Abstract
We have used a PCR assay based on the use of degenerate primers in order to characterize an internal fragment (sodAint) representing approximately 85% of the genes encoding the manganese-dependent superoxide dismutase in various streptococcal type strains (S. acidominimus, S. agalactiae, S. alactolyticus, S. anginosus, S. bovis, S. constellatus, S. canis, S. cricetus, S. downei, S. dysgalactiae, S. equi subsp. equi, S. equi subsp. zooepidemicus, S. equinus, S. gordonii, S. iniae, S. intermedius, S. mitis, S. mutans, S. oralis, S. parasanguis, S. pneumoniae, S. porcinus, S. pyogenes, S. salivarius, S. sanguis, S. sobrinus, S. suis, S. thermophilus, and S. vestibularis). Phylogenetic analysis of these sodAint fragments yields an evolutionary tree having a topology similar to that of the tree constructed with the 16S rRNA sequences. We have shown that clinical isolates could be identified by determining the positions of their sodAint fragments on the phylogenetic tree of the sodAint fragments of the type species. We propose this method for the characterization of strains that cannot be assigned to a species on the basis of their conventional phenotypic reactions.
The genus Streptococcus could be taxonomically divided into six major clusters which included at least 31 species (4, 8, 17, 18, 32, 34–36). These are (i) the pyogenic group, which includes S. agalactiae, S. canis, S. dysgalactiae, S. equi, S. iniae, S. porcinus, and S. pyogenes; (ii) the bovis group, which includes S. bovis, S. equinus, and S. alactolyticus; (iii) the salivarius group, which includes S. salivarius, S. thermophilus, and S. vestibularis; (iv) the mutans group, which includes S. cricetus, S. downei, S. mutans, and S. sobrinus; (v) the anginosus group (also referred to as the milleri group), which includes S. anginosus, S. constellatus, and S. intermedius; and (vi) the mitis group, which includes S. mitis, S. oralis, S. pneumoniae, S. sanguis, S. parasanguis, and S. gordonii. No single system of classification suffices for the differentiation of this heterogeneous group of organisms. Instead, classification depends on a combination of features including patterns of hemolysis observed on blood agar plates, antigenic composition, growth characteristics, biochemical reactions, and more recently, genetic analysis (3, 14, 18, 28).
In clinical laboratories, the current means of identification of streptococci rely on phenotypic tests such as those developed for the API ID 32 Strep system. However, the potential problems inherent to the use of phenotypic tests are that not all strains within a given species may be positive for a common trait (3, 18) and that the same strain may exhibit biochemical variability (15, 30). Moreover, small alterations in the realization of a test may give false results. Consequently, the routine technique based on phenotypic tests do not allow for an unequivocal identification of certain streptococcal species, in particular, those belonging to the milleri, the mutans, and the mitis groups (2, 3, 10, 18, 19). Nucleic acid-based technologies such as DNA hybridization (1, 16, 29) or amplification of selected targets (25, 27, 33) have been developed in recent years to complement and improve the identification of streptococci. We previously described a PCR assay based on the use of degenerate primers which enabled amplification of an internal fragment representing approximately 85% of the sodA gene encoding a manganese-dependent enzyme (manganese-dependent superoxide dismutase [Mn-SOD]) in various gram-positive bacteria including streptococci and enterococci (24). This gene has been identified as a target for the identification of mycobacteria at the species level by PCR (37), and we investigated in this study the sequencing of the sodA PCR product as an approach to the genotypic identification of 29 different streptococcal species including those constituting the milleri, mitis, and mutans groups.
(A report of this work was presented at the XIIIth Lancefield International Symposium [16 to 20 September 1996, Paris, France].)
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The main characteristics of the streptococcal strains used in this study, including the type strains, are listed in Tables 1 and 2. All strains were grown at 37°C on Columbia horse blood agar (bio-Mérieux, Marcy l’Etoile, France) in an anaerobic atmosphere. Phenotypic identifications were performed with the rapid ID 32 Strep System (API-bio-Mérieux, Marcy l’Etoile, France) according to the manufacturer’s instructions. The API profiles were interpreted from the computer database for identification.
TABLE 1.
Streptococcal type strains used in this study
| Straina | Other designationb | Relevant characteristics | sodAint accession no. |
|---|---|---|---|
| S. acidominimus CIP 82.4T | NCDO 2025 | Type strain | Z95892 |
| S. agalactiae CIP 103227T | ATCC 13813 | Type strain | Z95893 |
| S. alactolyticus CIP 103244T | ATCC 43077 | Type strain | Z95894 |
| S. anginosus CIP 102921T | ATCC 33397 | Type strain | Z95895 |
| S. bovis CIP 102302T | ATCC 33317 | Type strain | Z95896 |
| S. canis CIP 103223T | ATCC43496 | Type strain | Z99175 |
| S. constellatus CIP 103247T | ATCC 27823 | Type strain | Z95897 |
| S. cricetus CIP 102510T | ATCC 19642 | Type strain | Z95898 |
| S. downei CIP 103222T | ATCC 33748 | Type strain | Z95899 |
| S. dysgalactiae CIP 102914T | ATCC 43078 | Type strain | Z95900 |
| S. equinus CIP 102504T | ATCC 9812 | Type strain | Z95903 |
| S. equi subsp. equi CIP 102910T | ATCC 33398 | Type strain | Z95901 |
| S. equi subsp. zooepidemicus CIP 103228T | ATCC 43079 | Type strain | Z95902 |
| S. gordonii CIP 105258T | ATCC 10558 | Type strain | Z95905 |
| S. iniae CIP 102508T | ATCC 29178 | Type strain | Z99176 |
| S. intermedius CIP 103248T | ATCC 27335 | Type strain | Z95908 |
| S. mitis CIP 103335T | NCTC 12261 | Type strain | Z95909 |
| S. mutans CIP 103694 | ATCC 35668 | Quality control strain for API product | Z95910 |
| S. oralis CIP 10922T | ATCC 35037 | Type strain | Z95911 |
| S. parasanguis CIP 104372T | ATCC 15910 | Type strain | Z95913 |
| S. pneumoniae CIP 102911T | ATCC 33400 | Type strain | Z95914 |
| S. porcinus CIP 103218T | ATCC 43138 | Type strain | Z99177 |
| S. pyogenes CIP 56.41T | ATCC 12344 | Type strain | Z95915 |
| S. salivarius CIP 102509T | ATCC 19645 | Type strain | Z95916 |
| S. sanguis CIP 55.1328T | ATCC 10556 | Type strain | Z95918 |
| S. sobrinus CIP 103230T | ATCC 33478 | Type strain | Z95919 |
| S. suis CIP 103217T | ATCC 43765 | Type strain | Z95920 |
| S. thermophilus CIP 102303T | ATCC 19258 | Type strain | Z95921 |
| S. vestibularis CIP 103363T | ATCC 49124 | Type strain | Z95922 |
CIP, Collection de l’Institut Pasteur.
NCDO, National Collection of Dairy Organisms; ATCC, American Type Culture Collection; NCTC, National Collection of Type Cultures.
TABLE 2.
Comparative identification of various streptococcal strains
| Straina | Relevant characteristicsb | Species identification
|
sodAint accession no. | |
|---|---|---|---|---|
| ID 32 Strepc | sodAintd | |||
| S. agalactiae | ||||
| CIP 82.45 | Lancefield group B (ATCC 12403) | ND | S. agalactiae (98.6) | Z99178 |
| NEM318 | Lancefield group B | ND | S. agalactiae (97.9) | Z99179 |
| NEM1317 | Lancefield group B | ND | S. agalactiae (98.4) | Z99180 |
| Milleri group | ||||
| NEM1164 | Nontypeable, β-hemolytic | S. constellatus (99.6) | S. anginosus (97) | Z99181 |
| NEM1166 | Nontypeable, β-hemolytic | S. anginosus (99.8) | S. anginosus (97.4) | Z99182 |
| NEM1124 | Lancefield group F, β-hemolytic | S. constellatus (99.9) | S. constellatus (98.1) | Z99183 |
| NEM1162 | Lancefield group F, β-hemolytic | S. constellatus (99.6) | S. constellatus (99.1) | Z99184 |
| NEM1165 | Nontypeable | S. constellatus (93.1) | S. constellatus (98.9) | Z99185 |
| NEM1275 | Nontypeable | S. constellatus (63.9) | S. constellatus (99.1) | Z99186 |
| MG19 | Nontypeable | Milleri group | S. constellatus (99.1) | Z99187 |
| Mitis group | ||||
| CIP 103221 | ATCC 33399 | S. gordonii (?) | S. gordonii (95.9) | Z99189 |
| BM120 | Strain Challis | S. gordonii (99.9) | S. gordonii (98.2) | Z99188 |
| NEM666 | Nontypeable | ? | S. gordonii (95.9) | Z99190 |
| NEM1222 | Nontypeable | S. mitis (95.6) | S. mitis (97.8) | Z99192 |
| NEM1126 | Lancefield group C | S. mitis (59.5) | S. mitis (97.8) | Z99191 |
| S. mutans | ||||
| GS-5 | ND | S. mutans (99.8) | D01037 | |
| NEM1163 | Nontypeable | S. mutans (99.9) | S. mutans (99.5) | Z99193 |
| S. oralis | ||||
| CIP 103216 | ATCC 10557 | S. oralis (93.7) | Z99194 | |
| NEM1121 | Lancefield group C | S. salivarius (99.8) | S. oralis (96.1) | Z99195 |
| NEM895 | Nontypeable | ? | S. parasanguis (97.8) | Z99196 |
| S. pneumoniae | ||||
| NEM667 | Serotype 11 | ND | S. pneumoniae (100) | Z99246 |
| NEM1278 | Serotype 23F | ND | S. pneumoniae (99.8) | Z99204 |
| NEM1279 | Serotype 23F, Penr | ND | S. pneumoniae (100) | Z99205 |
| NEM1280 | Serotype 23F, Penr | ND | S. pneumoniae (100) | Z99206 |
| NEM1251 | Serotype 16 | ND | S. pneumoniae (100) | Z99201 |
| NEM1252 | Serotype 18 | ND | S. pneumoniae (100) | Z99202 |
| NEM1253 | Serotype 6 | ND | S. pneumoniae (100) | Z99203 |
| NEM1122 | Serotype 23F, Optr | ? | S. pneumoniae (99.8) | Z99200 |
| S. pyogenes | ||||
| BM105 | Lancefield group A | ND | S. pyogenes (100) | Z49247 |
| HSC5 | Lancefield group A | ND | S. pyogenes (100) | U43776 |
| S. salivarius | ||||
| CIP 102505 | ATCC 13419 | S. salivarius (?) | S. salivarius (96.3) | Z99197 |
| NEM1250 | Lancefield group D | S. salivarius (99.9) | S. salivarius (98.6) | Z99198 |
| NEM1257 | Nontypeable | S. salivarius (99.5) | S. salivarius (96.3) | Z99199 |
Data for all strains except S. agalactiae CIP 82.45 (24), S. agalactiae NEM318 (11), MG19 (5), S. gordonii BM120 (20), S. mutans GS-5 (21), S. pneumoniae NEM667 (24), S. pyogenes BM105 (24), and S. pyogenes HSC5 (13) are from this work.
Optor and Penr, resistance to optochin and penicillin, respectively.
The rapid ID 32 Strep system (API-bio-Mérieux) was used according to the manufacturer’s instructions to identify isolates to the species level. The API profiles were interpreted from the computer database for identification. The numbers in parentheses indicate the API identification percentage. ND, not determined; ?, undetermined.
The species identification was based on the phylogenic position of the sodAint fragment of the strain studied relative to those of the type strains, as indicated in Fig. 1. The numbers in parentheses indicate the percent identity of the sodAint fragment with that of the corresponding type strain.
DNA manipulations.
Rapid extraction of bacterial genomic DNA was performed as described previously (6), and primers d1 (5′-CCITAYICITAYGAYGCIYTIGARCC-3′) and d2 (5′-ARRTARTAIGCRTGYTCCCAIACRTC-3′) were used to amplify an internal fragment representing approximately 85% of the sodA genes of the bacterial strains. PCRs were performed with a Gene Amp System 9600 instrument (Perkin-Elmer Cetus, Roissy, France) in a final volume of 50 μl containing 250 ng of DNA as template, 0.25 μM (each) primer, 200 μM (each) deoxynucleoside triphosphate, and 1 U of Taq DNA polymerase in a 1× amplification buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2). The PCR mixtures were denatured (3 min at 95°C) and were then subjected to 35 cycles of amplification (90 s of annealing at 37°C, 90 s of elongation at 72°C, and 30 s of denaturation at 95°C) and to a final elongation cycle of 72°C for 10 min. The PCR products were resolved by electrophoresis on a 1% agarose gel stained with ethidium bromide.
Cloning and sequencing.
Amplification products were purified on a Sephadex S-200 column (Pharmacia, Uppsala, Sweden) and were ligated into the pUC18-SmaI dephosphorylated vector by using the Sure-clone ligation kit (Pharmacia, Uppsala, Sweden). Recombinant plasmids were analyzed by colony-PCR as follows. Twelve randomly chosen clones were amplified by using the universal −21 (5′-GTAAAACGACGGCCAGT-3′) and reverse (5′-AACAGCTATGACCATG-3′) primers in a final volume of 50 μl containing 103 bacteria, 0.1 μM (each) primer, 200 μM (each) deoxynucleoside triphosphate, and 1 U of Taq DNA polymerase in a 1× amplification buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2). The PCR mixtures were denatured (10 min at 95°C) and were then subjected to 30 cycles of amplification (90 s of annealing at 45°C, 1 min of elongation at 72°C, and 1 min of denaturation at 95°C). Colony-PCR products were directly sequenced after purification on a Sephadex S-400 column (Pharmacia). The entire nucleotide sequences of both strands of two cloned amplicons obtained from independent PCRs were determined by using the dideoxy chain termination method of Sanger with the dye primer cycle sequencing ready reaction kit on a Genetic ABI PRISM 310 Sequencer Analyzer (Perkin-Elmer, Applied Biosystem Division, Roissy, France).
Direct sequencing of the sodAint PCR products with either of the degenerate oligonucleotides d1 and d2 was performed with the dRhodamine dye terminator sequencing kit (Perkin-Elmer, Applied Biosystem Division), as follows. After purification on a Centricon-100 Concentrator column, 200 ng of the PCR product was mixed with 8 μl of terminator reaction mixture and 10 pmol of primer in a final volume of 20 μl, and the mixture was subjected to the following thermal cycling: 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min (which was repeated for 25 cycles).
Sequence analysis.
The nucleotide sequences were analyzed with Perkin-Elmer software (Sequence Analysis, Sequence Navigator, and Autoassembler). Multiple alignment of sod genes was carried out by the CLUSTAL X program (31). The construction of the unrooted phylogenetic tree was performed by the neighbor-joining method (26).
Nucleotide sequence accession numbers.
The sequences were submitted to the EMBL gene bank and were assigned the accession numbers listed in Tables 1 and 2.
RESULTS AND DISCUSSION
Amplification and sequencing of sodAint from various streptococcal type strains.
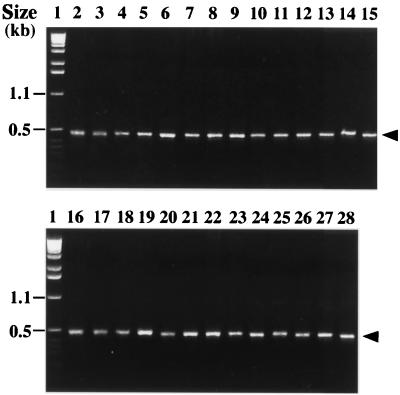
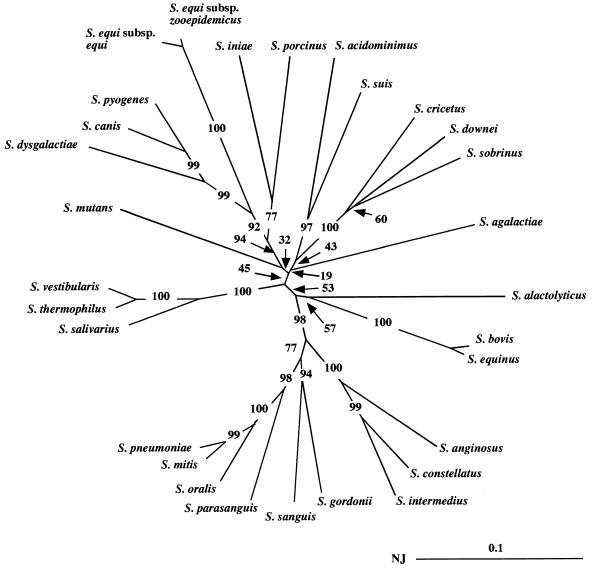
By using the primers d1 and d2 in a PCR assay, we amplified an internal fragment representing approximately 85% of the sodA gene encoding a manganese-dependent enzyme (Mn-SOD) in 29 type strains of streptococci (S. acidominimus, S. agalactiae, S. alactolyticus, S. anginosus, S. bovis, S. canis, S. constellatus, S. cricetus, S. downei, S. dysgalactiae, S. equi subsp. equi, S. equi subsp. zooepidemicus, S. equinus, S. gordonii, S. iniae, S. intermedius, S. mitis, S. mutans, S. oralis, S. parasanguis, S. pneumoniae, S. porcinus, S. pyogenes, S. salivarius, S. sanguis, S. sobrinus, S. suis, S. thermophilus, and S. vestibularis). A single amplification product having the expected size of 480 bp was observed with all streptococcal species (Fig. 1 shows the results of part of this analysis). The nucleotide sequences of the sodAint fragments from these type strains were determined following cloning into pUC18 (Table 1). Analysis of the corresponding deduced amino acid sequences (data not shown) revealed that they all possessed three histidyl residues and one aspartyl residue that supposedly serve as metal ligands at positions characteristic of Mn- or Fe-SODs (22, 23). Moreover, they all contain in the vicinity of the active site four other residues characteristic of Mn-SODs, which suggests that the corresponding enzymes are activated by Mn ions (23). We therefore concluded that the PCR products cloned and sequenced were actual sodAint fragments. Multiple alignment of the streptococcal sodAint sequences was carried out by the CLUSTAL X program (31), and an unrooted phylogenetic tree was constructed by the neighbor-joining method (26). Domains I and II corresponding to the degenerate oligonucleotides d1 and d2, respectively, and alignment gaps were not taken into consideration for calculations. The topology of the phylogenetic tree obtained (Fig. 2) was evaluated by bootstrap analyses to give the degree of confidence intervals for each node on the phylogenetic tree. The confidence values are given at the branches, which show possibly monophyletic clades of related organisms separated at each node. It is generally accepted that the monophyly of a clade can be accepted if the clade occurs in more than 95% of the bootstrapped trees (9). If this critical value is used, the sodAint-based phylogenetic positions of the streptococcal species studied here were in general agreement with those inferred from an analysis of their 16S rRNA sequences (4, 17), with the following remarkable exceptions: S. agalactiae did not cluster with the species constituting the pyogenic group, and S. mutans was genetically separate from S. cricetus, S. downei, and S. sobrinus. Pairwise comparison of two given streptococcal species always revealed a lower percentage of sequence identity between their sodAint fragments than between their 16S RNAs (Table 3 and data not shown). For example, the sequence identities of the 16S RNAs of type strains of S. mitis, S. oralis, and S. pneumoniae are greater than 99% (17), whereas those of their sodAint fragments vary from 92% (S. mitis versus S. oralis and S. oralis versus S. pneumoniae) to 96.6% (S. mitis versus S. pneumoniae) (Table 3). These results indicate that the sodA gene might constitute a more discriminative target sequence than the 16S RNA for differentiating closely related bacterial species. On the other hand, it is worth noting that the close structural relationship (98.9% identity) observed between the sodAint fragments of S. equi subsp. equi and S. equi subsp. zooepidemicus is consistent with their association in a single species (7).
FIG. 1.
Amplification of streptococcal type strains with the primers d1 and d2 and separation of the amplicons by 1% agarose gel electrophoresis. Lanes: 1, 1-kb ladder (Gibco, BRL); 2, S. acidominimus; 3, S. agalactiae; 4, S. alactolyticus; 5, S. anginosus; 6, S. bovis; 7, S. constellatus; 8, S. cricetus; 9, S. downei; 10, S. dysgalactiae; 11, S. equi subsp. equi; 12, S. equi subsp. zooepidemicus; 13, S. equinus; 14, S. gordonii; 15, S. intermedius; 16, S. mitis; 17, S. mutans; 18, S. oralis; 19, S. parasanguis; 20, S. pneumoniae; 21, S. porcinus; 22, S. pyogenes; 23, S. salivarius; 24, S. sanguis; 25, S. sobrinus; 26, S. thermophilus; 27, S. suis; 28, S. vestibularis. Arrowheads, 480-bp amplicon.
FIG. 2.
Phylogenetic unrooted tree showing relationships among the sodAint fragments from various streptococcal type strains. The tree was established from an analysis of the sequences listed in Table 1 by using the neighbor-joining method (26). The value on each branch is the estimated confidence limit (expressed as a percentage) for the position of the branch as determined by bootstrap analysis. The scale bar (neighbor-joining [NJ] distance) represents 10% differences in nucleotide sequences.
TABLE 3.
Identity matrix based pairwise comparisons of sodAint fragments of streptococcal type strains
| Straina | % Identity with the following strainb:
|
|||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | |
| 1. S. acidominimus | 71.3 | 67.8 | 71.7 | 68.0 | 69.9 | 69.9 | 70.8 | 73.3 | 67.6 | 65.3 | 66.0 | 69.9 | 72.6 | 70.8 | 68.5 | 69.7 | 72.6 | 70.8 | 66.9 | 69.0 | 70.3 | 68.0 | 72.0 | 69.7 | 70.1 | 79.5 | 73.3 | 73.3 |
| 2. S. agalactiae | 69.0 | 72.0 | 72.6 | 71.7 | 73.1 | 71.3 | 71.5 | 71.5 | 66.7 | 66.7 | 74.0 | 73.3 | 71.7 | 71.0 | 69.7 | 74.5 | 69.9 | 72.4 | 70.1 | 71.3 | 70.8 | 74.5 | 73.1 | 74.0 | 74.7 | 72.4 | 72.6 | |
| 3. S. alactolyticus | 73.1 | 74.9 | 69.7 | 76.1 | 66.4 | 69.4 | 69.7 | 66.2 | 65.7 | 75.2 | 72.0 | 66.9 | 73.1 | 70.8 | 70.6 | 70.6 | 71.0 | 71.0 | 69.7 | 70.3 | 74.5 | 72.2 | 68.3 | 68.3 | 72.2 | 71.3 | ||
| 4. S. anginosus | 71.7 | 67.1 | 86.4 | 69.4 | 68.5 | 65.5 | 66.9 | 66.7 | 72.2 | 80.5 | 65.3 | 83.4 | 75.9 | 69.7 | 74.3 | 75.9 | 75.2 | 66.9 | 66.7 | 69.2 | 76.6 | 66.7 | 72.4 | 70.8 | 70.3 | |||
| 5. S. bovis | 72.0 | 70.3 | 72.9 | 70.6 | 68.7 | 66.4 | 66.4 | 97.9 | 70.6 | 68.0 | 69.9 | 72.9 | 74.0 | 71.7 | 73.3 | 72.2 | 68.5 | 70.6 | 77.5 | 69.4 | 69.7 | 70.3 | 75.9 | 76.3 | ||||
| 6. S. canis | 64.1 | 73.1 | 73.1 | 85.1 | 78.9 | 79.5 | 72.4 | 65.5 | 78.9 | 63.0 | 67.8 | 76.8 | 64.6 | 66.7 | 67.6 | 74.0 | 92.2 | 74.0 | 64.6 | 72.9 | 67.8 | 72.0 | 71.3 | |||||
| 7. S. constellatus | 65.5 | 66.4 | 66.0 | 62.3 | 62.1 | 71.5 | 81.8 | 66.9 | 90.1 | 76.8 | 68.7 | 75.6 | 78.2 | 76.1 | 69.2 | 66.7 | 70.3 | 79.8 | 65.3 | 71.5 | 71.5 | 70.6 | ||||||
| 8. S. cricetus | 84.1 | 70.3 | 68.0 | 68.7 | 73.3 | 70.1 | 72.2 | 64.8 | 71.0 | 75.6 | 70.3 | 70.6 | 70.8 | 69.2 | 72.0 | 75.9 | 69.0 | 84.1 | 74.7 | 73.6 | 73.3 | |||||||
| 9. S. downei | 70.1 | 71.0 | 70.8 | 70.1 | 65.7 | 73.1 | 63.9 | 68.3 | 74.0 | 66.9 | 66.2 | 68.7 | 69.7 | 71.3 | 73.8 | 68.7 | 85.5 | 76.8 | 73.1 | 73.1 | ||||||||
| 10. S. dysgalactiae | 74.9 | 74.9 | 69.0 | 62.8 | 77.5 | 64.6 | 66.9 | 69.9 | 66.4 | 67.6 | 67.1 | 72.4 | 86.9 | 72.0 | 65.3 | 73.1 | 65.7 | 73.1 | 71.5 | |||||||||
| 11. S. equi subsp. equi | 98.9 | 65.7 | 63.0 | 71.3 | 63.2 | 63.7 | 70.1 | 63.7 | 64.1 | 63.7 | 68.0 | 76.1 | 69.7 | 63.4 | 68.7 | 66.9 | 66.7 | 66.4 | ||||||||||
| 12. S. equi subsp. zooepidemicus | 65.7 | 63.0 | 71.3 | 63.0 | 64.1 | 69.7 | 64.1 | 64.8 | 64.1 | 68.3 | 77.0 | 69.9 | 62.5 | 69.0 | 66.2 | 66.7 | 66.4 | |||||||||||
| 13. S. equinus | 71.7 | 68.5 | 71.7 | 72.6 | 74.7 | 72.2 | 73.3 | 71.7 | 69.2 | 70.3 | 77.2 | 70.1 | 69.2 | 69.9 | 75.9 | 76.8 | ||||||||||||
| 14. S. gordonii | 64.1 | 81.1 | 80.5 | 70.3 | 79.8 | 80.9 | 80.2 | 66.7 | 63.9 | 73.3 | 85.7 | 68.7 | 72.9 | 73.3 | 73.1 | |||||||||||||
| 15. S. iniae | 63.9 | 66.0 | 74.0 | 66.4 | 65.7 | 66.9 | 77.0 | 79.3 | 72.4 | 65.3 | 70.3 | 71.0 | 70.8 | 70.1 | ||||||||||||||
| 16. S. intermedius | 75.6 | 67.4 | 76.1 | 77.2 | 74.3 | 68.5 | 63.2 | 68.5 | 77.0 | 62.3 | 70.1 | 70.6 | 70.1 | |||||||||||||||
| 17. S. mitis | 68.5 | 91.7 | 85.7 | 96.6 | 67.1 | 66.7 | 76.3 | 78.9 | 70.8 | 73.3 | 75.6 | 75.2 | ||||||||||||||||
| 18. S. mutans | 67.4 | 67.6 | 68.5 | 74.3 | 75.2 | 78.4 | 68.7 | 72.0 | 74.5 | 74.7 | 75.6 | |||||||||||||||||
| 19. S. oralis | 85.3 | 92.0 | 66.9 | 63.0 | 73.6 | 79.5 | 69.4 | 73.3 | 74.0 | 73.6 | ||||||||||||||||||
| 20. S. parasanguis | 85.7 | 65.3 | 64.8 | 70.8 | 79.3 | 68.5 | 72.2 | 70.6 | 70.8 | |||||||||||||||||||
| 21. S. pneumoniae | 66.4 | 66.7 | 75.4 | 78.2 | 71.3 | 73.6 | 75.2 | 74.9 | ||||||||||||||||||||
| 22. S. porcinus | 74.9 | 71.0 | 66.4 | 69.4 | 71.3 | 73.1 | 72.0 | |||||||||||||||||||||
| 23. S. pyogenes | 74.5 | 63.7 | 72.6 | 66.7 | 71.5 | 70.3 | ||||||||||||||||||||||
| 24. S. salivarius | 72.2 | 75.6 | 73.3 | 89.7 | 90.1 | |||||||||||||||||||||||
| 25. S. sanguis | 67.4 | 72.0 | 72.4 | 71.7 | ||||||||||||||||||||||||
| 26. S. sobrinus | 75.4 | 77.0 | 75.6 | |||||||||||||||||||||||||
| 27. S. suis | 72.0 | 72.9 | ||||||||||||||||||||||||||
| 28. S. thermophilus | 96.8 | |||||||||||||||||||||||||||
| 29. S. vestibularis | ||||||||||||||||||||||||||||
The main characteristics of the strains are listed in Table 1.
The strain numbers correspond to the strains identified by the numbers on the left.
Species identification of streptococcal clinical isolates by sequencing the sodAint gene.
The design of species-specific oligonucleotides enabling the amplification of a given target DNA constitutes an interesting molecular approach for the identification of bacterial species by PCR (12, 37). The two major problems inherent to these techniques are that (i) species-specific oligonucleotides often cannot be designed for closely related species, and (ii) the number of PCRs necessary for the identification of one isolate is proportional to the number of bacterial type species that should be considered. Taking into consideration the fact that cloning and sequencing techniques are increasingly used as routine techniques in medical microbiology laboratories, we propose the sequencing of the sodAint fragment as a molecular approach to the identification of streptococcal species. In this system, the identification of a clinical isolate is determined by the position of its sodAint fragment on the phylogenetic tree of the sodAint fragments of the type species (Fig. 2). To test the functionality of this approach, various typeable and nontypeable streptococcal isolates were identified by using the ID 32 Strep and/or the sodAint systems (Table 2). We also include in this study the sequences of sodAint fragments of known streptococcal species present in the databases. The results obtained with the sodAint system indicated that, as expected, the two group A and the three group B streptococcal strains studied were identified as S. pyogenes and S. agalactiae, respectively. When the streptococcal strains that were unambiguously identified to the species level by the phenotypic method (API identification percentage, ≥99.9), the sodAint-based identification method gave the same results (Table 2). Some discrepancies were observed for the strains the species of which were determined with an API identification percentage of less than 99.9. This was the case for NEM1164 and NEM1121, which were identified with the ID 32 Strep system as S. constellatus and S. salivarius, respectively, but which were identified with the sodAint system as S. anginosus and S. oralis, respectively (Table 2). The sequence of the sodAint fragment of NEM1164 displays 97 and 86% identity with the sequences of the type strains of S. anginosus and S. constellatus, respectively. The sequence of the sodAint fragment of NEM1121 displays 96.1 and 73.6% identity with the sequences of the type strains of S. oralis and S. anginosus, respectively. On the basis of these sequence distances, we considered the sodAint-based identification of NEM1164 (S. anginosus) and NEM1121 (S. oralis) to be more reliable than the ID 32 Strep system-based identification. Interestingly, certain strains (NEM1275, MG19, NEM666, NEM1126, and NEM895) were identified to the species level with the sodAint system but not with ID 32 Strep system (Table 2). Finally, it is important that the intraspecies divergence between the sodAint fragments may vary greatly depending upon the species considered, conceivably because of differences in sequence divergence rates. Consequently, it is not possible to delineate streptococcal species on the basis of the level of DNA homology. In the case of S. oralis, the levels of intraspecies divergence of the sodAint fragments can exceed those observed between the sodAint fragments of the type strains of S. oralis, S. mitis, and S. pneumoniae. These results might suggest that the species S. oralis, as defined, is genomically heterogeneous. Surprisingly, the sodAint fragments from unrelated S. pneumoniae strains were found to display the highest level of intraspecies sequence identity (>99%), which suggests that transformation is not a source of sequence heterogeneity for the sodA gene, at least in pneumococci.
Concluding remarks.
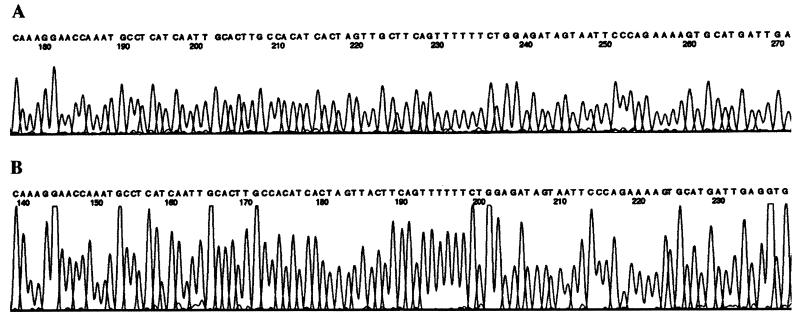
We have described a method that enables the reliable identification to the species level of groupable and nongroupable streptococci. It consists of (i) a PCR carried out with a single pair of degenerate oligonucleotides for amplification of a streptococcal sodAint fragment, (ii) molecular cloning of the resulting amplicon into an Escherichia coli vector, and (iii) sequencing of the insert on both DNA strands. Sequencing of a streptococcal sodAint fragment by using this procedure necessitates 72 h following receipt of the bacterial strain; however, we anticipate further simplification and/or automation of various steps of this method. For example, based on the observation that a single abundant PCR product was obtained by using the degenerate sod primers, whatever the species tested (Fig. 1), we successfully tried to sequence directly both strands of the amplified DNA with either of the degenerate PCR primers (Fig. 3 shows part of the results of that analysis). Removal of the cloning step greatly enhances the applicability of the procedure and reduces the delay required for bacterial identification to 24 h. This method might be useful in reference laboratories for the characterization strains that could not be assigned to a species on the basis of their conventional phenotypic reaction. It provides a convenient alternative to the DNA-DNA hybridization method, which constitutes the reference technique for the identification of strains to the species level. We are expanding the applicability of this approach by determining the nucleotide sequence of sodAint fragments from other species of streptococci as well as from other related genera such as Abiotrophia, Enterococcus, Gemella, Leuconostoc, and Pediococcus.
FIG. 3.
Electropherograms showing part of the nucleotide sequence of sodAint from S. porcinus. Sequence reactions were carried out with a pUC18ΩsodAint recombinant plasmid with the −21 dye primer sequencing kit (A) and sodAint PCR product with the degenerate oligonucleotide d2 and the dRhodamine dye terminator sequencing kit (B).
ACKNOWLEDGMENTS
We thank C. Bizet for the gift of streptococcal type strains (CIP); A. Buu-Hoï, L. Gutman, N. Fortineau, and O. Gaillot for gifts of clinical isolates; and E. Abachin for technical assistance.
This work was supported by the Institut Pasteur and by the University Paris V.
REFERENCES
- 1.Adnan S, Li N, Miura H, Hashimoto Y, Yamamoto H, Ezaki T. Covalently immobilized DNA plate for luminometric DNA-DNA hybridization to identify viridans streptococci in under 2 hours. FEMS Microbiol Lett. 1993;106:139–142. doi: 10.1111/j.1574-6968.1993.tb05949.x. [DOI] [PubMed] [Google Scholar]
- 2.Ahmet Z, Warren M, Houang E T. Species identification of members of the Streptococcus milleri group isolated from the vagina by ID 32 Strep system and differential phenotypic characteristics. J Clin Microbiol. 1995;33:1592–1595. doi: 10.1128/jcm.33.6.1592-1595.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beighton D, Hardie J M, Whiley A. A scheme for the identification of viridans streptococci. J Med Microbiol. 1991;35:367–372. doi: 10.1099/00222615-35-6-367. [DOI] [PubMed] [Google Scholar]
- 4.Bentley R W, Leigh J A, Collins M D. Intrageneric structure of Streptococcus based on comparative analysis of small-subunit rRNA sequences. Int J Syst Bacteriol. 1991;41:487–494. doi: 10.1099/00207713-41-4-487. [DOI] [PubMed] [Google Scholar]
- 5.Clermont D, Horaud T. Identification of chromosomal antibiotic resistance genes in Streptococcus anginosus (“S. milleri”) Antimicrob Agents Chemother. 1990;34:1685–1690. doi: 10.1128/aac.34.9.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Lamballerie X, Zandotti C, Vignoli C, Bollet C, de Micco P. A one-step microbial DNA extraction method using “Chelex 100” suitable for gene amplification. Res Microbiol. 1992;143:785–790. doi: 10.1016/0923-2508(92)90107-y. [DOI] [PubMed] [Google Scholar]
- 7.Farrow J A E, Collins M D. Taxonomic studies on streptococci of serological groups C, G, and L and possibly related taxa. Syst Appl Microbiol. 1984;5:483–493. [Google Scholar]
- 8.Farrow J A E, Kruze J, Phillips B A, Bramley A J, Collins M D. Taxonomic studies on Streptococcus bovis and Streptococcus equinus: description of Streptococcus alactolyticus sp. nov. Syst Appl Microbiol. 1984;5:467–482. [Google Scholar]
- 9.Felsenstein J. Confidence limits on phylogeny and approach using the boostrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 10.Flynn C E, Ruoff K L. Identification of Streptococcus milleri group isolates to the species level with a commercially available rapid test. J Clin Microbiol. 1995;33:2704–2706. doi: 10.1128/jcm.33.10.2704-2706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaillot, O., C. Poyart, P. Berche, and P. Trieu-Cuot. Molecular characterization and expression analysis of the superoxide dismutase gene from Streptococcus agalactiae. Gene, in press. [DOI] [PubMed]
- 12.Garnier F, Gerbaud G, Courvalin P, Galimand M. Identification of clinically relevant viridans group streptococci to the species level by PCR. J Clin Microbiol. 1997;35:2337–2341. doi: 10.1128/jcm.35.9.2337-2341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson C M, Caparon M G. Insertional inactivation of Streptococcus pyogenes sod suggests that prtF is regulated in response to a superoxide signal. J Bacteriol. 1996;178:4688–4695. doi: 10.1128/jb.178.15.4688-4695.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardie J M. Genus Streptococcus. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey’s manual of systematic bacteriology. Baltimore, Md: The Williams & Wilkins Co; 1986. pp. 1043–1071. [Google Scholar]
- 15.Hillman J D, Andrews S W, Painetr S, Stashenko P. Adaptative changes in a strain of Streptococcus mutans during colonization of the human oral cavity. Microb Ecol Health Dis. 1989;2:231–239. [Google Scholar]
- 16.Jacobs J A, Schot C S, Bunschoten A E, Schouls L M. Rapid species identification of “Streptococcus milleri” strains by line blot hybridization: identification of a distinct 16S rRNA population closely related to Streptococcus constellatus. J Clin Microbiol. 1996;34:1717–1721. doi: 10.1128/jcm.34.7.1717-1721.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawamura Y, Hou X-G, Sultana F, Miura H, Ezaki T. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int J Syst Bacteriol. 1995;45:406–408. doi: 10.1099/00207713-45-2-406. [DOI] [PubMed] [Google Scholar]
- 18.Kilian M, Mikkelsen L, Henrichsen J. Taxonomic studies of viridans streptococci: description of Streptococcus gordonii sp. nov. and emended descriptions of Streptococcus sanguis (White and Niven 1946), Streptococcus oralis (Bridge and Sneath 1982), and Streptococcus mitis (Andrewes and Horder 1906) Int J Syst Bacteriol. 1989;39:471–484. [Google Scholar]
- 19.Kilpper-Bälz R, Williams B L, Lutticken R, Schleifer K H. Relatedness of ‘Streptococcus miller’ with Streptococcus anginosus an Streptococcus constellatus. Syst Appl Microbiol. 1984;5:494–500. [Google Scholar]
- 20.Le Bouguénec C, Horaud T, Bieth G, Colimon R, Dauguet C. Translocation of antibiotic resistance markers of a plasmid-free Streptococcus pyogenes (group A) strain into different streptococcal hemolysin plasmids. Mol Gen Genet. 1984;194:377–387. doi: 10.1007/BF00425548. [DOI] [PubMed] [Google Scholar]
- 21.Nakayama K. Nucleotide sequence of Streptococcus mutans superoxide dismutase gene and isolation of insertion mutants. J Bacteriol. 1992;174:4928–4934. doi: 10.1128/jb.174.15.4928-4934.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker M W, Balke C C F. Crystal structure of manganese superoxide dismutase from Bacillus stearothermophilus at 2.4 Å resolution. J Mol Biol. 1988;199:649–661. doi: 10.1016/0022-2836(88)90308-7. [DOI] [PubMed] [Google Scholar]
- 23.Parker M W, Blake C C F. Iron- and manganese-containing superoxide dismutases can be distinguished by analysis of their primary structures. FEBS Lett. 1988;229:377–382. doi: 10.1016/0014-5793(88)81160-8. [DOI] [PubMed] [Google Scholar]
- 24.Poyart C, Berche P, Trieu-Cuot P. Characterization of superoxide dismutase genes from gram-positive bacteria by polymerase chain reaction using degenerate primers. FEMS Microbiol Lett. 1995;131:41–45. doi: 10.1016/0378-1097(95)00232-t. [DOI] [PubMed] [Google Scholar]
- 25.Rudney J D, Larson C J. Use of restriction fragment polymorphism analysis of rRNA genes to assign species to unknown clinical isolates of oral viridans streptococci. J Clin Microbiol. 1994;32:437–443. doi: 10.1128/jcm.32.2.437-443.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 27.Saruta K, Matsunaga T, Hoshina S, Kono M, Kitahara S, Kanemoto S, Sakai O, Machida K. Rapid identification of Streptococcus pneumoniae by PCR amplification of ribosomal DNA spacer region. FEMS Microbiol Lett. 1995;132:165–170. doi: 10.1111/j.1574-6968.1995.tb07827.x. [DOI] [PubMed] [Google Scholar]
- 28.Schleifer K H, Kilpper-Bälz R. Molecular and chemotaxonomic approaches to the classification of streptococci, enterococci, and lactococci: a review. Syst Appl Microbiol. 1987;10:1–19. [Google Scholar]
- 29.Schmidhuber S, Ludwig W, Schleifer K H. Construction of a DNA probe for the specific identification of Streptococcus oralis. J Clin Microbiol. 1988;26:1042–1044. doi: 10.1128/jcm.26.5.1042-1044.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tardif G, Sulavik M C, Jones G W, Clewell D B. Spontaneous switching of the sucrose-promoted colony phenotype in Streptococcus sanguis. Infect Immun. 1989;57:3945–3948. doi: 10.1128/iai.57.12.3945-3948.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whiley R A, Beighton D. Emended descriptions and recognition of Streptococcus constellatus, Streptococcus intermedius, and Streptococcus anginosus as distinct species. Int J Syst Bacteriol. 1991;41:1–5. doi: 10.1099/00207713-41-1-1. [DOI] [PubMed] [Google Scholar]
- 33.Whiley R A, Duke B, Hardie J M, Hall L M C. Heterogeneity among 16S-23S rRNA intergenic spacers of species within the ‘Streptococcus milleri group.’. Microbiology. 1995;141:1461–1467. doi: 10.1099/13500872-141-6-1461. [DOI] [PubMed] [Google Scholar]
- 34.Whiley R A, Fraser H Y, Douglas C W I, Hardie J M, Williams A M, Collins M D. Streptococcus parasanguis sp. nov., an atypical viridans Streptococcus from human clinical specimens. FEMS Microbiol Lett. 1990;68:115–122. doi: 10.1111/j.1574-6968.1990.tb04133.x. [DOI] [PubMed] [Google Scholar]
- 35.Whiley R A, Hardie J M. Streptococcus vestibularis sp. nov. from the human oral cavity. Int J Syst Bacteriol. 1988;38:335–339. doi: 10.1099/00207713-41-4-543. [DOI] [PubMed] [Google Scholar]
- 36.Whiley R A, Russell R R B, Hardie J M, Beighton D. Streptococcus downeii sp. nov. for strains previously described as Streptococcus mutans serotype H. Int J Syst Bacteriol. 1988;38:25–29. [Google Scholar]
- 37.Zolg J W, Philippi-Schulz S. The superoxide dismutase gene, a target for detection and identification of mycobacteria by PCR. J Clin Microbiol. 1994;32:2801–2812. doi: 10.1128/jcm.32.11.2801-2812.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]