Abstract
There is a geographic distribution of Mycobacterium tuberculosis strains with various rpoB gene mutations that account for rifampin resistance. We studied 17 rifampin-resistant clinical isolates from patients in Greece to identify rpoB mutations. The aim of our study was the evaluation of a commercially available line probe assay kit (INNO-LiPA Rif. TB) to detect rpoB mutations and rifampin resistance. The results obtained with the commercially available assay were compared to those obtained by automated DNA sequence analysis of amplified PCR products. Randomly amplified polymorphic DNA (RAPD) analyses of the isolates were also performed. The overall concordance of the line probe assay with phenotypic rifampin susceptibility test was 94%. Three distinct rpoB mutations in codons Ser531, His526, and Asp516 were correctly identified with the kit, but mutations in external regions and insertions were detected only by automated DNA sequence analysis. The changes in codons Ser531 and His526 accounted for the majority of rifampin resistance, as previously described for isolates from other geographic areas. The results obtained by RAPD analyses of the isolates suggested that clonally related M. tuberculosis strains can have subclones bearing distinct mutant rpoB alleles. We conclude that this line probe assay kit, which is fast and with which tests are easy to perform, can be used for the rapid detection of rifampin resistance in M. tuberculosis before the availability of results by conventional methods and for epidemiological studies but that negative results obtained by this method do not rule out rifampin resistance.
Tuberculosis has once again emerged as a significant public health problem in the United States and Europe. This phenomenon is at least partially due to the growing number of human immunodeficiency virus-infected patients and to immigration from areas where tuberculosis is endemic (4, 11).
The upsurge of tuberculosis has been accompanied by a rise in the frequency of Mycobacterium tuberculosis strains resistant to one or more primary antituberculosis drugs. In the United States about 13% of new cases are due to strains resistant to at least one primary antituberculosis drug, and 3.2% of isolates are resistant to both izoniazid and rifampin (10). In Greece, the incidence of resistance to rifampin in 1995 was 1.8%. In the same year, the incidence of multidrug-resistant (MDR) M. tuberculosis strains was 1.02% (unpublished data).
Early diagnosis of the disease and rapid identification of resistance to primary antituberculosis agents are essential for efficient treatment and control of MDR strains. Detection of M. tuberculosis infection and susceptibility testing with antituberculosis drugs currently take more than 1 month. The application of molecular biology-based methods has led to marked progress in diagnostics (13), and identification of the genetic basis of antimicrobial resistance has been used to develop rapid tests for the determination of drug susceptibility (10).
Resistance to rifampin, a key component of antituberculosis therapy, is conferred by the rpoB gene (15), which encodes the β subunit of RNA polymerase, an oligomeric enzyme responsible for RNA synthesis. Specific mutations in the rpoB gene result in drug resistance by reducing the rifampin binding affinity for RNA polymerase (9). Different molecular biology-based methods have therefore been applied to the detection of rifampin-resistant strains by detecting mutations in the rpoB gene. Detection of rifampin resistance should also identify MDR strains, nearly all of which are resistant to rifampin. More than 90% of rifampin-resistant M. tuberculosis strains from different countries appear to harbor specific point mutations located in a 69-bp nucleotide region of the rpoB gene (6, 12, 15). These mutations can be characterized by PCR–single-strand conformation polymorphism analysis (12), single-tube heminested PCR (14), dideoxy fingerprinting (4), line probe assay (1, 3), and automated DNA sequencing analysis (6).
In this study, we used the line probe assay, which is based on the reverse hybridization principle (1) and which is available as a kit (INNO-LiPA Rif. TB), to characterize point mutations in a hypervariable (hot-spot) region of the rpoB genes of 17 rifampin-resistant M. tuberculosis isolates from the Reference Center of Tuberculosis in Athens, Greece. By this commercially available assay, the rifampin resistance region of the rpoB gene was amplified with specific biotinylated primers and the amplified biotinylated DNA material was hybridized with specific oligonucleotide probes immobilized as parallel lines on membrane-based strips. The results of the commercially available assay were compared to the results obtained by an automated DNA sequence analysis of amplified PCR products. Randomly amplified polymorphic DNA (RAPD) analyses were used for the epidemiological study of the isolates.
MATERIALS AND METHODS
M. tuberculosis strains.
The 17 rifampin-resistant clinical isolates used in this study were isolated in the Reference Center for Tuberculosis in Athens, Greece, over a 2-year period (1995 to 1996). All patients were inhabitants of Athens, and 14 of the patients had previously received antituberculosis therapy, while the treatment history was unknown for the other 3 patients. The initial antituberculosis therapy included isoniazid, rifampin, and pyrazinamide, and all isolates included in this study were obtained before this regimen was changed according to the susceptibility data (2). The proportional method with 7H11 medium had been used to test resistance to the primary antituberculosis drugs. All isolates were resistant to 1 μg of rifampin per ml. Ten of the isolates were resistant to rifampin alone, two were resistant to streptomycin and rifampin, one was resistant to streptomycin, rifampin, and ethambutol, one was resistant to isoniazid, rifampin, and ethambutol, one was resistant to streptomycin, isoniazid, and rifampin, and the remaining two were resistant to streptomycin, isoniazid, ethambutol, and rifampin (Table 1). Isolates were considered resistant to isoniazid when they were resistant to at least 0.2 μg of isoniazid per ml. They were considered resistant to streptomycin and ethambutol when they were resistant to 5 μg of streptomycin per ml and 2 μg of ethambutol per ml, respectively.
TABLE 1.
Identification of mutations in rpoB gene found in rifampin-resistant M. tuberculosis strains isolated from Greek patients with tuberculosis
| Clinical isolate | Resistance patterna | INNO-LIPA Rif. TB pattern (amino acid change) | Sequencing results | RAPD pattern |
|---|---|---|---|---|
| N322 | Str/Eth/Rif | R5 (Ser531→Leu) | Ser 531→Leu | B1a |
| N299 | Rif | R5 (Ser531→Leu) | Ser 531→Leu | A1b |
| N836 | Str/Rif | R5 (Ser531→Leu) | Ser 531→Leu | F4e |
| N298 | Rif | R5 (Ser531→Leu) | Ser 531→Leu | A1a |
| N127 | Str/Rif | R5 (Ser531→Leu) | Ser 508→Thr | A1a |
| Ser 531→Leu | ||||
| N98 | Str/Eth/Inh/Rif | S2 (Asp516→Val) | Asp 516→Val | |
| R5 (Ser531→Leu) | Ser 531→Leu | A1a | ||
| Phe 505→Leu | ||||
| N365 | Rif | Wild type | Ser 508→Thr | B1a |
| N389 | Rif | R4b (His526→Asp) | His 526→Asp | B1a |
| N452 | Rif | S5 | Insertion | C1c |
| N545 | Rif | R5 (Ser531→Leu) | Ser 531→Leu | E3a |
| N217 | Str/Inh/Rif | R2 (Asp516→Val) | Asp 516→Val | A1a |
| N939 | Rif | S5 | Ser 508→Thr | G5f |
| N200 | Str/Eth/Inh/Rif | R5 (Ser531→Leu) | Ser 531→Leu | A1a |
| N634 | Rif | S5 | No mutation detected | ND |
| N17 | Rif | R5 (Ser531→Leu) | Ser 531→Leu | ND |
| N457 | Rif | R4b (His526→Asp) | His 526→Asp | D2d |
| N946 | Eth/Inh/Rif | R4b (His526→Asp) | His 526→Arg | G6f |
Str, streptomycin; Eth, ethambutol; Rif, rifampin; Inh, isoniazid.
Sample preparation for PCR.
M. tuberculosis was grown on Lowenstein-Jensen slopes. Distinct colonies were inoculated into Middlebrook 7H9 broth containing 10% oleic acid-albumin-dextrose-catalase enrichment. After centrifugation, mycobacterial DNA was extracted from the pellet by using a DNA extraction reagent containing Chelex anion-exchange resin (Perkin-Elmer, Branchburg, N.J.) as described previously (8). Briefly, the pellet was suspended in distilled water, and the Chelex reagent was added to the suspensions; after boiling for 25 min and centrifugation, the supernatants were used for analysis.
Detection of mutations.
The line probe assay kit, INNO-LiPA Rif. TB (Innogenetics, Biodynamiki, Athens, Greece), was used according to the manufacturer’s instructions. The rifampin resistance-determining region of the rpoB gene was amplified with specific biotinylated primers, as indicated by the manufacturer, by using 1 U of AmpliTaq DNA polymerase per reaction mixture (Roche Diagnostic Systems, F. Hoffmann-La Roche Ltd.) in a thermocycler (Omnigene; Hybaid, Middlesex, England). Amplified biotinylated DNA material was then hybridized with specific oligonucleotide probes immobilized as parallel lines on membrane-based strips, provided with the kit. After hybridization, alkaline phosphatase-labeled streptavidin was added and bound to any biotinylated hybrid previously formed. Incubation with 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium chromogen resulted in a purple or brown precipitate.
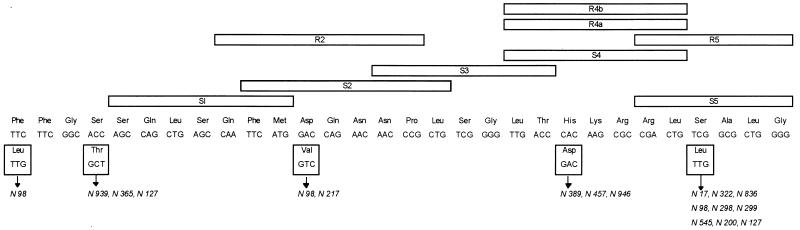
The presence of M. tuberculosis in the sample was detected with the M. tuberculosis complex-specific probe. The reactivity of an amplified fragment with the wild-type S probes S1, S2, S3, S4, and S5 was used to detect the mutations that lead to rifampin resistance in M. tuberculosis. If a mutation was present, the corresponding probe was prevented from hybridizing in the stringent hybridization and wash conditions that were used. Furthermore, four probes (R-type probes) were specifically designed to hybridize to mutant sequences of the four most frequently observed mutations: R2 (Asp516 to Val) R4a (His526 to Tyr), R4b (His528 to Asp), and R5 (Ser531 to Leu) (Table 2; Fig. 1).
TABLE 2.
Hybridization patterns obtained with the wild-type S and specific R probes of the INNO-LiPA Rif. TB compared with sequencing results
| Clinical isolate | INNO-LIPA Rif. TB pattern | Sequencing results (amino acid change detected only by sequencing) | RAPD patterna |
|---|---|---|---|
| N322 | R5 | Concordance | B1a |
| N299 | R5 | Concordance | A1b |
| N836 | R5 | Concordance | |
| N298 | R5 | Concordance | A1a |
| N127 | R5 | Concordance (Ser508→Thr) | A1a |
| N98 | S2 | Concordance | |
| R5 | Concordance (Phe505→Leu) | A1a | |
| N365 | Wild type | Ser508→Thr | |
| N389 | R4b | Concordance | |
| N452 | S5 | Insertion | |
| N545 | R5 | Concordance | |
| N217 | R2 | Concordance | A1a |
| N939 | S5 | Ser508→Thr | |
| N200 | R5 | Concordance | A1a |
| N634 | S5 | No mutation detected | |
| N17 | R5 | Concordance | |
| N457 | R4b | Concordance | |
| N946 | R4b | Concordance |
Distinct RAPD patterns for isolates with the same mutations in the rpoB gene and the same RAPD patterns with distinct mutations in the rpoB gene are shown.
FIG. 1.
Frequency of mutations of the M. tuberculosis rpoB gene in 17 rifampin-resistant isolates. To permit comparison between our data and those of others, we used the rpoB codon numbering system used by Telenti et al. (12). The positions of the Inno-LiPA Rif. TB probes are also indicated.
In conclusion, when all of the wild-type S probes gave a positive signal and when all of the R probes reacted negatively, the M. tuberculosis isolate was considered susceptible to rifampin. When at least one negative signal with the wild-type S probes was obtained, the isolate was rifampin resistant (INNO-LiPA S patterns). When the resistance to rifampin was due to one of the four most frequently observed mutations described above, a positive reaction was obtained with one of the four R probes, always accompanied by a negative reaction with the corresponding S probe (INNO-LiPA R patterns).
Automated DNA sequencing analyses of sense and antisense strands of PCR products were performed comparatively with all of the isolates. The sequences of the oligonucleotide primers flanking the rpoB hypervariable region consisted of primer TR9 (5′-TCGCCGCGATCAAGGAGT-3′) and primer TR8 (5′-TGCACGTCGCGGACCTCCA-3′) (7). These primers generated 157-bp amplicons for sequence analysis.
M. tuberculosis reference strain H37Rv was used as a control in the INNO-LiPA Rif. TB assays and automated DNA sequence analyses.
RAPD analysis.
RAPD analysis was used for molecular typing of the 17 rifampin-resistant strains of M. tuberculosis. For RAPD analysis the isolates were grown and DNA was extracted as described above for the INNO-LiPA Rif. TB assay. Three arbitrary primers were used for RAPD analysis: A1245 (5′-GCCGCCGAAACGATCTAC-3′), B1245 (5′-AGGTGGCGTCGAGGAAGAC-3′), and Leg2 (5′-CTGGCTTCTTCCAGCTTCA-3′). The primers and the PCR conditions have been described elsewhere (8). Briefly, PCR amplifications were performed in a total volume of 50 μl. The PCR mixture consisted of 50 mM Tris-HCl (pH 8.5), 17 mM (NH4)2SO4, 2 mM MgCl2, 6.7 μM EDTA, 10 mM β-mercaptoethanol, 0.1 mg of bovine serum albumin per ml, 0.01% gelatin, 200 μmol of each deoxynucleotide triphosphate, and 0.5 U of Taq DNA polymerase (Perkin-Elmer Cetus) per reaction mixture. Ten microliters of the DNA that eluted from the Chelex resin was added. The arbitrary primers were used at a concentration of 100 pmol. The reaction mixtures were overlaid with 100 μl of paraffin oil and were incubated for 5 min at 94°C. A total of 36 cycles (92°C for 1 min, 35°C for 1 min, and 72°C for 2 min) were used. The final cycle was followed by an elongation step of 7 min at 72°C. The DNA fragments were then separated by 2% agarose gel electrophoresis and were visualized by ethidium bromide staining. The gels were photographed, and the band patterns were compared visually. Band staining intensity was not considered a discriminatory factor, and profiles were considered unrelated if they differed by more than one band.
The association of RAPD patterns obtained with the three arbitrary primers was used to compare the isolates (the patterns obtained with the A1245 primer are indicated by capital letters, the patterns obtained with the B1245 primer are indicated by numbers, and the patterns obtained with the Leg2 primer are indicated by small letters).
RESULTS AND DISCUSSION
The probe specific for M. tuberculosis complex nucleotide sequences included on each strip of the line probe kit was positive for each of the 17 rifampin-resistant isolates and for the control strain M. tuberculosis H37Rv.
Three distinct nucleotide substitutions (TCG to TTG change in codon 531, CAC to GAC change in codon 526, and GAC to GTC change in codon 516) accounting for resistance in 13 of 17 isolates (76%) were identified by the kit R probes specific for these mutations (INNO-LiPA patterns R5, R4b, and R2 respectively; Table 1). The hybridization patterns obtained with the R probes correlated well with the sequencing results (Tables 1 and 2). However, one isolate (isolate N946) was correctly identified as having a point mutation in codon 526 with the line probe kit (INNO-LiPA pattern R4b), but there was a CAC to CGC change in the sequence and not a CAC to GAC change, as suggested by the kit (Table 1).
Four of 17 isolates (23%) yielded hybridization patterns with at least one negative signal with the wild-type S probes (INNO-LiPA S patterns). Only one hybridization pattern with the S probes matched the sequencing results (that for isolate N98, with an S2 pattern; Tables 1 and 2). Of the remaining three isolates, one (isolate N452, with an S5 pattern) had a 1-bp insertion, one (isolate N939, with an S5 pattern) had a nucleotide substitution outside the region detected by the probe provided with the line probe kit (codon 508), and one (isolate N634, also with an S5 pattern) had no detectable mutation (Tables 1 and 2).
Two isolates (isolates N127 and N98) had combinations of two and three point mutations, respectively, in noncontiguous codons (12%), and the line probe kit did not reveal the nucleotide substitutions in codons 505 and 508, which are outside the region covered by the probes provided with the kit (Tables 1 and 2).
The overall concordance of the line probe kit results and the results of phenotypic rifampin susceptibility testing was 94% in our study. Indeed, isolate N365 was rifampin resistant by phenotypic methods and was found by sequence analysis to have a mutation outside the region covered by the probes provided with the line probe assay kit; consequently, it was identified as susceptible by the commercially available assay (Table 1). It is noteworthy that 90% concordance of the line probe kit results and the results of phenotypic rifampin susceptibility testing has also been reported by others (3).
The concordance of the line probe kit results and the sequencing results was poorer. While a 100% concordance was observed between the results obtained with specific R probes and by sequencing, a concordance of only 25% was found between results of assays with wild-type S probes and by sequencing (Tables 1 and 2). All discordant results were observed with the S5 probe. The S5 probe is a wild-type probe, and its location is presented in Fig. 1. We do not know why discordant results were frequent with the S5 probe. A possible explanation could be a problem in the choice of the location and/or fabrication of this probe, but we did not retrieve from the manufacturer the data needed for testing this hypothesis.
The combined sequence and line probe kit results suggest that in Greece, as in other countries, the TCG to TTG (Ser to Leu) change in codon 531 is relatively abundant (53%). About 12% of the isolates had the CAC to GAC change at codon 526, and a further 6% had a CAC to CGC change at codon 526. About 18% of the isolates had a ACC to GCT change at codon 508. About 12% of the isolates had a GAC to GTC change at codon 516, and about 6% of the isolates had a TTC to TTG change at codon 505 (Fig. 1). Changes at codons 531, 526, and 516 have been extensively reported previously. Changes in codons 508 and 505 are infrequent, but the ACC to GCT change at codon 508 (7) and the TTC to TTG change at codon 505 (5) have also been described by others. The high incidence of these two infrequent changes in our isolates is in contrast to data from other geographic locations and can be explained by either geographic variations in the frequency of particular rpoB mutations or a sample bias.
RAPD analysis has clearly shown that isolates with the same rpoB alleles can have distinct RAPD profiles (e.g., isolates N322 and N299) and that isolates with the same RAPD profiles can have distinct rpoB alleles (e.g., N298, N217, and N200) (Table 2). Thus, clonally related M. tuberculosis strains can have subclones bearing distinct mutant rpoB alleles and the same mutant rpoB alleles can be borne by unrelated M. tuberculosis strains.
Sequence analysis identified no mutations in 1 of the 17 isolates tested (isolate N634), although this isolate was repeatedly resistant to rifampin. A similar phenomenon has been reported by others (6, 7, 12) and suggests that mutations located outside of the region of analysis can result in rifampin resistance. Another possibility is that for this resistant strain changes have occurred in genes whose products participate in antibiotic permeation or metabolism (6). However, this isolate was identified with the S5 probe of the line probe assay kit as being resistant (Table 1). It is noteworthy that the overall concordance of the assay with the S5 probe and sequence analysis was only 25%. However, sequence analysis could detect insertions, deletions, and mutations in regions external to the regions covered by the probes provided with the commercially available assay. It remains to be elucidated why the S-probe result was concordant with the phenotypic results for isolate N634, while the sequence analysis failed to detect the rifampin resistance.
In conclusion, the results obtained with the line probe kit do not always agree with those obtained by sequence analysis with regard to the exact mechanism of rifampin resistance, but they closely match those of phenotypic rifampin susceptibility testing and may be useful for the rapid screening of M. tuberculosis isolates for rifampin resistance and in epidemiological analyses. We have recently tested this commercially available assay with M. tuberculosis isolates from two human immunodeficiency virus-seropositive patients. Results were obtained in 2 days, and the phenotypic results confirmed that isolates were rifampin susceptible (data not shown). This assay is easy to perform, remains relatively expensive, and quickly provides results. However, because insertions, deletions, and mutations in external regions cannot be detected with the kit, negative results with the line probe assay kit do not rule out rifampin resistance.
REFERENCES
- 1.Beenhouwer H, Lhiang Z, Jannes G, Hijs W, Machtelinck L, Rossau R, Traore H, Portaels F. Rapid detection of rifampin resistance in sputum and biopsy speciments from tuberculosis patients by PCR and line probe assay. Tubercle Lung Dis. 1995;76:425–430. doi: 10.1016/0962-8479(95)90009-8. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Initial therapy for tuberculosis in the era of multidrug resistance: recommendations of the Advisory Council for the Elimination of Tuberculosis. Morbid Mortal Weekly Rep. 1993;42:1–8. [PubMed] [Google Scholar]
- 3.Cooksey R C, Morlock G P, Glickman S, Crawford J T. Evaluation of a line probe assay kit for characterization of rpoB mutations in rifampin-resistant Mycobacterium tuberculosis isolates from New York City. J Clin Microbiol. 1997;35:1281–1283. doi: 10.1128/jcm.35.5.1281-1283.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Felmlee T A, Liu Q, Whelen A C, Williams D, Sommer S S, Persing D H. Genotypic detection of Mycobacterium tuberculosis rifampin resistance: comparison of single-strand conformation polymorphism and dideoxy fingerprinting. J Clin Microbiol. 1995;33:1617–1623. doi: 10.1128/jcm.33.6.1617-1623.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heym B, Honore N, Truffot-Pernot C, Banerjee A, Schurra C, Jacobs W R, Jr, van Embden J D A, Grosset J H, Cole S T. Implications of multidrug resistance for the future of short course chemotherapy of tuberculosis: a molecular study. Lancet. 1994;344:293–298. doi: 10.1016/s0140-6736(94)91338-2. [DOI] [PubMed] [Google Scholar]
- 6.Kapur V, Li L-L, Iordanescu S, Hamrick M R, Wanger A, Kreiswirth B N, Musser J M. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase β subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J Clin Microbiol. 1994;32:1095–1098. doi: 10.1128/jcm.32.4.1095-1098.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim B-J, Kim S-Y, Park B-H, Lyu M-A, Park I-K, Bal G-H, Kim S-J, Cha C-Y, Kook Y-H. Mutations in the rpoB gene of Mycobacterium tuberculosis that interfere with PCR-single-strand conformation polymorphism analysis for rifampin susceptibility testing. J Clin Microbiol. 1997;35:492–494. doi: 10.1128/jcm.35.2.492-494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsiota-Bernard P, Waser S, Tassios P T, Kyriakopoulos A, Legakis N J. Rapid discrimination of Mycobacterium avium strains from AIDS patients by randomly amplified polymorphic DNA analysis. J Clin Microbiol. 1997;35:1585–1588. doi: 10.1128/jcm.35.6.1585-1588.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller L P, Crawford J T, Shinnick T M. The rpoB gene of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1994;38:805–811. doi: 10.1128/aac.38.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Musser J M. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin Microbiol Rev. 1995;8:496–514. doi: 10.1128/cmr.8.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sepkowitz K A, Raffalli J. Tuberculosis at the end of twentieth century. Eur J Clin Microbiol Infect Dis. 1994;13:902–907. doi: 10.1007/BF02111490. [DOI] [PubMed] [Google Scholar]
- 12.Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston M J, Matter L, Schopper K, Bodmer T. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 13.Wellstood S. Diagnostic mycobacteriology: current challenges and technologies. Lab Med. 1993;24:357–361. [Google Scholar]
- 14.Whelen A C, Felmlee T A, Hunt J M, Williams D L, Roberts G D, Stockman L, Persing D H. Direct genotypic detection of Mycobacterium tuberculosis rifampin resistance in clinical specimens by using single-tube heminested PCR. J Clin Microbiol. 1995;33:556–561. doi: 10.1128/jcm.33.3.556-561.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams D L, Waguespack C, Eisenach K, Crawford J T, Portaels F, Salfinger M, Nolan C M, Abz C, Sticht-Groh V, Gillis T P. Characterization of rifampin resistance in pathogenic mycobacteria. Antimicrob Agents Chemother. 1994;38:2380–2386. doi: 10.1128/aac.38.10.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]