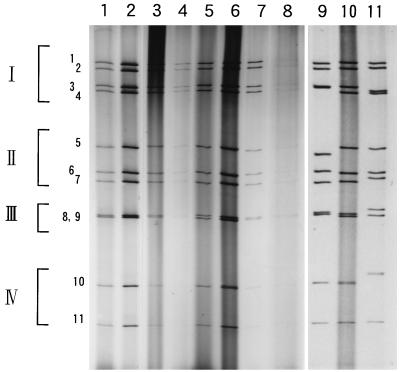
Only eight specimens which were obtained from five prefectures had enough volume from which viral dsRNA could be extracted for genome electropherotype analyses. The RNA segments were dissociated by PAGE with a 10% gel and were then visualized with silver nitrate. Figure 2 (lanes 1 to 8) shows the genome electropherotypes of the field isolates. Every strain exhibited the typical 4-3-2-2 profile of group C rotavirus. Although the strains were isolated in five individual prefectures, the electropherotypes of all strains were similar to each other.
FIG. 2.
Comparison of genome electropherotypes of CHRV field isolates. Lanes: 1, 93K332 strain (Chiba isolate); 2, 93K360 strain (Chiba isolate); 3, F522 strain (Fukui isolate); 4, 34803 strain (Tottori isolate); 5, T360 strain (Okayama isolate); 6, K9304 strain (Okayama isolate); 7, 93-48 strain (Saga isolate); 8, 93-42 strain (Saga isolate); 9, OK118 strain, which exhibits the pattern I electropherotype; 10, K9304 strain; 11, OK450 strain, which exhibits the pattern II electropherotype.
We previously demonstrated that the genome electropherotypes of CHRVs isolated in Okayama from 1988 to 1990 were classified into patterns I and II (6, 10). The electropherotype of the K9304 strain, which was selected as a representative of the isolates retrieved in 1993, was then compared with those of the OK118 (pattern I) and the OK450 (pattern II) strains in the same gel (Fig. 2, lanes 9 to 11). The RNA profile of K9304 was different from those of both OK118 and OK450, and so this electropherotype was tentatively designated pattern III. The electrophoretic mobilities of the 2nd, 3rd, 7th, 10th, and 11th segments of K9304 were similar to those of the corresponding segments of OK118, while the 1st, 4th, 5th, and 6th segments migrated to the same positions as those of OK450, suggesting that pattern III is a combined electropherotype of patterns I and II.
Analysis of the VP7 genes from CHRV field isolates.
The VP7 gene homologies of the field isolates were further analyzed by dot blot hybridization with VP7 gene probes from the K9304, OK118, and OK450 strains. Epidemiological data and the hybridization results for the 19 isolates used in this study are presented in Table 3. The VP7 genes of all isolates strongly reacted with the K9304 probe as well as the OK118 probe, while weak hybridization signals were observed with the OK450 probe. These results indicate that the VP7 genes of the isolates retrieved in 1993 have high levels of homology not only with each other but also with the VP7 gene of the OK118 strain, which was isolated in 1988.
TABLE 3.
Epidemiological data, genome patterns, and dot blot hybridization results for 19 CHRV isolates
| Strain | Date of isolation (mo/day/yr) | Age (yr) | Sexa | Prefecture | Genome pattern | Dot blot result with the following probe:
|
||
|---|---|---|---|---|---|---|---|---|
| K9304 | OK118 | OK450 | ||||||
| OK118 | 2/6/1988 | 6 | M | Okayama | I | + | + | − |
| OK450 | 2/8/1989 | 1 | F | Okayama | II | − | − | + |
| 93K332 | 3/23/1993 | 24 | F | Chiba | III | + | + | − |
| 93K360 | 4/27/1993 | 9 | M | Chiba | III | + | + | − |
| F522 | 4/30/1993 | 1 | M | Fukui | III | + | + | − |
| F618 | 4/24/1993 | 4 | F | Fukui | NDb | + | + | − |
| 34745 | 4/23/1993 | 15 | M | Tottori | ND | + | + | − |
| 34803 | 4/12/1993 | 1 | F | Tottori | III | + | + | − |
| T360 | 3/6/1993 | 3 | M | Okayama | III | + | + | − |
| K12 | 3/15/1993 | 6 | M | Okayama | ND | + | + | − |
| K9301 | 4/19/1993 | 8 | M | Okayama | ND | + | + | − |
| K9304 | 4/22/1993 | 4 | M | Okayama | III | + | + | − |
| K9306 | 4/26/1993 | 4 | F | Okayama | ND | + | + | − |
| KA63 | 3/3/1993 | 7 | M | Kagawa | ND | + | + | − |
| KA73 | 3/30/1993 | 5 | M | Kagawa | ND | + | + | − |
| S373 | 1/25/1993 | 1 | F | Shimane | ND | + | + | − |
| S891 | 2/2/1993 | 1 | M | Shimane | ND | + | + | − |
| 93-42 | 2/2/1993 | 1 | M | Saga | III | + | + | − |
| 93-48 | 2/23/1993 | 1 | M | Saga | III | + | + | − |
M, male; F, female.
ND, genome pattern could not be determined.
To confirm our findings, the VP7 gene of K9304 was cloned and sequenced. The VP7 gene of K9304 was 1,063 nucleotides in length and contained single open reading frame encoding 332 amino acids. The nucleotide and deduced amino acid sequences of the VP7 gene from K9304 were further compared with those of the genes from OK118 and OK450. As indicated in Table 4, a surprising level of sequence conservation was observed between K9304 and OK118 (more than 99.1%), whereas the overall nucleotide and amino acid identities between K9304 and OK450 were relatively low (95.6 and 96.7%, respectively).
TABLE 4.
Nucleotide and deduced amino acid sequence homologies of VP7 genes from three CHRV strains
| Strain | % Homology to the following straina:
|
Accession no. | ||
|---|---|---|---|---|
| K9304 | OK118 | OK450 | ||
| K9304 | 99.2 | 95.6 | AB004250 | |
| OK118 | 99.4 | 95.7 | D87543 | |
| OK450 | 96.7 | 96.7 | D87544 | |
The percent homologies of nucleotide sequences are given in boldface. Other data represent the percent homologies of deduced amino acid sequences.
DISCUSSION
This study, the first survey of CHRVs in various locations in Japan, indicated that CHRVs are widely distributed in Japan. The rates of CHRV positivity ranged from 2.7 to 13.3%, and the CHRV isolates were mainly detected in March and April. Moreover, CHRVs principally prevailed in children ages 3 to 8 years. These epidemiological features are clearly distinct from those of the human group A rotavirus. Oseto (16) previously carried out an epidemiological study of CHRVs over a 3-year period in Matsuyama City, Japan. Our observations were consistent with those reported by Oseto (16).
In this study, we screened only the human group A rotavirus-negative specimens for the presence of CHRV. Jiang et al. (9) have recently reported mixed infections with human group A and group C rotaviruses in the United States. We therefore examined by the RPHA test group A rotavirus-positive specimens (n = 52) that were collected in Okayama and Shimane (data not shown), but none of the specimens was positive for CHRV, indicating that the mixed infection might be rather rare. In fact, Jiang et al. (9) recognized only one mixed infection among 1,676 samples. However, screening of rotavirus infections must hereafter include screening for mixed infection.
The RPHA test could successfully detect CHRVs even in fecal specimens that were insufficient for immune electron microscopy or genome electropherotype analyses. To inspect the specificity of the RPHA test, the RPHA test-positive specimens were examined by our ELISA system (6), and all were determined to be positive. Recently, the PCR method has been applied to the detection of group C rotaviruses by Gouvea et al. (8). Although the sensitivity of the RPHA test is not comparable to that of the PCR method, the former test is faster and simpler and is more suitable for use for routine diagnosis in clinical settings.
In group A rotaviruses, genome electropherotyping of field isolates is a useful tool for obtaining epidemiological information about the origin of the isolates and diversity among these isolates, because each isolate reveals a unique genome profile (1, 22). The genome electropherotypes of the isolates retrieved in 1993 were surprisingly similar to each other, regardless of the prefectures from which the isolates were obtained. Moreover, the dot blot hybridization analysis showed that the VP7 genes of the isolates were highly homologous. These results strongly suggest that a large-scale outbreak of CHRV occurred during the winter of 1992 and 1993 in Japan. However, further comparative analysis of other genome segments will be required to confirm this hypothesis.
The electropherotype of the K9304 strain, which represented isolates retrieved in 1993, was compared with those of the pattern I and II strains in the same gel. Although K9304 revealed a distinct genome profile tentatively designated pattern III, this electropherotype seemed to be a combination of patterns I and II. The sequence analysis of the VP7 gene from K9304 also showed that the VP7 gene of K9304 was similar to that of the pattern I strain. These results indicate that the K9304 strain may be a reassortant virus between the pattern I and pattern II strains, because it has been reported that natural reassortants occurred between human group A rotaviruses strains belonging to different genogroups (13, 24). Quite recently, CHRVs have been successfully propagated in a continuous cell line (CaCo-2) (15, 21). To clarify the genetic and antigenic relationship among three strains with distinct electropherotypes, we are now attempting to adapt these strains to CaCo-2 cells.
ACKNOWLEDGMENTS
We thank S. Yamanishi, Kagawa Prefectural Institute of Public Health, for providing the fecal specimens. We are grateful to J. Nakamura and S. Nii, Department of Virology, Okayama University Medical School, for technical advice and helpful suggestions. We are also grateful to H. Ogura and T. Mori, Okayama Prefectural Institute for Environmental Science and Public Health, for critically reviewing the manuscript.
This work was partially supported by health science research grants from the Ministry of Health and Welfare, Japan.
REFERENCES
- 1.Ahmed M U, Urasawa S, Taniguchi K, Urasawa T, Kobayashi N, Wakasugi F, Islam A I M M, Sahikh H A. Analysis of human rotavirus strains prevailing in Bangladesh in relation to nationwide floods brought by the 1988 monsoon. J Clin Microbiol. 1991;29:2273–2279. doi: 10.1128/jcm.29.10.2273-2279.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bohl E H, Saif L J, Theil K W, Agnes A G, Cross R F. Porcine pararotavirus: detection, differentiation from rotavirus, and pathogenesis in gnotobiotic pigs. J Clin Microbiol. 1982;15:312–319. doi: 10.1128/jcm.15.2.312-319.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonsdorf C H V, Svensson L. Human serogroup C rotavirus in Finland. Scand J Infect Dis. 1988;20:475–478. doi: 10.3109/00365548809032493. [DOI] [PubMed] [Google Scholar]
- 4.Bridger J C, Pedley S, McCrae M A. Group C rotaviruses in humans. J Clin Microbiol. 1986;23:760–763. doi: 10.1128/jcm.23.4.760-763.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caul E O, Ashley C R, Darville J M, Bridger J C. Group C rotavirus associated with fatal enteritis in a family outbreak. J Med Virol. 1990;30:201–205. doi: 10.1002/jmv.1890300311. [DOI] [PubMed] [Google Scholar]
- 6.Fujii R, Kuzuya M, Hamano M, Yamada M, Yamazaki S. Detection of human group C rotaviruses by an enzyme-linked immunosorbent assay using monoclonal antibodies. J Clin Microbiol. 1992;30:1307–1311. doi: 10.1128/jcm.30.5.1307-1311.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gabbay Y B, D’Arc J, Mascarenas P, Linhares A C, Freitas R B. Atypical rotavirus among diarrhoeic children living in Belém, Brazil. Mem Inst Oswaldo Cruz. 1989;84:5–7. doi: 10.1590/s0074-02761989000100002. [DOI] [PubMed] [Google Scholar]
- 8.Gouvea V, Allen J R, Glass R I, Fang Z-Y, Bremont M, Cohen J, McCrea M A, Saif L J, Sinarachatanant P, Caul E O. Detection of group B and C rotaviruses by polymerase chain reaction. J Clin Microbiol. 1991;29:519–523. doi: 10.1128/jcm.29.3.519-523.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang B M, Dennehy P H, Spangenberger S, Gentsch J R, Glass R I. First detection of group C rotavirus in fecal specimens of children with diarrhea in the United States. J Infect Dis. 1995;172:45–50. doi: 10.1093/infdis/172.1.45. [DOI] [PubMed] [Google Scholar]
- 10.Kuzuya M, Fujii R, Hamano M, Nakamura J, Yamada M, Nii S, Mori T. Molecular analysis of outer capsid glycoprotein (VP7) genes from two isolates of human group C rotavirus with different genome electropherotypes. J Clin Microbiol. 1996;34:3185–3189. doi: 10.1128/jcm.34.12.3185-3189.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuzuya M, Fujii R, Hamano M, Nagabayashi T, Tsunemitsu H, Yamada M, Nii S, Mori T. Rapid detection of human group C rotaviruses by reverse passive hemagglutination and latex agglutination tests using monoclonal antibodies. J Clin Microbiol. 1993;31:1308–1311. doi: 10.1128/jcm.31.5.1308-1311.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsumoto K, Hatano M, Kobayashi K, Hasegawa A, Yamazaki S, Nakata S, Chiba S, Kimura Y. An outbreak of gastroenteritis associated with acute rotaviral infection in schoolchildren. J Infect Dis. 1989;160:611–615. doi: 10.1093/infdis/160.4.611. [DOI] [PubMed] [Google Scholar]
- 13.Nakagomi O, Nakagomi T. Molecular evidence for naturally occurring single VP7 gene substitution reassortant between human rotaviruses belonging to two different genogroups. Arch Virol. 1991;119:67–81. doi: 10.1007/BF01314324. [DOI] [PubMed] [Google Scholar]
- 14.Oishi I, Yamazaki K, Minekawa Y. An occurrence of diarrheal cases associated with group C rotavirus in adults. Microbiol Immunol. 1993;37:505–509. doi: 10.1111/j.1348-0421.1993.tb03243.x. [DOI] [PubMed] [Google Scholar]
- 15.Oseto M, Yamashita Y, Hattori M, Mori M, Inoue H, Ishimaru Y, Matsuno S. Serial propagation of human group C rotavirus in a continuous cell line (CaCo-2) J Clin Exp Med. 1994;168:177–178. . (In Japanese.) [Google Scholar]
- 16.Oseto M. Epidemiological study of group C rotavirus. J Jpn Assoc Infect Dis. 1990;64:1264–1273. doi: 10.11150/kansenshogakuzasshi1970.64.1264. . (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 17.Oseto M, Yamashita Y, Okuyama M, Kuwabara H, Inoue H. Detection of atypical rotaviruses by polyacrylamide gel electrophoresis. J Clin Exp Med. 1986;136:223–224. . (In Japanese.) [Google Scholar]
- 18.Penaranda M E, Cubitt W D, Sinarachatanant P, Taylor D N, Likanonsakul S, Saif L, Glass R I. Group C rotavirus infection in patients with diarrhea in Thailand, Nepal, and England. J Infect Dis. 1989;160:392–397. doi: 10.1093/infdis/160.3.392. [DOI] [PubMed] [Google Scholar]
- 19.Saif L J. Nongroup A rotaviruses. In: Saif L J, Theil K W, editors. Viral diarrhea of man and animals. Boca Raton, Fla: CRC Press, Inc.; 1990. pp. 73–95. [Google Scholar]
- 20.Saif L J, Bohl E H, Theil K W, Cross R F, House J A. Rotavirus-like, calicivirus-like, and 23-nm virus-like particles associated with diarrhea in young pigs. J Clin Microbiol. 1980;12:105–111. doi: 10.1128/jcm.12.1.105-111.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shinozaki K, Yamanaka T, Tokieda M, Shirasawa H, Simizu B. Isolation and serial propagation of human group C rotaviruses in a cell line (CaCo-2) J Med Virol. 1996;48:48–52. doi: 10.1002/(SICI)1096-9071(199601)48:1<48::AID-JMV8>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 22.Theil K W. Group A rotaviruses. In: Saif L J, Theil K W, editors. Viral diarrhea of man and animals. Boca Raton, Fla: CRC Press, Inc.; 1990. pp. 35–72. [Google Scholar]
- 23.Ushijima H, Honma H, Mukoyama A, Shinozaki T, Fujita Y, Kobayashi M, Oseto M, Morikawa M, Kitamura T. Detection of group C rotaviruses in Tokyo. J Med Virol. 1989;27:299–303. doi: 10.1002/jmv.1890270408. [DOI] [PubMed] [Google Scholar]
- 24.Ward R L, Nakagomi O, Knowlton D R, McNeal M M, Nakagomi T, Clemens J D, Sack D A, Schiff G M. Evidence for natural reassortants of human rotaviruses belonging to different genogroups. J Virol. 1990;64:3219–3225. doi: 10.1128/jvi.64.7.3219-3225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]