Abstract
In this article, we identify and characterize p600, a unique 600-kDa retinoblastoma protein- and calmodulin-binding protein. In the nucleus, p600 and retinoblastoma protein seem to act as a chromatin scaffold. In the cytoplasm, p600 and clathrin form a meshwork structure, which could contribute to cytoskeletal organization and membrane morphogenesis. Reduced expression of p600 with interference RNA abrogates integrin-mediated ruffled membrane formation and, furthermore, prevents activation of integrin-mediated survival pathways. Consequently, knockdown of p600 sensitizes cells to apoptosis induced by cell detachment. These findings provide mechanistic insight into the regulation of membrane-proximal events in tumorigenesis.
Keywords: apoptosis, cancer, calmodulin, retinoblastoma protein
Neoplastic transformation of human cells is a multistep process involving dysregulation of multiple cellular pathways, including control of cell cycle and cell death processes (1, 2). Viral oncoproteins, such as human papilloma virus (HPV) E7, adenovirus E1A, and simian virus 40 large-T antigen, have evolved to bind critical cellular targets involved in such processes to efficiently immortalize and transform cells (3). HPVs are small, nonenveloped viruses that contain double-stranded circular DNAs. High-risk HPVs, such as HPV-16, immortalize human and rodent cells in culture, which can subsequently convert to malignant growth either spontaneously or after exposure to other carcinogens (4). Among the virus-encoded factors, the E6 and E7 proteins are consistently expressed in HPV-associated cervical cancers. E6 forms a complex with the cellular tumor suppressor protein p53, leading to degradation of p53 by ubiquitin-dependent proteolysis. Similarly, E7 leads to degradation of the retinoblastoma-susceptibility gene product (retinoblastoma protein or RB), as well as its family members, including p107 and p130. Although dysregulation of RB family members could be an important role of HPV-16 E7, it is not sufficient for transforming activity. In addition to the RB-binding domain, the N-terminal domain of HPV-16 E7 has been shown to be essential for transformation (5). Thus, HPV-16 E7 could contribute to transformation by targeting RB-dependent and -independent pathways.
To gain insight into the molecular mechanisms of tumor suppression, we have purified RB complexes and identified p600 as an interacting protein for RB. Although RB localizes predominantly in the nucleus, p600 localizes to both nuclear and cytoplasmic compartments. In the cytoplasm, p600 is accumulated at the lamellipodia of the protrusion sites. Reduction of p600 expression suppresses formation of ruffled membranes, which play a crucial role in cell migration. As a consequence, reduction of p600 expression causes deficiencies in activating integrin-mediated survival pathways, sensitizing cells to apoptosis induced by cell detachment from the extracellular matrix. In this article, we discuss possible roles of p600 in both normal and cancer cells.
Materials and Methods
Purification of e-RB-Containing Complexes. The FLAG epitope tag was fused to the N terminus of RB (e-RB), and the fusion protein was stably expressed in HeLa cells by retroviral transduction. e-RB was purified from nuclear extracts by immunoprecipitation with anti-FLAG agarose, as described (6).
Calmodulin Binding Assays. Samples were fractionated by SDS/PAGE and blotted onto a nitrocellulose membrane. After blocking in 1% skim milk and TBST (20 mM Tris·HCl buffer, pH 8.0/100 mM NaCl/0.1% Tween 20), the membrane was incubated with 100 ng/ml biotinylated calmodulin in TBST with 0.1 mM CaCl2 or 10 mM EGTA at 22°C for 30 min. After washing with the same buffers, biotinylated calmodulin was detected with streptavidin–horseradish peroxidase (Amersham Pharmacia Biosciences).
Knockdown Experiments. For p600 knockdown, the human foreskin fibroblasts (hTERT-BJ1) (7) were transduced with the pSUPER.retro vector (8) containing the short hairpin RNA (shRNA) p600-6 cassette, and stably transduced subpopulations were selected with medium containing 4 μg/ml puromycin. The target 19-nt sequence of shRNA p600-6 is 5′-AATGATGAGCAGTCATCTA-3′. The empty pSUPER.retro was used as the control.
Apoptosis Analyses. The p600 knockdown and control fibroblasts were plated at 60% confluency. The medium was replaced 12 h after the plating, with no further replacements. For measuring apoptosis during cell growth, cells were used 3 days after plating. For serum starvation, cells at the confluent state were incubated further in serum-free medium for 3 days. For apoptotic analyses, live cells were costained with annexin V and Hoechst 33258 dye. The percentage of apoptotic cells was quantified by using fluorescence microscopic examination to determine the fraction of cells displaying annexin V positivity and nuclear fragmentation.
Microscopic Analyses. For immunofluorescent microscopy, cells were fixed with 4% paraformaldehyde for 10 min at 37°C. Cells were incubated with the primary antibodies, followed with appropriate secondary antibodies conjugated with Alexa Fluor 647 (Molecular Probes). Rabbit polyclonal anti-p600 antibodies were raised against p600 fragments containing residues 2656–3017 and 3362–3761, expressed in Escherichia coli. Sources for the other antibodies are as follows: inositol 1,4,5-trisphosphate receptor (IP3R) 1, IP3R2, and IP3R3 were from the Mikoshiba Laboratory (Department of Basic Medical Sciences, Institute of Medical Science, University of Tokyo, Tokyo) (9, 10); α-tubulin DM1A was from Sigma–Aldrich; RB G3-245, focal adhesion kinase (FAK), and clathrin heavy chain were from BD Pharmingen; phospho-FAKY397 was from Abcam (Cambridge, MA); and calmodulin was from Upstate Biotechnology (Lake Placid, NY). For staining of F-actin and DNA, cells were poststained with Alexa Fluor 647-conjugated phalloidin and PicoGreen (Molecular Probes), respectively. Stained cells were examined with a Nikon TE2000U inverted microscope/PerkinElmer ultraview spinning disk confocal unit. Confocal images were deconvoluted with autovisualize-3d (Autoquant Imaging, Troy, NY).
For scanning electron microscopy, cells were fixed with 2% glutaraldehyde in PBS for 1 h at room temperature. After extensive washing with PBS, cells were postfixed with 1% osmium tetroxide in PBS for 1 h. Subsequently, cells were dehydrated, dried, coated, and examined with a Zeiss LEO 1450VP scanning electron microscope.
Results
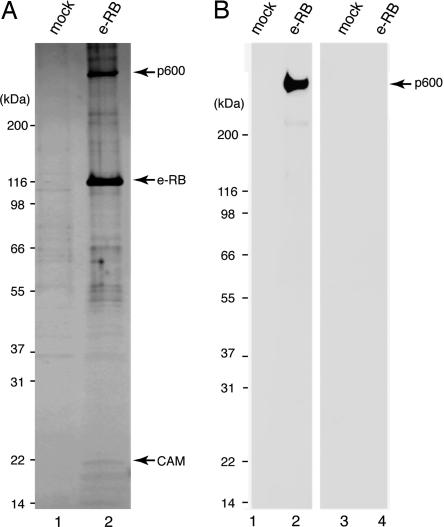
Cloning and Domain Mapping of p600. We have purified RB from HeLa cells that stably express (e-RB). Silver staining of the purified complex revealed that various proteins copurified specifically with e-RB (Fig. 1A). Mass spectrometric analyses of the purified complex revealed that among these associated factors were known RB-interacting proteins, including E2F-1, E2F-2, E2F-3, E2F-4, and DP-1. We have identified p600 as an RB-associated protein (Fig. 1 A). The peptide sequences of the 600-kDa band that were determined by mass spectrometry were used to identify EST clones to screen a cDNA library to isolate several overlapping clones. This screening permitted identification of an ORF encoding a 5,183-residue protein with an estimated molecular mass of 573,536 Da (GenBank accession no. AF348492). Potential p600 homologues were found in multicellular organisms, including Drosophila CALO/pushover and Arabidopsis BIG, but not in yeast. Although BIG mutations cause defects in polar-dependent transport of auxin (11), pushover mutations cause defects in regulating synaptic transmission at the neuromuscular junction (12). In these organisms, p600-related proteins may be involved in regulation of vesicle trafficking, although interactions of these proteins with RB have not been described.
Fig. 1.
Cloning and mapping p600. (A) Silver staining of the purified RB complex. e-RB was purified from e-RB-expressing HeLa cells by immunoprecipitation with anti-FLAG antibody (lane 2). As a control, mock purification was performed with nontransduced HeLa cells (lane 1). (B) p600 is a calmodulin-binding protein. The purified RB complex (lanes 2 and 4) and nontransduced control (lanes 1 and 3) were incubated with biotinylated calmodulin in buffer containing CaCl2 (lanes 1 and 2) or EGTA (lanes 3 and 4).
We also identified calmodulin in the RB complex by mass spectrometry (Fig. 1 A). To determine which components in the complex bind directly to calmodulin, we performed far-Western blotting. Purified RB complex was transferred to a nitrocellulose membrane and probed with biotinylated calmodulin in the presence (Fig. 1B, lanes 1 and 2) or absence (Fig. 1B, lanes 3 and 4) of Ca2+. The 600-kDa band was detected by Western blotting with horseradish peroxidase-conjugated streptavidin only when incubated in the presence of Ca2+, indicating that p600 binds directly to Ca2+-bound calmodulin. These data are consistent with the report of the isolation of Drosophila CALO/pushover in a screen for calmodulin-binding proteins (13).
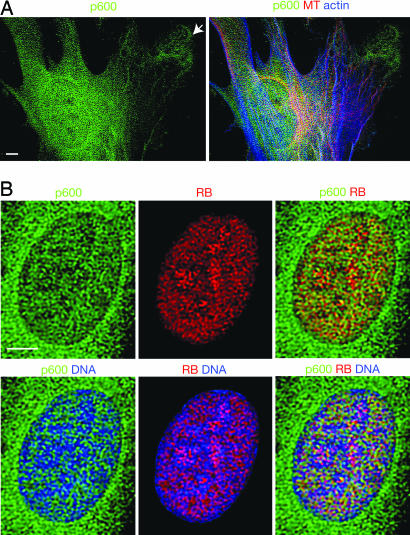
Nuclear and Cytoplasmic Localization of p600. To examine subcellular localization of p600, we performed immunostaining of human foreskin fibroblasts (hTERT-BJ1). Immunofluorescent microscopic images reveal that p600 localizes in continuous mesh patterns from the nucleus to the cytoplasm by perforating the nuclear envelope (Fig. 2A). The meshwork structure of p600 was observed by distinct fixation methods, including cold methanol fixation, as well as by staining with several different antibodies that recognize different domains of p600 (data not shown). Thus, the meshwork structure of p600 could reflect its localization in vivo. RB exists predominantly in the nucleus and localizes in the nucleus at close proximity with p600 (Fig. 2B). Nevertheless, RB seems to form patterns distinct from those of p600 rather than be strictly colocalized with it. Thus, p600 could be a structural component that regulates RB rather than a subunit of RB. DNA seems to be organized along the trenches formed by the p600/RB meshes, suggesting that p600 and RB act as a chromatin scaffold (Fig. 2B).
Fig. 2.
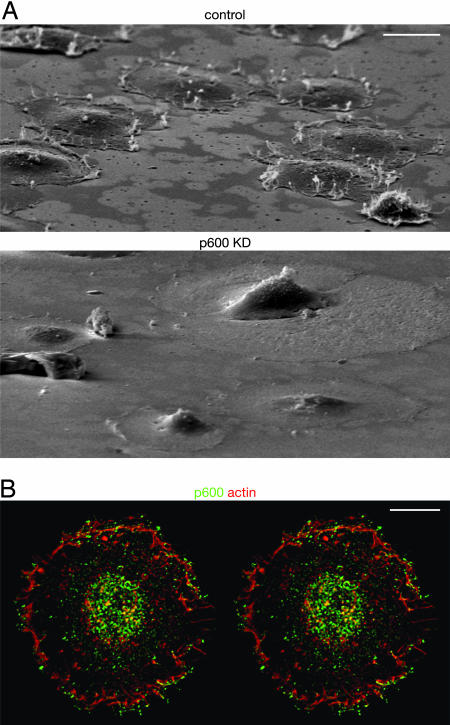
Immunolocalization of p600 and RB. (A) Localization of p600, F-actin, and microtubules (MT). In the cytoplasm, p600 is accumulated near the leading edge of protrusion sites (see arrow). (B) Localization of p600, RB, and DNA. The colors detecting stained materials are shown above the images. (Scale bars, 5 μm.)
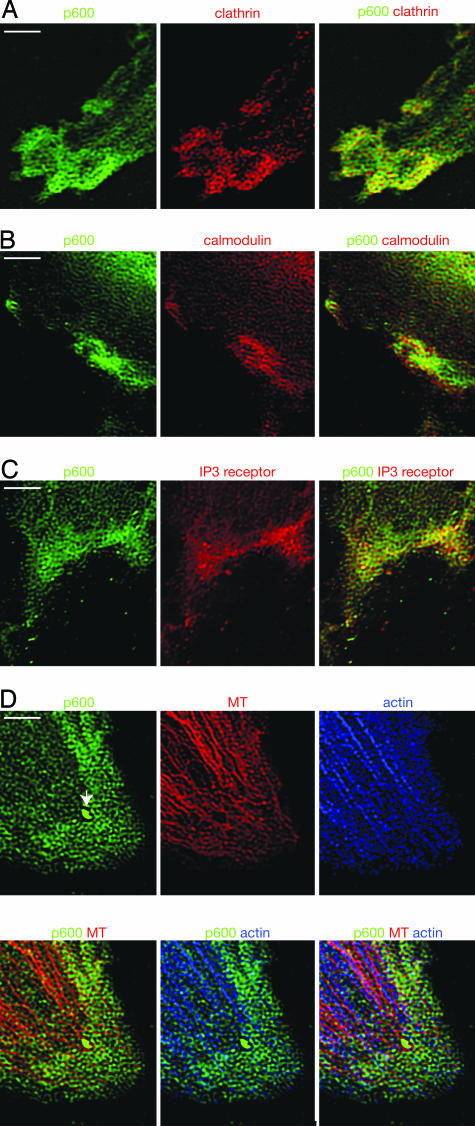
In the cytoplasm, p600 seems to concentrate at the leading edge of membrane structures in human fibroblasts known to be important for actin-based cell motility (Fig. 2 A, see arrow). In the course of this study, we also discovered that p600 and clathrin form an integrated structure near the leading edge. Clathrin is known to play a role in transporting membrane domains between intracellular membrane-bound compartments, including the trans-Golgi network and endosomes, by forming clathrin-coated membrane vesicles (14). Immunofluorescent staining shows that p600 and clathrin form similar mesh patterns that are spatially arranged at close proximity (Fig. 3A). Assembly of clathrin is believed to determine the curvature of the underlying membrane in vesicle formation, leading us to postulate that the p600–clathrin meshes may contribute to membrane morphogenesis near the leading edge. The image (Fig. 3A) suggests that p600 and clathrin do not associate in a fixed relationship to one another but rather associate to form diverse polygonal structures with distinct angles in different areas of the meshes. This flexibility of association between p600 and clathrin in the meshwork enables the structure to determine a variety of cell membrane shapes. These results suggest that p600 and clathrin play a structural role in membrane morphology.
Fig. 3.
Immunolocalization of p600 in the cytoplasm. (A) Localization of p600 and clathrin. (B) Localization of p600 and calmodulin. (C) Localization of p600 and IP3R. (D) Localization of p600, F-actin, and microtubules (MT). The newly developed surface extension is shown by the arrow. The colors detecting stained materials are shown above the images. (Scale bars, 5 μm.)
Increases in Ca2+ have a complex temporal and spatial arrangement (e.g., oscillations and waves) and have been implicated in regulating various aspects of cell adhesion, spreading, and motility (15, 16). Given that p600 is a calmodulin-binding protein, we hypothesized that p600 may play a role in Ca2+ signaling. In support of this model, calmodulin was found in close proximity to p600 in the ruffles (Fig. 3B). Moreover, IP3R was accumulated near the leading edge of ruffles and was spatially organized in close proximity to p600 (Fig. 3C). Because IP3R is a Ca2+-release channel that triggers an increase in intracellular Ca2+ concentration (17), our data suggest that p600 may act as a sensor of Ca2+, which is released through IP3R.
Extension of the leading edge is thought to be driven by polymerization of actin filaments at their fast-growing “barbed” ends, pushing the cell's boundary membrane outward (18). Our data, however, suggest that formation of the p600–clathrin meshwork precedes actin polymerization. It is intriguing that the patterns of actin and microtubules somewhat resemble the patterns of p600 and seem to develop underneath the p600 meshwork (Figs. 2 A and 3D). Near the leading edge, F-actin and microtubules seem to be deposited along with the p600 meshes (Fig. 3D). These results suggest that p600 meshes assist actin and microtubule structure development by serving as molds. p600 may function to activate specific Rho effectors that promote polymerization of actin (e.g., the neuronal Wiskott–Aldrich syndrome protein; N-WASP) or microtubules (e.g., mDia) at the specific sites of the p600 meshwork (19, 20). Such pinpoint activation of Rho effectors may determine position and direction of actin and microtubule polymerization. It is significant that newly developed surface extensions are extremely abundant in p600 (see the arrow in Fig. 3D) and clathrin (data not shown) localizations, even though F-actin and microtubules are not detectable (Fig. 3D). Taken together, our results suggest that the p600–clathrin mesh structure determines membrane morphology before development of actin and microtubule cytoskeletons.
p600 Is Required for Membrane Ruffle Formation. To determine whether a functional relationship exists between p600 and the membrane structures to which it localizes, we performed knockdown of p600 levels by expressing shRNAs that target p600 and examined cells for serum-induced membrane ruffling. The shRNA expression cassettes were introduced into BJ1 cells by retroviral infection, and stably transduced subpopulations were selected on the basis of their resistance to puromycin. The p600 level was reduced to ≈20% of normal levels in the shRNA-expressing cells (see Fig. 6A). Because its localization suggests a role for p600 in membrane morphogenesis (Fig. 4), we investigated how p600 knockdown affects ruffle formation, an actin polymerization-dependent process closely linked to cell-crawling activity. When control cells were spread on a fibronectin-coated matrix, the cells formed a number of ruffled membranes near the leading edge (Fig. 4A Upper). The stereographic image reveals that p600 accumulates near the outlines of the ruffles, especially at the top of ruffles (Fig. 4B). It is significant that knockdown of p600 caused serious defects in ruffle formation (Fig. 4A Lower), suggesting that p600 is an essential structural component for membrane morphogenesis. We have found by cell adhesion assays (21) that the p600 knockdown cells retain normal adhesion capability on fibronectin-coated matrix (data not shown). These results suggest that knockdown of p600 causes deficiency in integrin- and growth factor-induced signaling pathways rather than integrin–matrix interactions.
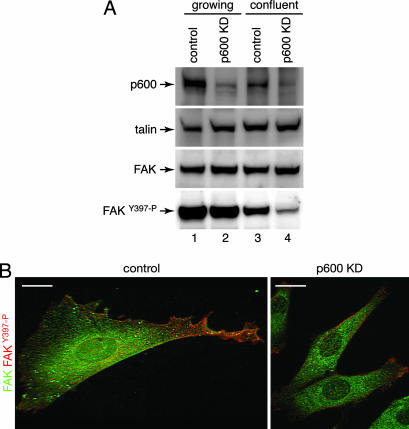
Fig. 6.
Involvement of p600 in integrin-FAK signaling. (A) Analysis of FAK phosphorylation by Western blotting. Cell lysates prepared from growing (lanes 1 and 2) and confluent (lanes 3 and 4) control (lanes 1 and 3) and p600 knockdown (KD) cells (lanes 2 and 4) were analyzed by Western blotting with the indicated antibodies. (B) Analysis of FAK phosphorylation by immunofluorescent microscopy. The control (Left) and p600 knockdown (Right) cells were costained with FAK (green) and phospho-FAKY397 (red) antibodies. (Scale bars, 10 μm.)
Fig. 4.
Knockdown (KD) of p600 causes deficiencies in ruffle formation. (A) Scanning electron microscopic images of the control (Upper) and the p600 knockdown (Lower) cells 20 min after plating on fibronectin-coated plates. (Scale bar, 20 μm.) (B) Immunolocalization of p600 (green) and fibrous actin (red). The image is represented as a stereographic view. (Scale bar, 10 μm.)
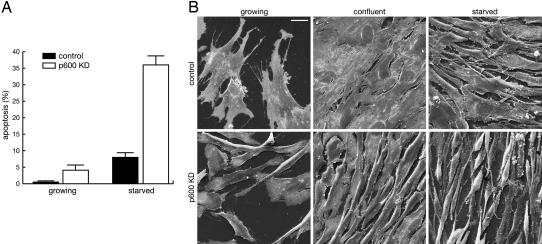
Knockdown of p600 Sensitizes Cells to Apoptosis. Although we routinely plate BJ1 cells at 30% confluency, the p600 knockdown cells could not propagate well under these conditions because of elevated cell death, suggesting a requirement for cell survival signals. When the knockdown cells were plated at higher density (e.g., 60% confluency), apoptosis was partially suppressed, and the cells propagated at a rate comparable to that of the control cells. Even under these conditions, however, the p600 knockdown cells underwent apoptosis at a higher frequency during proliferation than the control cells (Fig. 5A). When the confluent cells were grown in serum-free medium, more than one-third of the knockdown cells died by apoptosis.
Fig. 5.
Knockdown (KD) of p600 sensitizes cells to apoptosis. (A) Stimulation of apoptosis by p600 knockdown. Percentage of apoptotic cells induced during cell proliferation and serum starvation are shown in the control (black) and p600 knockdown (white) bars. Error bars are SDs. (B) Morphological changes induced by p600 knockdown. Scanning electron microscopic images of growing (Left), confluent (Center), and serum-starved (Right) states of the control (Upper) and p600 knockdown fibroblasts (Lower) are shown. Note that most apoptotic cells were removed during sample preparation because apoptosis was induced mostly after detachment from the extracellular matrix. (Scale bar, 20 μm.)
Scanning electron microscopic images show that the growing control cells are well spread and develop various kinds of surface extensions to explore their surroundings (Fig. 5B). In contrast, the p600 knockdown cells poorly develop surface extensions and have a significantly smaller contact area with the matrix. Morphological differences became more obvious when cells reached confluence. Although the control cells formed a flattened monolayer, the p600 knockdown cells develop lamellipodia poorly and maintain contact loosely with neighboring cells. As judged from the incorporation of bromodeoxyuridine, a significant fraction of p600 knockdown cells entered the S phase under these conditions, whereas most control cells remained quiescent (data not shown). Thus, knockdown of p600 causes defects in the entry into quiescence when cells reach confluence. After serum starvation, the knockdown cells form many tiny filopodia on the surface and seem to contact the matrix loosely (Fig. 5B). These results suggest that p600 is involved in cell–cell and cell–matrix interactions through dynamic amoeboid movements of the anterior lamellipodia.
Failure of FAK Activation by p600 Knockdown. The apoptosis phenotype suggests a role for p600 in activating integrin-mediated survival pathways. To examine this possibility, we determined the level of active FAK, an important regulator of cellular survival in response to extracellular signals, in the p600 knockdown cells. When integrins bind to appropriate ligands in the extracellular matrix, integrins near the leading edge form a cluster that leads to the recruitment and assembly of the focal adhesion complex. A key step in the assembly of this complex is activation of FAK by phosphorylation at Y397, enabling it to act as a docking site for the SH2 domain-containing proteins, such as the p85 regulatory subunit of phosphatidylinositol 3-kinase.
Western blot analyses reveal that the protein level of FAK was comparable or modestly increased in the knockdown cells (Fig. 6A). Likewise, talin, another component of the focal adhesion complex, also showed a modest increase in expression in the knockdown cells. It is notable that the level of FAK phosphorylated at Y397 was reduced in the p600 knockdown cells, especially in the confluent state, indicating the involvement of p600 in the FAK-signaling pathways.
Failure of FAK activation in the p600 knockdown cells was confirmed further by immunofluorescent microscopy. The control cells show that, although FAK is distributed broadly, it is preferentially phosphorylated at the lamellipodia (Fig. 6B). In the p600 knockdown cells, FAK was only weakly phosphorylated in the poorly developed lamellipodia. These results suggest that p600 has a role in activation of FAK in response to cell–matrix interactions.
Discussion
We have demonstrated that p600 plays a role in regulating integrin-mediated signaling and detachment-induced apoptosis. Diverse roles for p600 in both nuclear and cytoplasmic compartments could enable coordinate regulation of chromatin events and cytoskeleton development. In the nucleus, p600 seems to interact with RB and forms meshwork structures that closely localize to the nuclear matrix components, including lamin A and B23 (unpublished observations). This relationship between RB, p600, and nuclear architecture requires further investigation.
In the cytoplasm, p600 plays an essential role in integrin- and growth factor-induced membrane morphogenesis. Near the leading edge, p600 and clathrin form meshwork structures, which apparently link to the membrane morphogenesis. Bovine papilloma virus (BPV)-1 E6 has been shown to bind to the activating protein-1 clathrin adapter protein in Golgi membrane (22). Through a similar mechanism, BPV-1 E6 may modulate the p600–clathrin meshes to facilitate cell detachment from the matrix. The rise in Ca2+ represents a common activation pathway that results from triggering cells with various stimuli. Our data suggest that p600 could contribute to signal transduction mechanisms by serving as a sensor of Ca2+, which is released through the IP3R. In support of this view, IP3R has been shown to play a crucial role in membrane protrusion in nerve growth (22). We speculate that formation of the p600-clathrin meshes near the leading edge may be regulated by Ca2+ signaling.
Although we have isolated p600 as an RB-binding protein, the cellular localizations suggest that cytoplasmic functions of p600 are probably RB-independent. The RB-independent functions of p600 are supported by the molecular actions of HPV-16 E7. Through mutational analyses, two regions of HPV-16 E7 were identified as being essential for transformation: the CR2 domain, which targets the RB family proteins, and the N-terminal domain (5). Independent studies show that the N-terminal domain of HPV-16 E7 binds to p600 (23). Thus, this domain could target RB-independent functions of p600. The RB-independent role of p600 is further emphasized by the evidence that BPV-1 E7 binds to p600. BPV-1 and HPV-16 E7 share sequence similarity at the C-terminal region and within the extreme N-terminal sequences, but BPV-1 E7 lacks an RB-binding domain. It is significant that the conserved N-terminal domain of BPV-1 E7 binds to p600. Moreover, the BPV-1 E7 construct, which is incapable of nuclear entry, still retains transforming activity (24). Taken together, these results suggest that the cytoplasmic p600 is a common target for HPV-16 E7 and BPV-1 E7.
Although transforming factors often promote degradation of cellular factors, ectopic expression of BPV or HPV E7 does not affect p600 protein levels (23, 24). Thus, E7 binding to p600 may modulate its functions to promote transformation. p600 has been shown to be essential for anchorage-independent growth. Inhibition of p600 expression drastically suppresses E7-mediated transforming activity, determined by the ability to form colonies in soft agar (23, 24). Moreover, inhibition of p600 expression also prevents anchorage-independent growth of the HPV-16-positive CaSki cervical cancer line, without affecting growth in monolayer culture (23). Thus, E7 may bind to cytoplasmic p600 to protect apoptosis in the anchorage-independent growth.
Although it remains unclear whether p600 directly regulates apoptosis, an intriguing possibility is the action of p600 by ubiquitination. The sequence of p600 predicts the presence of the UBR box (residues 1660–1727), a recognition component of the ubiquitin-dependent N-end rule pathway. The N-end rule relates the in vivo half-life of a protein with the identity of its N-terminal residue, which is exposed after a proteolytic cleavage event (25). Although this pathway has not been well characterized in higher eukaryotes, DIAP1, a central anti-apoptotic factor in Drosophila, has been recently identified as a physiological substrate for the N-end rule pathway after caspase cleavage (26). Identifying physiological substrates for the N-end rule pathway would advance our understanding of the molecular actions of p600.
In summary, we have used a protein purification approach to identify an RB-interacting protein that seems to have significant RB-independent function in cell survival and growth factor-dependent membrane morphogenesis. That these functions are targeted by the viral oncoprotein HPV-16 E7 independently of RB binding strongly suggests a connection between events that occur at the leading edge of the plasma membrane and cellular transformation. Furthermore, these functions create a potentially new target for small molecule intervention in HPV-driven cervical carcinoma. Future studies into the mechanistic foundation for p600 function in primary and transformed cells may lend insight into processes that are required to suppress a broad range of malignancies, including those that are independent of viral infection.
Acknowledgments
We thank Roger Greenberg, David Livingston, Kyung-Won Huh, Joe DeMasi, Peter Howley, Karl Munger, and Ken Yamada for discussions; Katsuhiko Mikoshiba for providing IP3R antibodies; and Deborah Goff for editing the manuscript. We also thank the following members in core facilities: Rebecca Stearns (Electron Microscopy Facility, Harvard School of Public Health) and Lara Petrak and Jennifer Shuler (Nikon Image Center, Harvard Medical School). This work was supported, in part, by the Claudia Adams Barr Program (Y.N.) and a Human Frontier Science Program Research grant (to Y.N.).
Author contributions: Y.N. and S.J.K. designed research; Y.N., H. Konishi, A.V., H. Kurooka, K.I., J.-i.S., T.I., J.Q., and A.M.H. performed research; and Y.N. wrote the paper.
Abbreviations: HPV, human papilloma virus; RB, retinoblastoma protein; e-RB, FLAG-epitope-tagged RB; shRNA, short hairpin RNA; FAK, focal adhesion kinase; IP3R, inositol 1,4,5-trisphosphate receptor; BPV, bovine papilloma virus.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF348492).
References
- 1.Hahn, W. C. & Weinberg, R. A. (2002) Nat. Rev. Cancer 2, 331–341. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein, B. & Kinzler, K. W. (2004) Nat. Med. 10, 789–799. [DOI] [PubMed] [Google Scholar]
- 3.Nevins, J. R. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott Williams & Wilkins, Philadelphia), 4th Ed., pp. 245–283.
- 4.Muller, H. & Helin, K. (2000) Biochim. Biophys. Acta 1470, M1–M12. [DOI] [PubMed] [Google Scholar]
- 5.Jones, D. L., Thompson, D. A. & Munger, K. (1997) Virology 239, 97–107. [DOI] [PubMed] [Google Scholar]
- 6.Nakatani, Y. & Ogryzko, V. (2003) Methods Enzymol. 370, 430–444. [DOI] [PubMed] [Google Scholar]
- 7.Bodnar, A. G., Ouellette, M., Frolkis, M., Holt, S. E., Chiu, C. P., Morin, G. B., Harley, C. B., Shay, J. W., Lichtsteiner, S. & Wright, W. E. (1998) Science 279, 349–352. [DOI] [PubMed] [Google Scholar]
- 8.Brehm, A. & Kouzarides, T. (1999) Trends Biochem. Sci. 24, 142–145. [DOI] [PubMed] [Google Scholar]
- 9.Sugiyama, T., Furuya, A., Monkawa, T., Yamamoto-Hino, M., Satoh, S., Ohmori, K., Miyawaki, A., Hanai, N., Mikoshiba, K. & Hasegawa, M. (1994) FEBS Lett. 354, 149–154. [DOI] [PubMed] [Google Scholar]
- 10.Maeda, N., Niinobe, M. & Mikoshiba, K. (1990) EMBO J. 9, 61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gil, P., Dewey, E., Friml, J., Zhao, Y., Snowden, K. C., Putterill, J., Palme, K., Estelle, M. & Chory, J. (2001) Genes Dev. 15, 1985–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards, S., Hillman, T. & Stern, M. (1996) Genetics 142, 1215–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu, X. Z., Wes, P. D., Chen, H., Li, H. S., Yu, M., Morgan, S., Liu, Y. & Montell, C. (1998) J. Biol. Chem. 273, 31297–31307. [DOI] [PubMed] [Google Scholar]
- 14.Le Roy, C. & Wrana, J. L. (2005) Nat. Rev. Mol. Cell Biol. 6, 112–126. [DOI] [PubMed] [Google Scholar]
- 15.Berridge, M. J., Bootman, M. D. & Roderick, H. L. (2003) Nat. Rev. Mol. Cell Biol. 4, 517–529. [DOI] [PubMed] [Google Scholar]
- 16.Maghazachi, A. A. (2000) Int. J. Biochem. Cell Biol. 32, 931–943. [DOI] [PubMed] [Google Scholar]
- 17.Palazzo, A. F., Eng, C. H., Schlaepfer, D. D., Marcantonio, E. E. & Gundersen, G. G. (2004) Science 303, 836–839. [DOI] [PubMed] [Google Scholar]
- 18.Pantaloni, D., Le Clainche, C. & Carlier, M. F. (2001) Science 292, 1502–1506. [DOI] [PubMed] [Google Scholar]
- 19.Etienne-Manneville, S. & Hall, A. (2002) Nature 420, 629–635. [DOI] [PubMed] [Google Scholar]
- 20.Miki, H. & Takenawa, T. (2003) J. Biochem. (Tokyo) 134, 309–313. [DOI] [PubMed] [Google Scholar]
- 21.McClay, D. R. & Hertzler, P. L. (1988) in Current Protocols in Cell Biology, eds. Bonifacino, J. S., Dasso, M., Harford, J. B., Lippincott-Schwartz, J. & Yamada, K. M. (Wiley, New York), pp. 9.2.1–9.2.10.
- 22.Tong, X., Boll, W., Kirchhausen, T. & Howley, P. M. (1998) J. Virol. 72, 476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huh, K. W., Demasi, J., Ogawa, H., Nakatani, Y., Howley, P. M. & Munger, K. (2005) Proc. Natl. Acad. Sci. USA 102, 11492–11497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demasi, J., Huh, K. W., Nakatani, Y., Munger, K. & Howley, P. M. (2005) Proc. Natl. Acad. Sci. USA 102, 11486–11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varshavsky, A. (2003) Nat. Cell Biol. 5, 373–376. [DOI] [PubMed] [Google Scholar]
- 26.Ditzel, M., Wilson, R., Tenev, T., Zachariou, A., Paul, A., Deas, E. & Meier, P. (2003) Nat. Cell Biol. 5, 467–473. [DOI] [PubMed] [Google Scholar]