Abstract
A nested PCR method was developed for the detection of Coxiella burnetii in human serum samples. Two pairs of oligonucleotide primers were designed to amplify a 438-bp fragment of the com1 gene encoding a 27-kDa outer membrane protein of C. burnetii. The primers amplified the predicted fragments of 21 various strains of C. burnetii but did not react with DNA samples from other microorganisms. The 438-bp amplification products could be digested with restriction enzymes SspI and SalI. The utility of the nested PCR was evaluated by testing human serum samples. The com1 gene fragment was amplified from 135 (87%) of 155 indirect immunofluorescence test (IF)-positive serum samples and from 11 (11%) of 100 IF-negative serum samples. The nested PCR with primers targeted to the com1 gene appeared to be a sensitive, specific, and useful method for the detection of C. burnetii in serum samples.
Coxiella burnetii is the causative agent of acute Q fever and chronic endocarditis in humans (1). Acute Q fever is a flu-like illness which is self-limiting and which is easily treated with antibiotics when an appropriate diagnosis is made. Chronic Q fever is a severe disease that requires prolonged antibiotic therapy, because the infection can result in endocarditis (14, 18, 24) or granulomatous hepatitis (28). Rapid diagnosis of the disease is very important, because appropriate antibiotic treatment may lead to a better prognosis for individuals suffering from Q fever.
Routine diagnosis of Q fever is usually established by serological tests, since isolation of C. burnetii from patients is time-consuming, difficult, and hazardous. Serological methods, including the indirect immunofluorescence test (IF) (3, 6, 16), complement fixation test (5, 22, 23), enzyme-linked immunosorbent assay (21, 25, 26), and high-density particle agglutination test (17), can be used to detect antibodies to C. burnetii antigens. However, these serological tests have some limitations. Antibodies cannot be detected during the early stage of the infection, and it is difficult to discriminate between current and past infection by a test with a single serum sample, because antibodies often persist after the organisms disappear from the blood. Thus, serological tests offer only a retrospective diagnosis and are useless for the treatment of the afflicted patients.
Recently, PCR has become a useful tool for the detection of C. burnetii in clinical samples (10, 27, 29, 30, 31). The PCR appeared to be a very sensitive method for the laboratory diagnosis of Coxiella infection, able to detect DNA sequences in very small samples. Most recently, we demonstrated that the com1 gene encoding a 27-kDa outer membrane protein (OMP) was highly conserved among 21 strains of C. burnetii from a variety of clinical and geographical sources (32). The com1 gene is the genetic target for the detection of C. burnetii in clinical samples.
In the present study, we have developed a useful nested PCR assay based on the com1 gene sequence for the detection of C. burnetii in human serum samples.
MATERIALS AND METHODS
Microorganisms.
The microorganisms used in the study included 21 isolates of C. burnetii (strains Nine Mile VR 615, California 76 VR 614, Bangui VR 730, Ohio 314 VR 542, Henzerling VR 145, Priscilla, MAN, ME, GQ212, SQ217, and KoQ229 and 10 Japanese isolates) and 14 other bacterial isolates (Bordetella bronchiseptica GIFU 1127, Chlamydia pneumoniae TW183, Chlamydia psittaci GCP-1, Chlamydia trachomatis E, Escherichia coli C600, Haemophilus influenzae GIFU 3191, Klebsiella pneumoniae GIFU 2926, Legionella pneumophila SL94-1, L. pneumophila SL94-2, Mycoplasma pneumoniae, Oriertia tsutsugamushi Karp, O. tsutsugamushi Kato, O. tsutsugamushi Gilliam, and Streptococcus pneumoniae GIFU 8766.). The isolates of C. burnetii were propagated in Buffalo green monkey (BGM) cell cultures as described elsewhere (9).
Sera.
A total of 255 human serum samples were used in this study (155 IF-positive serum samples and 100 IF-negative serum samples) were selected from among 3,000 samples collected from 1,740 patients between September and December 1995. The patients were from the Gifu University Medical Faculty Hospital, where sera are randomly tested for antibodies to C. burnetii by IF (17). In addition, 50 serum samples from patients with pneumonia of viral or bacterial origin (influenza virus, parainfluenza virus, respiratory syncytial virus, C. psittaci, L. pneumophila, or M. pneumoniae) served as negative controls for the PCR.
DNA extraction.
DNA was extracted from the C. burnetii isolates as described previously (8). Briefly, the purified organisms from BGM cell cultures were suspended in TNE buffer (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA) and digested with proteinase K in the presence of 0.1% sodium dodecyl sulfate at 55°C for 60 min. DNA was extracted with phenol, phenol-chloroform, and chloroform; this was followed by ethanol precipitation. Dried under vacuum, the DNA was resuspended in TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). The DNA concentration and purity were determined by measuring the optical density at both 260 and 280 nm with a DNA calculator (GeneQuant II; Pharmacia Biotech), and the DNA was kept at −20°C.
Preparation of samples for PCR.
The serum samples used for PCR were prepared as described previously (10). Ten microliters of each serum sample was mixed with 40 μl of sample buffer (1% Nonidet P-40, 1% Tween 20, 10 mM Tris-HCl [pH 8.0]), the mixture was boiled for 10 min and then centrifuged at 12,000 × g for 5 min, and the supernatant was directly used for the PCR analysis.
Nucleotide primers.
All oligonucleotide primers were obtained from a commercial source (Rikaken Co., Ltd., Nagoya, Japan). The first primer system included primers Q3-Q5 and Q4-Q6, which were designed from the nucleotide sequence of the htpB gene encoding a 62-kDa protein and which were used to specifically amplify 501- and 325-bp fragments (10). The second primer system, including primers OMP1 (5′-AGT AGA AGC ATC CCA AGC ATT G-3′), OMP2 (5′-TGC CTG CTA GCT GTA ACG ATT G-3′), OMP3 (5′-GAA GCG CAA CAA GAA GAA CAC-3′), and OMP4 (5′-TTG GAA GTT ATC ACG CAG TTG-3′), was designed from the nucleotide sequence of the com1 gene encoding a 27-kDa OMP and was used to specifically amplify 501- and 438-bp fragments (7). These primers were designed from a conserved region of the com1 gene of C. burnetii on the basis of the gene sequences of 21 strains (32). The sequence specificities of these primers were checked by using the sequences in the GenBank database, and no homology with the sequences of other viral or bacterial organisms was detected by a search with the BLAST program.
The nested PCR was performed with serial 10-fold dilutions of total DNA (from 500 ng to 0.5 fg) extracted from the Nine Mile strain of C. burnetii to determine the minimum level of DNA detectable by the assay.
DNA amplification.
Amplification programs for the primers Q3-Q5 and Q4-Q6 were described previously (10). For the nested PCR with primers OMP1-OMP2 and OMP3-OMP4, the first amplification was performed in a total volume of 50 μl containing 5 μl of DNA sample, 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 200 μM (each) dATP, dCTP, dGTP, and dTTP, 0.5 μM primer OMP1, 0.5 μM primer OMP2, and 2 U of Taq DNA polymerase (Takara Shuzo, Co., Ltd., Shiga, Japan). The mixtures were overlaid with 2 drops of mineral oil. PCR was performed at 94°C for 3 min and then for 36 cycles of 94°C for 1 min, 54°C for 1 min, and 72°C for 1 min in a DNA thermal cycler (Perkin-Elmer GeneAmp PCR system 9600; Takara Biomedicals, Kyoto, Japan). In the second amplification, the reaction mixture and conditions were the same as those in the first amplification except for the primers and DNA templates. Primers OMP3 and OMP4 were used at 0.5 μM each, and 1 μl of the first amplification product was used as the DNA template. A positive control with 5 pg of C. burnetii DNA as the template and a negative control without DNA template were included in each PCR run.
Detection of PCR products.
The PCR-amplified products were examined by electrophoresis in a 1.5% agarose gel, stained with ethidium bromide (0.5 μg/ml), visualized under UV illumination (TM-20; UVP, Inc.) at 320 nm, and photographed.
Restriction endonuclease digestion.
The products were digested with restriction enzymes known to cut within the target sequence to confirm the identities of the amplified products. The 438-bp amplification products were digested with the restriction enzymes SspI and SalI. One SspI site and one SalI site were present in the amplified region of the com1 gene sequence of C. burnetii. These were compared with the amplification products of reference strains and human serum samples digested with SspI and SalI. The restriction products were also examined by electrophoresis and UV illumination for photography as described above.
RESULTS
Specificity of the nested PCR.
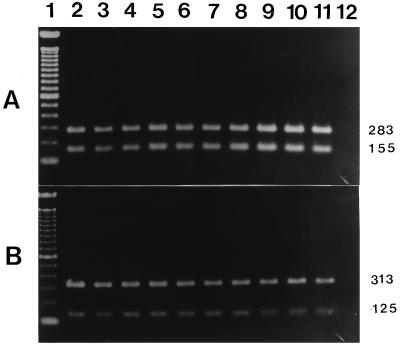
The primers OMP1-OMP2 and OMP3-OMP4 amplified the predicted products of the 501-bp DNA in the first amplification and the 438-bp DNA in the second amplification of PCR with DNA templates from all 21 of the isolates of C. burnetii used. No products were amplified when the DNAs from the 14 other microorganisms and negative controls were used. The specificities of the newly synthesized primers OMP1-OMP2 and OMP3-OMP4 were further demonstrated by digesting the amplified products from a reference strain of C. burnetii with the restriction enzymes SspI and SalI. Digestion of the first PCR products of 501 bp of DNA with SspI and SalI yielded 318- and 183-bp and 348- and 153-bp fragments, respectively. Digestion of the second PCR products of 438 bp of DNA with SspI and SalI yielded 283- and 155-bp and 313- and 125-bp fragments, respectively (Fig. 1).
FIG. 1.
Analysis of the restriction endonuclease profile of the 438-bp amplification products of 10 reference strains of C. burnetii. (A) The amplification products were digested with SspI, electrophoresed on agarose gels, and stained with ethidium bromide. Lane 1, molecular size markers (100-bp DNA ladder); lanes 2 to 8, seven reference strains (Nine Mile VR 615, Priscilla, MAN, ME, GQ212, SQ217, and KoQ229, respectively), lanes 9 to 11, three Japanese isolates (307, 605, and TK-1, respectively); lane 12, negative control. (B) The amplification products were digested with SalI. The samples in lanes 2 to 12 are the same as those in panel A. The numbers on the right are in base pairs.
Sensitivity of the nested PCR.
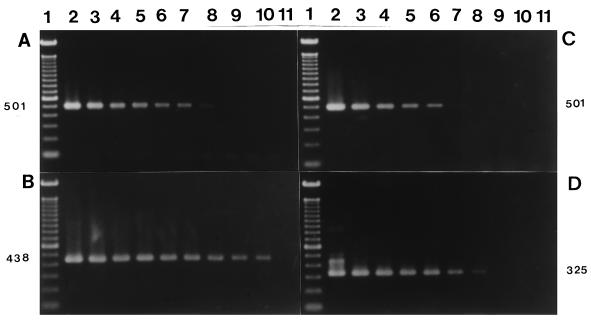
The primers OMP1-OMP2 and OMP3-OMP4 amplified the predicted products in reactions with about 5 fg of total DNA (corresponding to 1 organism) (Fig. 2A and B). The primers Q3-Q5 and Q4-Q6 amplified the predicted products in reactions with about 500 fg of total DNA (corresponding to 100 organisms) (Fig. 2C and D).
FIG. 2.
Sensitivity of the nested PCR with serial 10-fold dilutions of total DNA (from 500 ng to 0.5 fg) extracted from the Nine Mile VR 615 strain of C. burnetii. (A and B) Gel electrophoresis of first-round and second-round (B) PCR products amplified with primers OMP1-OMP2 and OMP3-OMP4. Lane 1, molecular size markers (100-bp DNA ladder); lanes 2 to 11, PCR products with serial 10-fold dilutions of total C. burnetii DNA (from 500 ng to 0.5 fg). (C and D) Gel electrophoresis of first-round (C) and second-round (D) PCR products amplified by primers Q3-Q5 and Q4-Q6. Lane 1, molecular size markers (100-bp DNA ladder); the DNA samples in lanes 2 to 11 are the same as those in panel A. The numbers on the sides are in base pairs.
Detection of C. burnetii DNA sequences in human serum samples.
The efficacies of the two primer systems for the detection of C. burnetii DNA in human serum samples were compared (Table 1). The primers Q3-Q5 and Q4-Q6 amplified the predicted products with DNA templates from 86 of 255 serum samples. The primers OMP1-OMP2 and OMP3-OMP4 amplified the predicted products with DNA templates from 146 of 255 serum samples (Fig. 3). Among sera positive by IF, 55.5% (86 of 155) were positive with primers Q3-Q5 and Q4-Q6, 87.1% (135 of 155) were positive with primers OMP1-OMP2 and OMP3-OMP4, while 31.6% (49 of 155) were positive with primers OMP1-OMP2 and OMP3-OMP4 but negative with primers Q3-Q5 and Q4-Q6. Among the IF-negative sera, none were positive with primers Q3-Q5 and Q4-Q6, but 11% (11 of 100) were positive with primers OMP1-OMP2 and OMP3-OMP4. The results indicate that the nested PCR with primers OMP1-OMP2 and OMP3-OMP4 detected 60 positive serum specimens negative by the nested PCR with primers Q3-Q5 and Q4-Q6.
TABLE 1.
Comparison of IF and nested PCR assay results with two sets of primers for detection of C. burnetii in human serum samples
| IF result | No. of serum samples with the indicated result by PCR with the following:
|
||||
|---|---|---|---|---|---|
| Total |
htpB genea
|
com1 geneb
|
|||
| Positive | Negative | Positive | Negative | ||
| Positive | 155 | 86 | 69 | 135 | 20 |
| Negative | 100 | 0 | 100 | 11 | 89 |
| Total | 255 | 86 | 169 | 146 | 109 |
Detected by nested PCR with primers Q3-Q5 and Q4-Q6.
Detected by nested PCR with primers OMP1-OMP2 and OMP3-OMP4.
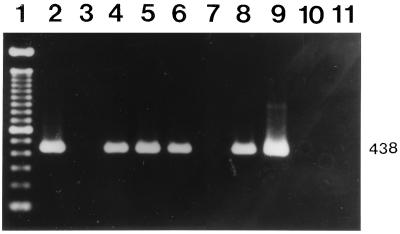
FIG. 3.
Detection of C. burnetii DNA in human sera by nested PCR with primers OMP1-OMP2 and OMP3-OMP4. An agarose gel electrophoretogram of amplified DNA after the nested PCR and ethidium bromide staining is shown. Lane 1, molecular size markers (100-bp DNA ladder); lanes 2 to 8, human serum samples (the serum samples in lanes 2, 4, 5, 6, and 8 were positive; the remaining samples were negative); lane 9, positive control (5 pg of purified C. burnetii DNA); lane 10, negative control serum; lane 11, reagent-negative control. The number on the right is in base pairs.
Positive results of the nested PCR with primers OMP1-OMP2 and OMP3-OMP4 were correlated with the IF antibody titer of the sera (Table 2). Among 155 IF-positive serum samples, 9, 35, 67, 25, and 19 samples with IF titers of ≥1:1,024, 1:256 to 1:512, 1:64 to 1:128, 1:32, and 1:16 were 100, 94.3, 85.1, 84, and 78.9% positive by nested PCR, respectively. High levels of antibody detected by IF were correlated with the presence of the com1 gene sequences in the sera.
TABLE 2.
Positive results by nested PCR compared with antibody titers by IF
| No. of serum samples | Antibody titer by IFa | No. (%) of PCR-positive serum samplesb |
|---|---|---|
| 9 | ≥1,024 | 9 (100) |
| 35 | 256–512 | 33 (94.3) |
| 67 | 64–128 | 57 (85.1) |
| 25 | 32 | 21 (84.0) |
| 19 | 16 | 15 (78.9) |
Antibody to strain Nine Mile VR 615 phase II antigen.
Nested PCR with primers OMP1-OMP2 and OMP3-OMP4.
DISCUSSION
A nested PCR with newly designed primers targeted to the com1 gene encoding a 27-kDa OMP was developed for the detection of C. burnetii. This nested PCR was demonstrated to be highly specific for 21 strains of C. burnetii and a useful method for the detection of the pathogen in human serum samples.
Although the PCR method has been used for the detection of C. burnetii by some researchers (10, 27, 29, 30, 31), our method is the first to use primers designed from the OMP gene, com1, for the detection of C. burnetii in human serum samples by nested PCR. The com1 gene sequence was chosen for the target of PCR amplification because we demonstrated in a previous report that it is highly conserved among 21 strains of C. burnetii from various clinical and geographical sources (32).
The sensitivity of the nested PCR with primers OMP1-OMP2 and OMP3-OMP4 was higher than that of the nested PCR with primers Q3-Q5 and Q4-Q6. Comparison of the sensitivities of the two primer systems in tests with human serum samples indicated that the nested PCR with primers OMP1-OMP2 and OMP3-OMP4 detected 60 positive serum specimens negative by the nested PCR with primers Q3-Q5 and Q4-Q6. Thus, the nested PCR with primers OMP1-OMP2 and OMP3-OMP4 was more sensitive than the nested PCR with primers Q3-Q5 and Q4-Q6 for the detection of C. burnetii in serum samples.
Comparison of the nested PCR results with those of IF indicated agreement for most of the serum samples, but discrepant results were found for some serum samples. We found that 11 of 100 IF-negative serum samples were PCR positive, probably resulting from the failure of IF to detect antibodies in serum samples collected during the early stage of infection. This possibility was further supported by the occurrence of C. burnetii in the serum samples obtained during the acute phase (2), during which antibodies were sometimes undetectable by IF (12, 22). We have also found that some serum samples from patients with the acute phase of Q fever were PCR positive but IF negative (10). These results suggest that the nested PCR is more sensitive than IF for the primary diagnosis of acute Q fever.
Antibodies against C. burnetii often persist for long periods after the organisms disappear from the blood of Q fever patients who are convalescing or receiving antibiotic therapy (4, 6). Musso and Raoult (15) also indicated that C. burnetii could not be isolated from the blood of similar patients by using cell culture. Hence, in our present study, the occurrence of 20 PCR-negative samples among 155 IF-positive serum samples may be explained by antibody persistence in convalescing patients who are devoid of C. burnetii at levels above the detection limits of the PCR assay. Therefore, the PCR results appear to indicate the presence or absence of the com1 gene fragment of C. burnetii in the blood, so PCR may be used to evaluate the efficacy of antibiotic therapy or optimize the antibiotic regimen for Q fever patients.
Our results also indicated that a high level of antibody detected by IF was correlated with the presence of C. burnetii DNA sequences in human serum samples. This observation is not surprising, since antibodies apparently do not play a direct role in resistance to C. burnetii infections (11) or prevent the occurrence of chronic disease (19, 20). Blood culture and serology can be positive at the same time in patients with acute Q fever, and C. burnetii can persist in patients for long periods, despite the presence of high levels of antibodies (15). Kazar et al. (13) also demonstrated that immune sera containing either phase II antibody or both phase I and phase II antibodies did not neutralize the organisms.
The results of this study suggest that the nested PCR with primers targeted to the com1 gene is highly specific and sensitive for the detection of C. burnetii, and it may be useful for laboratory diagnosis and assessment of the efficacy of antibiotic therapy for Q fever. Particularly in combination with IF, it may provide a more reliable yet quick means of diagnosing Q fever. We suggest that the clinical diagnosis of Q fever could be made on the basis of both the results of IF and the results of PCR for the detection of C. burnetii in serum samples.
ACKNOWLEDGMENTS
We thank Ted Meyers for helpful comments and suggestions concerning the manuscript.
This work was supported by a Grant-in-Aid for Developmental Scientific Research (grant 07306015) from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Baca O G, Paretsky D. Q fever and Coxiella burnetii: a model for host-parasite interactions. Microbiol Rev. 1983;47:127–149. doi: 10.1128/mr.47.2.127-149.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Derrick E H. The course of infection with Coxiella burnetii. Med J Aust. 1973;1:1051–1057. [PubMed] [Google Scholar]
- 3.Dupuis G, Peter O, Peacock M, Burgdorfer W, Haller E. Immunoglobulin responses in acute Q fever. J Clin Microbiol. 1985;22:484–487. doi: 10.1128/jcm.22.4.484-487.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dupuis G, Peter O, Pedroni D, Petite J. Clinical aspects observed during an epidemic of 415 cases of Q fever. Schweiz Med Wochenschr J Suisse Med. 1985;115:814–818. [PubMed] [Google Scholar]
- 5.Field P R, Hunt J G, Murphy A M. Detection and persistence of specific IgM antibody to Coxiella burnetii by enzyme-linked immunosorbent assay: a comparison with immunofluorescence and complement fixation tests. J Infect Dis. 1983;148:477–487. doi: 10.1093/infdis/148.3.477. [DOI] [PubMed] [Google Scholar]
- 6.Guigno D, Coupland B, Smith E G, Farrell I D, Desselberger U, Caul E O. Primary humoral antibody response to Coxiella burnetii, the causative agent of Q fever. J Clin Microbiol. 1992;30:1958–1967. doi: 10.1128/jcm.30.8.1958-1967.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendrix L R, Mallavia L P, Samuel J E. Cloning and sequencing of Coxiella burnetii outer membrane protein gene com1. Infect Immun. 1993;61:470–477. doi: 10.1128/iai.61.2.470-477.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendrix L R, Samuel J E, Mallavia L P. Differentiation of Coxiella burnetii isolates by analysis of restriction-endonuclease-digested DNA separated by SDS-PAGE. J Gen Microbiol. 1991;137:269–276. doi: 10.1099/00221287-137-2-269. [DOI] [PubMed] [Google Scholar]
- 9.Ho T, Htwe K K, Yamasaki N, Zhang G Q, Ogawa M, Yamaguchi T, Fukushi H, Hirai K. Isolation of Coxiella burnetii from dairy cattle and ticks, and some characteristics of the isolates in Japan. Microbiol Immunol. 1995;39:663–671. doi: 10.1111/j.1348-0421.1995.tb03254.x. [DOI] [PubMed] [Google Scholar]
- 10.Ho T, Kako N, Zhang G Q, Otsuka H, Ogawa M, Ochiai O, Nguyen Sa V, Yamaguchi T, Fukushi H, Nagaoka N, Akiyama M, Amano K, Hirai K. Q fever pneumonia in children in Japan. J Clin Microbiol. 1996;34:647–651. doi: 10.1128/jcm.34.3.647-651.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Humphres R C, Hinrichs D J. Role of antibody in Coxiella burnetii infection. Infect Immun. 1981;31:641–645. doi: 10.1128/iai.31.2.641-645.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunt J G, Field P R, Murphy A M. Immunoglobulin responses to Coxiella burnetii (Q fever): single-serum diagnosis of acute infection, using an immunofluorescence technique. Infect Immun. 1983;39:977–981. doi: 10.1128/iai.39.2.977-981.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kazar J, Brezina R, Kovacova E, Urvolgyi J. Testing in various systems of the neutralizing capacity of Q fever immune sera. Acta Virol. 1973;17:79–89. [PubMed] [Google Scholar]
- 14.Kimbrough R C D, Ormsbee R A, Peacock M, Rogers W R, Bennetts R W, Raaf J, Krause A, Gardner C. Q fever endocarditis in the United States. Ann Intern Med. 1979;91:400–402. doi: 10.7326/0003-4819-91-3-400. [DOI] [PubMed] [Google Scholar]
- 15.Musso D, Raoult D. Coxiella burnetii blood cultures from acute and chronic Q-fever patients. J Clin Microbiol. 1995;33:3129–3132. doi: 10.1128/jcm.33.12.3129-3132.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagaoka H, Akiyama M, Sugieda M, Nishio T, Akahane S, Hattori H, Ho T, Fukushi H, Hirai K. Isolation of Coxiella burnetii from children with influenza-like symptoms in Japan. Microbiol Immunol. 1996;40:147–151. doi: 10.1111/j.1348-0421.1996.tb03330.x. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen S V, Otsuka H, Zhang G Q, To H, Yamaguchi T, Fukushi H, Noma A, Hirai K. Rapid method for detection of Coxiella burnetii antibodies using high-density particle agglutination. J Clin Microbiol. 1996;34:2947–2951. doi: 10.1128/jcm.34.12.2947-2951.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noseda A, Liesnard C, Goffin Y, Thys J P. Q fever endocarditis: relapse five years after successful valve replacement for a first unrecognized episode. J Cardiovasc Surg. 1988;29:360–363. [PubMed] [Google Scholar]
- 19.Peacock M G, Fiset P, Ormsbee R A, Wisseman C L., Jr Antibody response in man following a small intradermal inoculation with Coxiella burnetii phase I vaccine. Acta Virol. 1979;23:73–81. [PubMed] [Google Scholar]
- 20.Peacock M G, Philip R N, Williams J C, Faulkner R S. Serological evaluation of O fever in humans: enhanced phase I titers of immunoglobulins G and A are diagnostic for Q fever endocarditis. Infect Immun. 1983;41:1089–1098. doi: 10.1128/iai.41.3.1089-1098.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peter O, Dupuis G, Bee D, Luthy R, Nicolet J, Burgdorfer W. Enzyme-linked immunosorbent assay for diagnosis of chronic Q fever. J Clin Microbiol. 1988;26:1978–1982. doi: 10.1128/jcm.26.10.1978-1982.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peter O, Dupuis G, Burgdorfer W, Peacock M. Evaluation of the complement fixation and indirect immunofluorescence tests in the early diagnosis of primary Q fever. Eur J Clin Microbiol. 1985;4:394–396. doi: 10.1007/BF02148690. [DOI] [PubMed] [Google Scholar]
- 23.Peter O, Dupuis G, Peacock M G, Burgdorfer W. Comparison of enzyme-linked immunosorbent assay and complement fixation and indirect fluorescent-antibody tests for detection of Coxiella burnetii antibody. J Clin Microbiol. 1987;25:1063–1067. doi: 10.1128/jcm.25.6.1063-1067.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raoult D, Urvolgyi J, Etienne J, Roturier M, Puel J, Chaudet H. Diagnosis of endocarditis in acute Q-fever by immunofluorescence serology. Acta Virol. 1988;32:70–74. [PubMed] [Google Scholar]
- 25.Schmeer N. Enzyme-linked immunosorbent assay (ELISA) for the demonstration of IgG1, IgG2, and IgM antibodies in bovine Q fever infection. Zentralbl Bakteriol Parasitenkd Infectionskr Hyg Abt 1 Orig. 1985;259:20–34. [PubMed] [Google Scholar]
- 26.Schmeer N, Muller H P, Baumgartner W, Wieda J, Krauss H. Enzyme-linked immunosorbent fluorescence assay and high-pressure liquid chromatography for analysis of humoral immune responses to Coxiella burnetii proteins. J Clin Microbiol. 1988;26:2520–2525. doi: 10.1128/jcm.26.12.2520-2525.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stein A, Raoult D. Detection of Coxiella burnetii by DNA amplification using polymerase chain reaction. J Clin Microbiol. 1992;30:2462–2466. doi: 10.1128/jcm.30.9.2462-2466.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weir W R C, Bannister B, Chambres S, Coke K D, Mistry H. Chronic Q fever associated with granulomatous hepatitis. J Infect. 1984;8:56–60. doi: 10.1016/s0163-4453(84)93354-1. [DOI] [PubMed] [Google Scholar]
- 29.Willems H, Thiele D, Frolich-Ritter R, Krauss H. Detection of Coxiella burnetii in cow’s milk using the polymerase chain reaction (PCR) Zentralbl Veterinaermed B. 1994;41:580–587. doi: 10.1111/j.1439-0450.1994.tb00267.x. [DOI] [PubMed] [Google Scholar]
- 30.Willems H, Thiele D, Krauss H. Plasmid based differentiation and detection of Coxiella burnetii in clinical samples. Eur J Epidemiol. 1993;9:411–418. doi: 10.1007/BF00157399. [DOI] [PubMed] [Google Scholar]
- 31.Yuasa Y, Yoshiie K, Takasaki T, Yoshida H, Oda H. Retrospective survey of chronic Q fever in Japan by using PCR to detect Coxiella burnetii DNA in paraffin-embedded clinical samples. J Clin Microbiol. 1996;34:824–827. doi: 10.1128/jcm.34.4.824-827.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang, G. Q., H. To, T. Yamaguchi, H. Fukushi, and K. Hirai. Differentiation of Coxiella burnetii by sequence analysis of the gene (com1) encoding a 27-kDa outer membrane protein. Microbiol. Immunol., in press. [DOI] [PubMed]