Abstract
PCR was performed on DNA extracts derived from clinical serum samples submitted for human herpesvirus 6 (HHV-6) serological examination. To detect amplified HHV-6 products, a hybridization-based microtiter plate assay (PCR ELISA; Boehringer Mannheim) was used. The assay system was found to be rapid, specific, and sensitive. Approximately three copies of a plasmid-based HHV-6 sequence could be detected, and no cross amplification was observed with HHV-7 genomic DNA. There was no correlation found between HHV-6 DNA detection and serological status in clinical serum samples from individuals more than 2 years old. On the other hand, in serum samples from infants less than 2 years old, a high rate of detection of HHV-6 DNA was observed in those who lacked immunoglobulin G and M antibodies to HHV-6 (55%). In this regard, PCR of serum DNA extracts may be used as a sensitive indicator of active HHV-6 infection in infants prior to their seroconversion.
Primary infection with human herpesvirus 6 (HHV-6) causes exanthem subitum (ES), which is characterized by fever lasting approximately 3 days followed by a rash (3, 41). Seroprevalence among the population is greater than 90% (22), suggesting that HHV-6 is ubiquitous in the population. Seroconversion occurs early in life following the usually benign clinical course of ES (28, 33). However, more serious outcomes of HHV-6 infection have been known to occur (26), therefore illustrating the need for a rapid and sensitive diagnostic system. Currently the most common means of laboratory diagnosis of HHV-6 is by antigen detection from cultured specimens or antibody detection. PCR amplification of a variety of clinical samples, such as peripheral blood mononuclear cells (PBMC) or saliva (1, 8, 16), cerebrospinal fluid (42), urine (27), or tissue biopsy (32, 39) has shown variable detection rates of HHV-6 DNA. As HHV-6 establishes latency in various sites such as PBMC and salivary glands (20, 23, 32), the detection of viral DNA in these clinical samples provides no information to differentiate between active HHV-6 infection and latent or persistently shed virus. In this regard, plasma or serum samples are more suitable for the detection of HHV-6 DNA. In this report we describe the evaluation of a nonradioactive hybridization microtiter plate assay for detection of amplified DNA from clinical serum samples.
MATERIALS AND METHODS
Clinical serum samples.
One hundred and five serum samples submitted from various provinces of Canada for routine HHV-6 serological analysis were included in the present study. Stored serum samples (−20°C) were chosen retrospectively based solely upon their anti-HHV-6 immunoglobulin G (IgG) or IgM titer. All sera were numerically encoded by a different investigator and were then analyzed sequentially by number. The investigator analyzing specimens for HHV-6 DNA was unaware of the age and clinical history of the individual from whom the serum sample was taken. Serum stored for as long as 5 years was included; however, the majority of serum samples used in the present study were received within the past year. Twenty-four serum samples, including one pooled serum sample from apparently healthy adult donors, collected by the Canadian Red Cross, were included as a control. These 24 samples were submitted from donors without any apparent symptoms to the Laboratory Centre for Disease Control as control samples in a measles-rubella serosurvey.
Clinical histories including symptoms or condition associated with presentation to the physician, date of birth, and date of serum collection were available for each serum sample. For this study, data from samples without a symptom description were excluded.
Virus and cell culture.
The GS strain of HHV-6 was grown in HSB2 cells for a source of infected tissue culture supernatant by the method of Parker and Weber (29). HSB2 cells, a T-cell line (ATCC CCL 120.1), were cultured in Iscove’s modified Dulbecco’s medium supplemented with 10% fetal bovine serum and 2% penicillin-streptomycin (Gibco Laboratories, Burlington, Ontario, Canada).
Antibody detection.
IgG and IgM antibodies to HHV-6 were detected by an enzyme-linked immunosorbent assay (ELISA) by the method of Parker and Weber (29). In brief, HHV-6 antigen was prepared from virus-infected HSB2 cells. Infected cells were manually lysed, and the clarified lysate was used to coat microtiter plates (Immulon 2; Dynatech, Mississauga, Ontario, Canada). Control antigen was prepared from uninfected HSB2 cells in an identical manner. Following overnight incubation at room temperature to coat the wells, wells were blocked with 1% bovine serum albumin in phosphate-buffered saline (PBS).
For antibody testing, serum was diluted in PBS containing 1% bovine serum albumin and 0.05% Tween 20 (PBS-BT) and added to wells for a 1-h incubation at 37°C. For IgM determination, serum samples were pretreated with RF-Absorbant (Hoechst-Behring, Kanata, Ontario, Canada) to absorb interfering IgG and rheumatoid factor. The wells were then washed with PBS containing 0.15% Tween 20, alkaline phosphatase-conjugated goat anti-human IgG or IgM (Kirkegaard-Perry, Burlington, Ontario, Canada) diluted in PBS-BT was then added to each well, and the plate was incubated for 1 h at 37°C. The plate was then washed, and p-phenylnitrophosphate substrate (Sigma, St. Louis, Mo.) in 10% diethanolamine buffer was added. Absorbance of the wells was measured over the range of 405 to 650 nm after approximately 45 min of color development at room temperature.
The negative cutoff value was calculated as the absorbance reading equal to the mean absorbance of three values from negative-control sera. Sera were considered positive for IgM or IgG if the absorbance value was greater than 3 standard deviations (SD) above the negative cutoff value. Samples were considered indeterminate if the absorbance value fell between the negative cutoff value and 3 SD above the negative cutoff. Samples that were indeterminate were retested. In order to standardize all results, each test included a standard curve of diluted serum from an anti-HHV-6 IgM and IgG high responder.
Extraction of DNA.
DNA extraction from serum samples and tissue culture supernatant controls was performed by the methods detailed in Kramvis et al. (21), with some modifications. Serum (150 μl) was extracted with a proteinase K-sodium dodecyl sulfate (SDS) extraction mix (267 μg of proteinase K per ml, 1% SDS, 2.5 mM EDTA, 25 mM sodium acetate) by incubation at 70°C for 2 h. DNA was then purified by phenol-chloroform extraction and ethanol precipitation. The resulting DNA pellet was resuspended in 20 μl of sterile water and incubated at 95°C for 10 min in order to reduce or inactivate PCR inhibitory factors (12). DNA extracts were then allowed to cool and were amplified immediately. HHV-6-infected tissue culture supernatant served as a positive extraction control, while tissue culture supernatant from uninfected cells was used as a negative extraction control.
Amplification of HHV-6 DNA.
Serum DNA extracts were selectively amplified for HHV-6 DNA by PCR using sequence-specific primers for a 249-bp portion of the large-tegument protein gene of HHV-6 by the method of Collandre et al. (7): HHV6A 5′ GATCCGACGCCTACAAACAC 3′; HHV6B, 5′ TACCGACATCCTTGACATATTAC 3′. This primer set has previously been shown to be specific for HHV-6 and to not amplify DNA from other members of the herpesvirus family including cytomegalovirus (CMV), Epstein-Barr virus, varicella-zoster virus, and herpes simplex viruses 1 and 2 (7, 26, 34, 39). PCRs were performed in a final volume of 40 μl, which included 6.5 μl of DNA extract, 1 U of Taq DNA polymerase (Boehringer Mannheim, Laval, Quebec, Canada), 0.25 μM (each) primer, 200 μM digoxigenin (DIG)-deoxynucleoside triphosphate labelling mix, (Boehringer Mannheim), 10 mM Tris HCl (pH 8.3), 1.5 mM MgCl2, and 50 mM KCl. Thermocycling conditions were as follows: (i) an initial denaturation step of 2.5 min at 94°C; (ii) 35 cycles, with 1 cycle consisting of 30 s of denaturation at 94°C, 30 s of annealing at 62°C, and 50 s of extension at 72°C; and (iii) a final extension step of 5 min at 72°C. PCR was performed in a Perkin-Elmer GeneAmp 9600 thermocycler with 200-μl thin-walled reaction tubes. The resulting amplified products are labelled with DIG by incorporation of DIG-labelled dUTP nucleotides during the amplification procedure.
For a positive control, an 830-bp portion of the large-tegument protein gene was cloned in the pCRII TA cloning vector (Invitrogen, San Diego, Calif.). The 830-bp segment was amplified from DNA extracted from HHV-6-infected HSB2 cells, using primers A and C as described by Collandre et al. (7): HHV6A as above and HHV6C, 5′ CGGTGTCACACAGCATGAACTCTC 3′. HHV6C is upstream of the HHV6B sequence; therefore, the 249-bp target sequence forms a part of the 830-bp segment and can be amplified by HHV6A and HHV6B. For evaluation of assay sensitivity, serial dilutions of plasmid copies (HHV6AC) were used to spike serum samples previously shown to be consistently negative by the assay procedure. Spiked sera were extracted as described above, and the DNA was amplified by PCR.
To prevent possible PCR contamination and false-positive results, many precautions were taken. Each step of the assay procedure, including reagent premix preparation, DNA extraction, amplification, and detection, was performed in a different room. Reserved lab coats, pipettors, aerosol-barrier tips, tubes, and reagents were kept in each room, and fresh gloves were worn at all times with frequent changes. A face mask was also worn during addition of DNA to amplification reaction tubes. Negative controls included a DNA-minus control (H2O) and a non-HHV-6 DNA control. At least two negative controls were used for analysis of up to 10 clinical samples. To prevent false-negative results, amplification reaction premixes were prepared and aliquoted, and DNA extracts giving negative PCR results were spiked with approximately 100 HHV6AC plasmid copies to test for PCR inhibitors (8, 18, 24). All DNA extracts were amplified in duplicate or triplicate.
Detection of amplified products.
Amplified products were detected by a commercially available liquid hybridization-based microtiter plate assay (PCR ELISA; Boehringer Mannheim). The manufacturer’s recommendations were followed, with several steps changed for optimal sensitivity. A biotin end-labelled HHV-6 oligonucleotide probe, prepared according to the sequence given by Aubin et al. (4) (5′ GGCTGATTAGGATTAATAGGAGA 3′) was diluted to a final concentration of 8 pmol/ml to serve as a capture probe during hybridization. Thirty microliters of DIG-labelled amplicon was assayed per well, and hybridization occurred for 3 h at 37°C with shaking of the microtiter plate. Hybridized products which bound to streptavidin-coated microtiter plate wells via the biotin-labelled capture probe were then detected by standard ELISA procedures. An anti-DIG peroxidase-labelled conjugate antibody and peroxidase substrate permit a colorimetric reaction dependent upon the presence and amount of amplified product. All plates were measured for absorbance at 405 nm following a 30-min color development incubation at 37°C.
To compare the sensitivity of detection by the microtiter plate assay, amplified products were also electrophoretically separated on 4% NuSieve agarose (FMC Bioproducts, Guelph, Ontario, Canada) gels and stained with ethidium bromide.
Antigen capture ELISA.
Detection of HHV-6 antigen in serum was performed by use of a commercially available HHV-6 antigen capture ELISA kit (Advanced Biotechnologies Inc., Columbia, Md.). The ELISA kit permits direct detection of HHV-6 antigen in human serum through antigen capture by an HHV-6-specific monoclonal antibody. Serum samples, tissue culture control samples and the kit positive control were analyzed in duplicate, while the kit negative control was run in triplicate for each test. Captured antigen was detected by a peroxidase-conjugated anti-HHV-6 monospecific antibody, according to the manufacturer’s instructions. The cutoff value was the absorbance reading equal to twice the mean absorbance of three values from negative-control wells. Optical density (OD) ratios were then calculated by dividing the absorbance of each sample well by the cutoff value. OD ratios greater than or equal to 1.0 were considered positive, while those less than or equal to 0.75 were considered negative. Any OD ratios falling between the two values were considered indeterminate, and these samples were then reassayed.
Statistical analysis.
The Chi-square test was used to analyze the significance of differences in serology and DNA detection in the age groups.
RESULTS
Clinical samples.
One-hundred and forty-seven serum samples were chosen retrospectively based upon their HHV-6 serology and processed for DNA detection by PCR ELISA. Of these samples, 105 had reference to patient symptoms on their clinical history and so were included in the final data analysis. Fifty-six samples were HHV-6 IgG negative. Forty-nine samples were HHV-6 IgM indeterminate or positive. Sixty-seven of the patient clinical histories included a mention of fever, febrile episodes, or rash or roseola, while the other clinical histories mentioned the following (number of individuals shown in parentheses): spina bifida (1), hepatosplenomegaly (1), cellulitis (1), pancytopenia (1), pancreatitis (1), transplant (7), encephalitis (2), surgery (1), chronic fatigue (11), renal failure (1), lymphadenopathy (2), respiratory or gastrointestinal symptoms (3), malaria (1), weight loss (1), arthritis (1), lethargy (2), and nephrotic syndrome (1). Forty-four samples were from individuals less than 2 years old (mean, 11 months; median, 11 months; range, 2 months to 1 year 9 months), and 61 samples were from individuals older than 2 years (mean, 20 years 1.5 months; median, 14 years 9 months; range, 2 years 3 months to 50 years 7 months).
Specificity and sensitivity of PCR ELISA.
Amplification of DNA extracts resulted in a 249-bp band when detected by agarose gel electrophoresis and ethidium bromide staining (Fig. 1, lane 4). By ELISA detection, a sample was determined to be negative for HHV-6 DNA if the OD value was less than the background cutoff value. The background cutoff value was calculated as the mean of all negative-control OD values plus 3 SD of the mean. The background cutoff value was generally within the range of 0.010 to 0.050 OD units. A sample was considered positive for HHV-6 DNA if the OD value was greater than twice the background cutoff value, while OD values which fell between the background cutoff value and twice this value were considered indeterminate. All reagent control and kit control OD values were found to fall within the kit manufacturer’s guidelines.
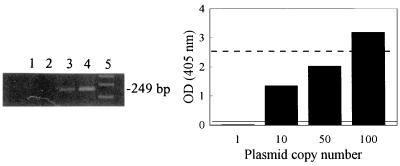
FIG. 1.
Relationship between agarose gel electrophoresis detection of HHV-6 amplicons and the corresponding OD of amplicons as obtained by PCR ELISA. Increasing numbers of HHV6AC plasmid copies were amplified by using the conditions described in Materials and Methods. (Left) Amplified products from a representative experiment were separated by agarose gel electrophoresis and visualized by ethidium bromide staining (lane 1, 1 plasmid copy; lane 2, 10 plasmid copies; lane 3, 50 plasmid copies; lane 4, 100 plasmid copies; lane 5, DNA size markers). The size of the amplified HHV-6 product is shown to the right of the gel (249 bp). (Right) Amplified products were also detected by PCR ELISA as described in Materials and Methods, and the OD values at 405 nm for the products are shown graphically. Each bar represents the mean OD value of replicate experiments. The ELISA negative and positive cutoff values were 0.029 and 0.058, respectively. The positive cutoff value is indicated as the solid line in the graph. The broken line represents the approximate visual cutoff of detection by gel electrophoresis.
Amplification of DNA extracts was performed in duplicate, and agreement among duplicate runs was obtained in all cases except six. In the cases where discrepant results were obtained between runs, the sample was reextracted and the DNA was amplified in duplicate again. All 6 reextracted samples gave concordant results. Ten samples which gave an indeterminate ELISA result were found to be negative upon repeated PCR, except for four samples which became positive. No PCR inhibitors were encountered following extraction, as all DNA-negative samples which were spiked with approximately 100 HHV6AC plasmid copies became positive for DNA following PCR ELISA.
The specificity of the primers used for HHV-6 was demonstrated by a lack of amplification of HHV-7 DNA (data not shown). When DNA extracted from HHV-7-infected HSB2 cells (a kind gift of Jodi Black, Centers for Disease Control and Prevention, Atlanta, Ga.) was amplified with the HHV-6 primer set used in the present study, no HHV-7 amplicon band was observed following gel electrophoresis. HHV-7 DNA was confirmed to be present in the extract by amplification with HHV-7-specific primers. DNA extracted from CMV (Ad169 and Davis strains)-infected MRC-5 cells was also observed to not be amplified by the HHV-6 primers used (data not shown).
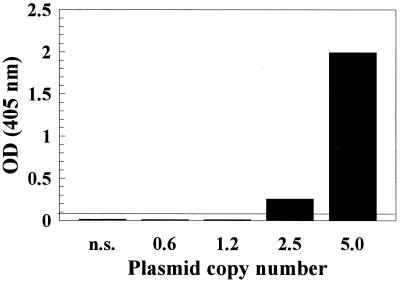
The sensitivity of DNA detection by PCR ELISA was approximately 30- to 50-fold greater than detection by electrophoresis and ethidium bromide staining. As observed in Fig. 1, the lower limit of visual detection by gel electrophoresis was between 50 and 100 plasmid copies, which was equivalent to between 2 and 3 OD units. The PCR ELISA, on the other hand, had a positive cutoff OD value of 0.058, therefore increasing detection sensitivity by 30- to 50-fold over detection by gel electrophoresis. The limit of detection following extraction, amplification, and detection by PCR ELISA was investigated by extraction and amplification of HHV-6-negative serum spiked with serial dilutions of HHV6AC plasmid copies. Experiments were run in duplicate, and the mean OD values for each plasmid dilution extract were plotted (Fig. 2). Detection down to approximately three plasmid copies (approximately 0.015 fg of HHV6AC plasmid) was observed, which is comparable or better than detection limits previously published for HHV-6 DNA in serum and PBMC (1, 8, 10, 13, 14, 31, 34, 38, 40). HHV-6 DNA was detected in 21% (5 of 24) of control sera from apparently healthy adults, which is similar to the percentage of healthy adults having HHV-6 antigen-positive sera (21 to 25%) (25).
FIG. 2.
PCR ELISA lower limit of detection of HHV-6 DNA extracted from serum. Serum negative for HHV-6 DNA was spiked with increasing numbers of HHV6AC plasmid. Following extraction procedures and amplification of extracted DNA, amplified products were detected by PCR ELISA. OD values of amplified DNA from each plasmid dilution extract are shown graphically and represent mean values from repeated experiments. The ELISA negative and positive cutoff OD values were 0.045 and 0.090, respectively. The positive cutoff value is indicated as the solid line in the graph. n.s., nonspiked serum.
DNA detection in clinical serum samples.
Serological and DNA detection data were compiled according to the age of the individual (Table 1) to illustrate differences in the two age groups. The majority of serum samples from infants less than 2 years old were found to lack HHV-6 IgG and IgM, while most older patients had evidence of seroconversion (P < 0.001). We observed significant differences in the rate of DNA detection in the two age groups, such that the rate of HHV-6 DNA detection was higher with the majority of infant samples (less than 2 years old) than with samples from older patients (P < 0.001). More importantly, within the infant serum group, we observed a high rate of DNA detection among samples lacking antibody to HHV-6. Specifically, 55% (16 of 29) of infant serum samples lacking IgG and IgM to HHV-6 were HHV-6 DNA positive. Of those samples lacking IgG, 63% (26 of 41) were DNA positive, while 55% (16 of 29) of IgM-negative samples were DNA positive.
TABLE 1.
Detection of HHV-6 DNA according to age and serological status of individualsa
| Age and sero- logical status | No. of samples (%)
|
||
|---|---|---|---|
| Total | DNA+ | DNA− | |
| <2 yrs (n = 44) | |||
| IgG+ | 3 (7) | 0 (0) | 3 (100) |
| IgG− | 41 (93) | 26 (63) | 15 (37) |
| IgM+ | 15 (34) | 10 (67) | 5 (33) |
| IgM− | 29 (66) | 16 (55) | 13 (45) |
| ≥2 yrs (n = 61) | |||
| IgG+ | 46 (75) | 12 (26) | 34 (74) |
| IgG− | 15 (25) | 4 (27) | 11 (73) |
| IgM+ | 47 (77) | 13 (28) | 34 (72) |
| IgM− | 14 (23) | 3 (21) | 11 (79) |
Symbols: +, positive; −, negative.
Seven individuals were noted to have symptoms of seizure, convulsion, meningitis, or encephalitis included on their clinical history. Two of these individuals were HHV-6 DNA negative (ages, 4 months and 37 years), while the remaining five were infants less than two years old, lacking IgG to HHV-6 but positive for HHV-6 DNA in their serum. Therefore, we observed an occurrence of 19% (5 of 26) of infants infected with HHV-6 experiencing neurological symptoms, which although in the range of previous reports (5, 13) may not necessarily constitute a causal role for HHV-6 in the neurological symptoms experienced by these infants.
Detection of HHV-6 antigen in serum.
The detection capability of the PCR ELISA for HHV-6 was compared to the detection of HHV-6 antigen in the same sample by use of a commercially available HHV-6 antigen capture ELISA kit. All kit control wells gave absorbance readings equivalent to those recommended by the manufacturer for proper quality control. As the HHV-6 antigen is inherently unstable in serum (25), relatively fresh serum samples which had been recently submitted to our laboratory for HHV-6 serological analysis were chosen. Of 27 samples tested, 22 (81%) had concordant results for antigen and DNA detection. Six samples were both DNA and antigen positive, while 16 samples were both DNA and antigen negative. Of the five remaining samples, three of those in which HHV-6 antigen was detected were DNA negative, while two samples had detectable HHV-6 DNA but no antigen.
DISCUSSION
Current methods for routine diagnosis of HHV-6 infection include detection by cultivation of PBMC or by detection of a significant increase in antibody titer to HHV-6. Both methods require at least 1 week for detection, and in the case of serological analysis, a second blood sample must be taken. The presence of a high IgM titer is also construed as evidence of a recent infection or reactivation; however, 4 to 5% of healthy adults have IgM antibodies to HHV-6 at any one time (13, 36) and IgM antibody can persist for months following infection (29). Also, there is the possibility that anti-HHV-6 IgM increases during Epstein-Barr virus or CMV infection or reactivation (11, 15).
Detection of viral DNA by PCR offers some hope for a rapid, specific, and sensitive approach to diagnosing active HHV-6 infection. The use of various cellular clinical specimens for PCR increases the risk of detecting latent or persistently shed virus, particularly if the clinical specimens are PBMC or saliva samples. In these cases, the presence of active HHV-6 infection is difficult to interpret without quantitative analysis (6, 35). Several studies have investigated the detection of HHV-6 DNA in serum (14, 34), and it is apparent that serum or plasma offers a suitable alternative to cellular samples for PCR diagnosis of a current HHV-6 infection. The present study illustrates that the PCR ELISA method allows for specific and sensitive detection of HHV-6 DNA in clinical serum samples.
In our study, 93% of serum samples from infants less than 2 years old were found to lack IgG to HHV-6; however, approximately one-third of these infants had evidence of anti-HHV-6 IgM. This response pattern is indicative of primary infection with HHV-6 in these infants. At birth, infants carry maternally derived anti-HHV-6 IgG up until approximately 6 months of age, after which infection with HHV-6 usually occurs (14, 30, 33). Antibody levels then increase over time, such that HHV-6 seroprevalence ranges up to 100% in children and adults (22). This prevalence was also observed in our study, with 75% of individuals older than 2 years having anti-HHV-6 IgG.
Our study showed that with most individuals there is no correlation between the presence of anti-HHV-6 antibody and HHV-6 DNA in the serum. This finding is in agreement with previous studies which investigated antibody titer and the presence of DNA in PBMC (8, 16, 31, 40). More significantly, however, we found that this lack of correlation appears to be age dependent. Approximately 70% of all samples from individuals older than 2 years were DNA negative regardless of their serological status. On the other hand, approximately 60% of all samples tested from infants less than two years old were DNA positive which corresponded to a lack of IgG to HHV-6 in these samples. This difference in DNA detection between the two age groups was found to be significant (P < 0.001).
In this regard, PCR ELISA appears highly suited for diagnosis of ES in infants. PCR is better suited than antibody detection in this case, as several weeks are required for paired samples to show IgG antibody increases and IgM is often not observed until a week or more has passed following symptom onset in infants (19). This window of time presents problems for diagnosis of HHV-6 by currently available standard serological tests. Our observation of a high rate of detection of HHV-6 DNA in infant sera lacking antibody to HHV-6 suggests that PCR ELISA has great potential value for the early detection of infection during this critical window period. PCR ELISA is also ideal for analysis of large sample numbers, it is amenable for analysis of most fluid or tissue samples, it is suitable for automation, and results can be achieved within a day and a half. Most importantly though, observation of HHV-6 DNA in serum provides an accurate diagnosis of active HHV-6 infection (34), while its presence in PBMC or saliva samples may indicate only latent or persistently shed virus (1). Serum is also the most common type of specimen collected from patients, and only a small volume is required for DNA extraction.
The detection of HHV-6 DNA in clinical serum samples and tissue culture controls by PCR ELISA was compared to the detection of HHV-6 antigen in the same samples by use of a commercially available HHV-6 antigen capture ELISA (Advanced Biotechnologies, Inc.). We found an 81% concordance between the two detection procedures which was equivalent to the 75% concordance between antigen detection in sera and specimen cultivation assays, as described in the antigen capture kit insert. Therefore, PCR ELISA was able to sensitively differentiate the presence of HHV-6 DNA in serum at a rate comparable to the rate of detection of viral antigen in the same serum sample. However, PCR ELISA may have certain advantages over the detection of HHV-6 antigen, including the increased stability of sample DNA over sample antigen (25), and the potential capability to differentiate HHV-6 strains using strain-specific primer or probe sequences (4).
The low incidence of DNA-positive samples from individuals older than 2 years suggests that analysis by PCR may not be as instructive as culture or serology for diagnosis of reinfection or reactivation. We may observe a higher rate of DNA detection in the infant samples due to infants possibly suffering a higher viral load during primary infection. Another possible reason for our observing HHV-6 DNA in infant sera was the lack of antibody to HHV-6 in these samples. Ninety-three percent of all infant samples lacked anti-HHV-6 IgG, and only one-third had anti-HHV-6 IgM. It has been shown that a direct relationship exists between the disappearance of HHV-6 from the blood and the presence of increasing antibody levels (2, 37). Therefore, free virus in serum may be more difficult to detect in seroconverted children and adults than in infants undergoing primary infection following loss of protective maternal antibodies.
We observed that 21% of control sera from apparently healthy adults were HHV-6 DNA positive. Other groups (9, 14, 34), have not observed amplifiable HHV-6 DNA in sera from healthy controls. It has been shown in numerous studies that HHV-6 prevalence in peripheral blood samples from healthy donors ranges widely, up to 90% (1, 9, 31, 40) and that a positive result is dependent upon the number of cells tested (8, 10, 16). It is also known that a high HHV-6 load can persist in the blood of an apparently healthy individual (6) and that up to 25% of healthy donors have HHV-6 antigen-positive sera (25). Therefore, the differences encountered with the present study may result from different rates of active HHV-6 infection within the population, differences in the amount of DNA tested, or differences in sensitivity among PCR assays. Our detection limit of approximately three plasmid copies by PCR ELISA may have permitted a more sensitive evaluation of the prevalence of HHV-6 in apparently healthy, asymptomatic adult donors.
HHV-6 has been associated with severe infection in infants resulting in convulsive seizures or encephalitis (13, 26, 30). In cases such as these, rapid and accurate diagnosis of HHV-6 by PCR may be important for implementing possible treatment options such as foscarnet or ganciclovir to which HHV-6 is susceptible (17). In this manner, detection of HHV-6 DNA in serum by PCR ELISA may be an effective tool for diagnosis of HHV-6 infection in infants.
ACKNOWLEDGMENTS
We acknowledge the DNA Core Facility, Laboratory Centre for Disease Control, Ottawa, Ontario, Canada for primer synthesis.
C. Osiowy is supported by a Visiting Fellowship in Canadian Government Laboratories.
REFERENCES
- 1.Aberle S W, Mandl C W, Kunz C, Popow-Kraupp T. Presence of human herpesvirus 6 variants A and B in saliva and peripheral blood mononuclear cells of healthy adults. J Clin Microbiol. 1996;34:3223–3225. doi: 10.1128/jcm.34.12.3223-3225.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asano Y, Nakashima T, Yoshikawa T, Suga S, Yazaki T. Severity of human herpesvirus-6 viremia and clinical findings in infants with exanthem subitum. J Pediatr. 1991;118:891–895. doi: 10.1016/s0022-3476(05)82200-0. [DOI] [PubMed] [Google Scholar]
- 3.Asano Y, Yoshikawa T, Suga S, Kobayashi I, Nakashima T, Yazaki T, Kajita Y, Ozaki T. Clinical features of infants with primary human herpesvirus 6 infection (exanthem subitum, roseola infantum) Pediatrics. 1994;93:104–108. [PubMed] [Google Scholar]
- 4.Aubin J, Agut H, Collandre H, Yamanishi K, Chandran B, Montagnier L, Huraux J. Antigenic and genetic differentiation of the two putative types of human herpesvirus 6. J Virol Methods. 1993;41:223–234. doi: 10.1016/0166-0934(93)90129-f. [DOI] [PubMed] [Google Scholar]
- 5.Caserta M T, Hall C B, Schnabel K, McIntyre K, Long C, Costanzo M, Dewhurst S, Insel R, Epstein L G. Neuroinvasion and persistence of human herpesvirus 6 in children. J Infect Dis. 1994;170:1586–1589. doi: 10.1093/infdis/170.6.1586. [DOI] [PubMed] [Google Scholar]
- 6.Clark D A, Ait-Khaled M, Wheeler A C, Kidd I M, McLaughlin J E, Johnson M A, Griffiths P D, Emery V C. Quantification of human herpesvirus 6 in immunocompetent persons and post-mortem tissues from AIDS patients by PCR. J Gen Virol. 1996;77:2271–2275. doi: 10.1099/0022-1317-77-9-2271. [DOI] [PubMed] [Google Scholar]
- 7.Collandre H, Aubin J, Agut H, Bechet J, Montagnier L. Detection of HHV-6 by the polymerase chain reaction. J Virol Methods. 1991;31:171–180. doi: 10.1016/0166-0934(91)90155-s. [DOI] [PubMed] [Google Scholar]
- 8.Cone R W, Huang M W, Ashley R, Corey L. Human herpesvirus 6 DNA in peripheral blood cells and saliva from immunocompetent individuals. J Clin Microbiol. 1993;31:1262–1267. doi: 10.1128/jcm.31.5.1262-1267.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuende J I, Ruiz J, Civeira M P, Prieto J. High prevalence of HHV-6 DNA in peripheral blood mononuclear cells of healthy individuals detected by nested-PCR. J Med Virol. 1994;43:115–118. doi: 10.1002/jmv.1890430203. [DOI] [PubMed] [Google Scholar]
- 10.DiLuca D, Dolcetti R, Mirandola P, DeRe V, Secchiero P, Carbone A, Boiocchi M, Cassai E. Human herpesvirus 6: a survey of presence and variant distribution in normal peripheral lymphocytes and lymphoproliferative disorders. J Infect Dis. 1994;170:211–215. doi: 10.1093/infdis/170.1.211. [DOI] [PubMed] [Google Scholar]
- 11.Fox J D, Ward P, Briggs M, Irving W, Stammers T G, Tedder R S. Production of IgM antibody to HHV6 in reactivation and primary infection. Epidemiol Infect. 1990;104:289–296. doi: 10.1017/s095026880005946x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frickhofen N, Young N S. A rapid method of sample preparation for detection of DNA viruses in human serum by polymerase chain reaction. J Virol Methods. 1991;35:65–72. doi: 10.1016/0166-0934(91)90086-f. [DOI] [PubMed] [Google Scholar]
- 13.Hall C B, Long C E, Schnabel K C, Caserta M T, McIntyre K M, Costanzo M A, Knott A, Dewhurst S, Insel R A, Epstein L G. Human herpesvirus-6 infection in children. A prospective study of complications and reactivation. N Engl J Med. 1994;331:432–438. doi: 10.1056/NEJM199408183310703. [DOI] [PubMed] [Google Scholar]
- 14.Huang L, Kuo P, Lee C, Chen J, Liu M, Yang C. Detection of human herpesvirus-6 DNA by polymerase chain reaction in serum or plasma. J Med Virol. 1992;38:7–10. doi: 10.1002/jmv.1890380103. [DOI] [PubMed] [Google Scholar]
- 15.Irving W L, Cunningham A L. Serological diagnosis of infection with human herpesvirus type 6. Br Med J. 1990;300:156–159. doi: 10.1136/bmj.300.6718.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarrett R F, Clark D A, Josephs S F, Onions D E. Detection of human herpesvirus-6 DNA in peripheral blood and saliva. J Med Virol. 1990;32:73–76. doi: 10.1002/jmv.1890320113. [DOI] [PubMed] [Google Scholar]
- 17.Jones C A, Isaacs D. Human herpesvirus-6 infections. Arch Dis Child. 1996;74:98–100. doi: 10.1136/adc.74.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimberlin D W, Lakeman F D, Arvin A M, Prober C G, Corey L, Powell D A, Burchett S K, Jacobs R F, Starr S E, Whitley R J the National Institute of Allergy and Infectious Diseases Collaborative Antiviral Study Group. Application of the polymerase chain reaction to the diagnosis and management of neonatal herpes simplex virus disease. J Infect Dis. 1996;174:1162–1167. doi: 10.1093/infdis/174.6.1162. [DOI] [PubMed] [Google Scholar]
- 19.Kondo K, Hayakawa Y, Mori H, Sato S, Kondo T, Takahashi K, Minamishima Y, Takahashi M, Yamanishi K. Detection by polymerase chain reaction amplification of human herpesvirus 6 DNA in peripheral blood of patients with exanthem subitum. J Clin Microbiol. 1990;28:970–974. doi: 10.1128/jcm.28.5.970-974.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo K, Kondo T, Okuno T, Takahashi M, Yamanishi K. Latent human herpesvirus 6 infection of human monocytes/macrophages. J Gen Virol. 1991;72:1401–1408. doi: 10.1099/0022-1317-72-6-1401. [DOI] [PubMed] [Google Scholar]
- 21.Kramvis A, Bukofzer S, Kew M C. Comparison of hepatitis B virus DNA extractions from serum by the QIAamp blood kit, GeneReleaser, and the phenol-chloroform method. J Clin Microbiol. 1996;34:2731–2733. doi: 10.1128/jcm.34.11.2731-2733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levy J A, Ferro F, Greenspan D, Lennette E T. Frequent isolation of HHV-6 from saliva and high seroprevalence of the virus in the population. Lancet. 1990;335:1047–1050. doi: 10.1016/0140-6736(90)92628-u. [DOI] [PubMed] [Google Scholar]
- 23.Luppi M, Marasca R, Barozzi P, Ferrari S, Ceccherini-Nelli L, Batoni G, Merelli E, Torelli G. Three cases of human herpesvirus-6 latent infection: integration of viral genome in peripheral blood mononuclear cell DNA. J Med Virol. 1993;40:44–52. doi: 10.1002/jmv.1890400110. [DOI] [PubMed] [Google Scholar]
- 24.Lyall E G H, Cubie H A. Human herpesvirus-6 DNA in the saliva of paediatric oncology patients and controls. J Med Virol. 1995;47:317–322. doi: 10.1002/jmv.1890470405. [DOI] [PubMed] [Google Scholar]
- 25.Marsh S, Kaplan M, Asano Y, Hoekzema D, Komaroff A L, Whitman J E, Jr, Ablashi D V. Development and application of HHV-6 antigen capture assay for the detection of HHV-6 infections. J Virol Methods. 1996;61:103–112. doi: 10.1016/0166-0934(96)02075-7. [DOI] [PubMed] [Google Scholar]
- 26.McCullers J A, Lakeman F D, Whitley R J. Human herpesvirus 6 is associated with focal encephalitis. Clin Infect Dis. 1995;21:571–576. doi: 10.1093/clinids/21.3.571. [DOI] [PubMed] [Google Scholar]
- 27.McElhinney L M, Cooper R J, Morris D J. Multiplex polymerase chain reaction for human herpesvirus-6, human cytomegalovirus, and human β-globin DNA. J Virol Methods. 1995;53:223–233. doi: 10.1016/0166-0934(95)00019-q. [DOI] [PubMed] [Google Scholar]
- 28.Okuno T, Takahashi K, Balachandra K, Shiraki K, Yamanishi K, Takahashi M, Baba K. Seroepidemiology of human herpesvirus 6 infection in normal children and adults. J Clin Microbiol. 1989;27:651–653. doi: 10.1128/jcm.27.4.651-653.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parker C A, Weber J M. An enzyme-linked immunosorbent assay for the detection of IgG and IgM antibodies to human herpesvirus type 6. J Virol Methods. 1993;41:265–276. doi: 10.1016/0166-0934(93)90017-l. [DOI] [PubMed] [Google Scholar]
- 30.Pruksananonda P, Hall C B, Insel R A, McIntyre K, Pellett P E, Long C E, Schnabel K C, Pincus P H, Stamey F R, Dambaugh T R, Stewart J A. Primary human herpesvirus 6 infection in young children. N Engl J Med. 1992;326:1445–1450. doi: 10.1056/NEJM199205283262201. [DOI] [PubMed] [Google Scholar]
- 31.Ranger-Rogez S, Vidal E, Labrousse F, Riche A, Vidal J, Collineau M, Liozon F, Denis F. Large-scale study suggests no direct link between human herpesvirus-6 and primary Sjogren’s syndrome. J Med Virol. 1995;47:198–203. doi: 10.1002/jmv.1890470303. [DOI] [PubMed] [Google Scholar]
- 32.Sada E, Yasukawa M, Ito C, Takeda A, Shiosaka T, Tanioka H, Fujita S. Detection of human herpesvirus 6 and human herpesvirus 7 in the submandibular gland, parotid gland, and lip salivary gland by PCR. J Clin Microbiol. 1996;34:2320–2321. doi: 10.1128/jcm.34.9.2320-2321.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saxinger C, Polesky H, Eby N, Grufferman S, Murphy R, Tegtmeir G, Parekh V, Memon S, Hung C. Antibody reactivity with HBLV (HHV-6) in U.S. populations. J Virol Methods. 1988;21:199–208. doi: 10.1016/0166-0934(88)90066-3. [DOI] [PubMed] [Google Scholar]
- 34.Secchiero P, Carrigan D R, Asano Y, Benedetti L, Crowley R W, Komaroff A L, Gallo R C, Lusso P. Detection of human herpesvirus 6 in plasma of children with primary infection and immunosuppressed patients by polymerase chain reaction. J Infect Dis. 1995;171:273–280. doi: 10.1093/infdis/171.2.273. [DOI] [PubMed] [Google Scholar]
- 35.Secchiero P, Zella D, Crowley R W, Gallo R C, Lusso P. Quantitative PCR for human herpesvirus 6 and 7. J Clin Microbiol. 1995;33:2124–2130. doi: 10.1128/jcm.33.8.2124-2130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suga S, Yoshikawa T, Asano Y, Nakashima T, Yazaki T, Fukuda M, Kojima S, Matsuyama T, Ono Y, Oshima S. IgM neutralizing antibody responses to human herpesvirus-6 in patients with exanthem subitum or organ transplantation. Microbiol Immunol. 1992;36:495–506. doi: 10.1111/j.1348-0421.1992.tb02047.x. [DOI] [PubMed] [Google Scholar]
- 37.Suga S, Yazaki T, Kajita Y, Ozaki T, Asano Y. Detection of human herpesvirus 6 DNAs in samples from several body sites of patients with exanthem subitum and their mothers by polymerase chain reaction assay. J Med Virol. 1995;46:52–55. doi: 10.1002/jmv.1890460112. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka-Taya K, Kondo T, Mukai T, Miyoshi H, Yamamoto Y, Okada S, Yamanishi K. Seroepidemiological study of human herpesvirus-6 and -7 in children of different ages and detection of these two viruses in throat swabs by polymerase chain reaction. J Med Virol. 1996;48:88–94. doi: 10.1002/(SICI)1096-9071(199601)48:1<88::AID-JMV14>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 39.Wang H, Chen M, Berneman Z N, Delgado G, DiPaolo J A. Detection of human herpesvirus-6 in paraffin-embedded tissue of cervical cancer by polymerase chain reaction. J Virol Methods. 1994;47:297–305. doi: 10.1016/0166-0934(94)90026-4. [DOI] [PubMed] [Google Scholar]
- 40.Wilborn F, Schmidt C A, Zimmerman R, Brinkmann V, Neipel F, Siegert W. Detection of herpesvirus type 6 by polymerase chain reaction in blood donors: random tests and prospective longitudinal studies. Br J Haematol. 1994;88:187–192. doi: 10.1111/j.1365-2141.1994.tb04995.x. [DOI] [PubMed] [Google Scholar]
- 41.Yamanishi K, Okuno T, Shiraki K, Takahashi M, Kondo T, Asano Y, Kurata T. Identification of human herpesvirus-6 as a casual agent for exanthem subitum. Lancet. 1988;8:1065–1067. doi: 10.1016/s0140-6736(88)91893-4. [DOI] [PubMed] [Google Scholar]
- 42.Yoshikawa T, Nakashima T, Suga S, Asano Y, Yazaki T, Kimura H, Morishima T, Kondo K, Yamanishi K. Human herpesvirus-6 DNA in cerebrospinal fluid of a child with exanthem subitum and meningoencephalitis. Pediatrics. 1992;89:888–890. [PubMed] [Google Scholar]