Abstract
We isolated a hitherto undescribed microorganism from a patient with endocarditis. The microscopic appearance, a negative catalase reaction, and growth as satellite colonies next to Staphylococcus epidermidis suggested that this microorganism is a member of the genus Abiotrophia, formerly known as nutritionally variant streptococci. However, the clinical isolate described herein differed markedly from the known Abiotrophia spp., A. adiacens and A. defectiva, in terms of its (i) biochemical properties, (ii) restricted growth temperature range, (iii) whole-cell lysate polypeptide profile, and (iv) unique nutritional requirements. In contrast to the type strains of A. adiacens and A. defectiva, which used l-cysteine and pyridoxal hydrochloride as growth factors, the growth of the clinical isolate was only supported by l-cysteine hydrochloride and not by pyridoxal hydrochloride when the organism was tested in Todd-Hewitt or casein-soy peptone broth. Comparative 16S rRNA gene sequence analysis revealed that the microorganism was a member of the genus Abiotrophia and was most closely related to A. adiacens (96.9% homology). Phenotypic and phylogenetic data are consistent with the assumption of a new species within the genus Abiotrophia, for which we propose the name Abiotrophia elegans sp. nov. The unique nutritional requirements of this strain are of importance for diagnostic laboratories. The media of blood culture systems supplemented only with pyridoxal hydrochloride as a growth factor may fail to promote the growth of A. elegans sp. nov., and thus, these systems might not detect this microorganism as a possible cause of endocarditis.
Nutritionally variant streptococci (NVS) were originally isolated by Frenkel and Hirsch (9) from patients with endocarditis and otitis media and were described as ungroupable viridans group streptococci that grow as satellite colonies around other bacteria. These organisms have been shown to grow in complex media supplemented with l-cysteine or pyridoxal hydrochloride (vitamin B6) (6, 9). NVS are members of the normal flora of the human pharynx and the human urogenital and intestinal tracts (13, 14). NVS are the causes of 5% of cases of streptococcal endocarditis, including most of the so-called blood culture-negative cases of endocarditis. Moreover, NVS have been isolated from patients with a variety of other infectious diseases (3, 13).
The taxonomic position of NVS remained unclear until Bouvet et al. (3) performed DNA-DNA hybridization studies and named the NVS Streptococcus adjacens and Streptococcus defectivus. In 1995 Kawamura et al. (11) determined the 16S rRNA sequences of the type strains of Streptococcus adjacens and Streptococcus defectivus and placed them in the new genus Abiotrophia as Abiotrophia adiacens and Abiotrophia defectiva, respectively, because of their levels of low sequence homology to other Streptococcus species.
CASE REPORT
A 38-year-old patient was admitted to the Hospital Dritter Orden in Munich, Germany, presenting with clinical signs of acute endocarditis (aortic valve). Four consecutive blood cultures revealed growth of a gram-positive bacterium. The patient was treated empirically with piperacillin-tacobactam and gentamicin. Despite the antimicrobial therapy, the patient’s fever persisted and two more blood cultures were positive for the gram-positive bacterium. Therefore, vancomycin was added to the treatment protocol. However, the endocarditis had a rapid course, signs of valvular insufficiency appeared, and an early surgical intervention was necessary. The aortic valve was replaced, and the antimicrobial therapy was continued for a further 10 days. The patient was released from the hospital without fever after 14 days. The blood culture isolate was then characterized in more detail.
MATERIALS AND METHODS
Bacterial strains.
A. defectiva SC10 (ATCC 49176), A. adiacens GaD (ATCC 49175), and A. elegans B1333 (clinical isolate) were grown an Schaedler agar base (Difco Laboratories GmbH, Augsburg, Germany) supplemented with 7% defibrinated sheep blood (Oxoid, Unipath Ltd., Basingstoke, England) at 37°C.
Biochemical differentiation.
Tests for tolerance to bile esculin and 6.5% NaCl were performed by the method of Facklam (8). Biochemical testing was performed with the API 20 Strep identification system (bio-Merieux Ltd., Marcy l’Etoile, France) as described by Bouvet et al. (5), with modifications, and values were obtained after 4 h and 1 and 7 days of incubation at 37°C.
Growth characteristics and nutritional requirements.
Microorganisms grown on Schaedler blood agar at 37°C for 24 h were resuspended in Todd-Hewitt broth (THB; Difco) to an optical density at 600 nm (OD600) of 1.0. The bacterial suspensions were diluted 1:100 in THB (resulting in an OD600 of <0.01) with or without the following growth factors: pyridoxal hydrochloride (final concentration, 0.001%) and l-cysteine hydrochloride, d-alanine, and l-alanine (final concentrations, 0.01%; Sigma-Aldrich Chemie GmbH, Deisenhofen, Germany). After incubation at 37°C for 24 h the OD600s of the liquid cultures were determined. OD600 values of <0.04 were defined as negative (no growth), and OD600 values of >0.15 were considered positive (multiplication). Possible microbial contaminations of positive cultures were excluded by plating 10 μl of the culture broth on blood agar plates lacking growth factors. In addition, bacterial strains were characterized by satellite tests on Trypticase soy agar (TSA; Oxoid, Unipath Ltd.) as described by Bouvet et al. (4), with minor modifications. Fresh bacterial cultures (30 μl of a suspension with an OD600 of 1.0) were streaked onto the surfaces of TSA plates. Then, filter discs containing 5 μl of 5 mM l-cysteine, d-alanine, l-alanine, and 0.1% pyridoxal hydrochloride, respectively, were placed onto the streaked cultures. After 48 h of incubation at 37°C, the growth of small colonies around the filter discs indicated a requirement for the respective nutrition factor.
Growth (colony formation) on Schaedler blood agar plates incubated at different temperatures was determined after 48 h. All tests were repeated at least three times to check their reproducibilities.
Chromophore assay.
The presence of a pH-dependent chromophore was tested as described by van de Rijn and Bouvet (18), with the modification that l-cysteine-supplemented THB was used as the growth medium.
Whole-cell lysates.
Strains grown on Schaedler blood agar plates for 24 h at 37°C were removed with a loop, washed twice in phosphate-buffered saline (pH 7.5), and resuspended in the same volume of 2× Laemmli sample buffer containing 10% 2-mercaptoethanol. The samples were boiled for 10 min and centrifuged at 10,000 × g for 30 s, and the supernatant was analyzed by sodium dodecyl sulfate (SDS)–11% polyacrylamide gel electrophoresis (PAGE) (12).
16S rRNA gene sequence analysis.
The amplification and direct sequencing of the gene encoding the 16S rRNA was done as described previously (7), but with some modifications. Universal primers corresponding to the Escherichia coli 16S rRNA gene from bp 8 to 28 and bp 1542 to 1522 were used for PCR amplification. For solid-phase DNA sequencing one of the oligonucleotides was biotinylated at the 5′ end. Dynabeads were used for the preparation of single-stranded DNA as recommended by the manufacturer (DYNAL GmbH, Hamburg, Germany). In addition to the oligonucleotides mentioned above, forward and reverse oligonucleotides corresponding to conserved regions of the 16S rRNA gene were used for sequencing (bp 341 to 361, 515 to 531, 785 to 804, 1101 to 1114, and 1391 to 1406). The PCR products were sequenced by the Taq DyeDideoxy terminator method with a 377XL DNA Sequencer (Applied Biosystems GmbH, Darmstadt, Germany). All sequence analyses were performed with the sequence analysis software package GCG (Genetics Computer Group, Inc., Madison, Wis.). Sequence data for comparisons were obtained from GenBank. A matrix of pairwise evolutionary distances between aligned sequences (similarity matrix) was constructed and was corrected by the method of Jukes and Cantor (10). A phylogenetic tree was created by the neighbor-joining method (15, 17).
Nucleotide sequence accession number.
The nucleotide sequence of the 16S rRNA gene of A. elegans B1333 have been deposited in the GenBank Data Library under accession no. AF016390.
RESULTS
Isolation and microbiological characterization of strain B1333.
Strain B1333 was isolated in a culture bottle (Oxoid) containing blood from a patient suffering from endocarditis. Small unpigmented colonies with α-hemolysis grew after 48 h of anaerobic incubation at 37°C on Schaedler blood agar plates. No bacterial growth was observed on TSA blood agar plates, and hardly any growth was observed on chocolate agar plates after incubation for 48 h in 5% CO2. Gram staining of colonies from Schaedler blood agar plates as well as from a dried droplet of blood cultured in bouillon showed an extremely pleomorphic gram-positive microorganism with small coccoid forms (about 0.5 μm in diameter) lying next to long elongated swollen forms (4 by 1.2 μm). The catalase and oxidase reactions of the colonies and the motility test were negative. A positive satellite test with growth on TSA blood agar plates next to Staphylococcus epidermidis resembled NVS, but biochemical characterization of the organism with the API 20 Strep identification system gave no acceptable result. The antibiotic susceptibility tests showed that the organism was sensitive to penicillin G and ampicillin (MICs of both drugs, <0.016 μg/ml). Agar diffusion testing on Schaedler-blood agar plates after 24 h of anaerobic incubation revealed that the organism was sensitive to all β-lactam antibiotics, vancomycin, erythromycin, and ciprofloxacin.
Sequencing of the 16S rRNA gene.
The 16S rRNA gene from strain B1333 was amplified by PCR with universal primers corresponding to bp 8 to 28 and 1544 to 1542 of the E. coli 16S rRNA gene. The PCR product was sequenced directly in both directions. Comparison of the sequence with those of other bacteria in the GenBank database indicated that the gram-positive microorganism B1333 was most related to A. adiacens (96.9% homology) (Table 1).
TABLE 1.
Distance matrix between aligned sequences of the 16S rRNA genes corrected by the method of Jukes and Cantora
| Species no. | Species | % Sequence differences for the following speciesb:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ||
| 1. | Leuconostoc mesenteroides | 0.0 | ||||||||||||
| 2. | Lactobacillus intestinalis | 17.9 | ||||||||||||
| 3. | Carnobacterium pisiciola | 15.8 | 15.1 | |||||||||||
| 4. | Streptococcus pneumoniae | 15.7 | 17.3 | 10.9 | ||||||||||
| 5. | Streptococcus mitis | 15.7 | 17.0 | 11.7 | 1.0 | |||||||||
| 6. | Streptococcus sanguis | 14.8 | 16.3 | 10.9 | 2.5 | 2.6 | ||||||||
| 7. | Streptococcus pyogenes | 15.9 | 15.2 | 11.2 | 5.3 | 5.1 | 4.2 | |||||||
| 8. | Streptococcus bovis | 15.8 | 15.7 | 12.1 | 5.0 | 5.4 | 4.0 | 4.0 | ||||||
| 9. | Streptococcus mutans | 16.7 | 15.2 | 12.6 | 6.0 | 6.5 | 4.8 | 6.2 | 4.9 | |||||
| 10. | Vagococcus salmoniarum | 15.8 | 15.8 | 6.4 | 12.0 | 12.4 | 11.4 | 11.9 | 12.1 | 12.9 | ||||
| 11. | Abiotrophia adiacens | 15.2 | 14.2 | 6.3 | 13.4 | 13.9 | 12.4 | 13.0 | 13.4 | 14.4 | 7.9 | |||
| 12. | Abiotrophia defectiva | 15.5 | 15.5 | 8.9 | 14.0 | 14.0 | 13.2 | 13.1 | 14.3 | 14.1 | 11.2 | 8.0 | ||
| 13. | Strain B1333 | 14.8 | 14.4 | 6.5 | 13.7 | 14.5 | 12.7 | 13.6 | 13.5 | 14.3 | 8.6 | 3.1 | 8.0 | 0.0 |
The method of Jukes and Cantor has been described previously (10).
The numbers in the subheads correspond to the species numbers listed in the first column.
Biochemical characterization and whole-cell lysate profile.
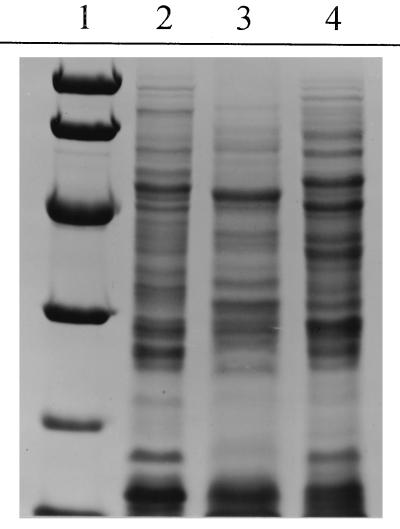
The biochemical characterization of strain B1333 was repeated as described by Bouvet et al. (5), with minor modifications for the differentiation of NVS, and the biochemical properties of B1333 were compared with those of the type strains of A. adiacens (ATCC 49175) and A. defectiva (ATCC 49176). The different biochemical properties of the strains are presented in Table 2. To confirm the distinction between strain B1333 and the two type strains of Abiotrophia, we analyzed the whole-cell lysate polypeptide profile by SDS-PAGE (Fig. 1). Corresponding to the results of the 16S rRNA gene sequencing, strain B1333 seems to be more related to A. adiacens than to A. defectiva but differed significantly from A. adiacens in more than six polypeptide bands.
TABLE 2.
Biochemical characterization of Abiotrophia spp.
| Characteristic | A. adiacens | A. defectiva | Strain B1333 |
|---|---|---|---|
| Enzyme production | |||
| Pyrrolidonyl-arylamidase | + | + | + |
| Alkaline phosphatase | − | − | − |
| α-Galactosidase | − | + | − |
| β-Galactosidase | − | + | − |
| β-Glucuronidase | + | − | − |
| β-Glucosidase | − | − | − |
| Leucine arylamidase | + | + | + |
| Hippurate hydrolysis | − | − | + |
| Acetoin production | − | − | − |
| Acidification of: | |||
| Trehalose | − | + | − |
| Inulin | + | − | − |
| Lactose | − | + | − |
| Raffinose | − | + | + |
| Starch | − | + | − |
| Glycogen | − | − | − |
FIG. 1.
Coomassie-stained SDS-polyacrylamide gels. Microorganisms grown on Schaedler blood agar plates were washed twice in phosphate-buffered saline, resuspended in sample buffer, boiled, and separated by SDS–11% PAGE. The polypeptide profiles were visualized by Coomassie staining. Lane 1, low-molecular-mass markers (94, 67, 43, 30, 20, and 14 kDa); lane 2, A. adiacens; lane 3, A. defectiva; lane 4, strain B1333.
Growth characteristics and nutritional requirements.
The pleomorphology of Abiotrophia cells upon Gram staining is a hallmark of Abiotrophia spp. and is influenced by the nutritional state of the organisms. Gram staining performed in parallel with freshly grown cultures (Schaedler blood agar) showed that the morphology of strain B1333 was much more variable than that of A. adiacens, whereas A. defectiva presented quite regular coccoid forms. This behavior prompted us to study the nutritional requirements of strain B1333 in comparison to those of the type strains of Abiotrophia. For standardization we determined the growth by measuring the OD600 in supplemented THB and compared the results with those of satellite tests performed on TSA, which recorded colony formation next to a nutritional factor. The results are presented in Table 3. Both test systems gave nearly identical results and demonstrated the known nutritional requirements for A. adiacens and A. defectiva. To our surprise, strain B1333 did not use vitamin B6 as a growth factor, but the type strains of Abiotrophia did. However, growth could be stimulated with l-cysteine hydrochloride. As suggested previously (1), vitamin B6 is used during cell wall biosynthesis by conversion of l-alanine to d-alanine. Therefore, we also tested whether these amino acids are growth factors. d-Alanine promoted the growth of A. adiacens but not that of A. defectiva or strain B1333. The effect of d-alanine was weak and was detectable only in supplemented THB, possibly because of the sensitivity of this test system.
TABLE 3.
Growth characteristics of Abiotrophia spp.
| Test | A. adiacens | A. defectiva | Strain B1333 |
|---|---|---|---|
| Tolerance to: | |||
| Bile esculin | − | − | − |
| 6.5% NaCl | − | − | − |
| Growth at: | |||
| 20°C | + | + | − |
| 27°C | + | + | + |
| 37°C | + | + | + |
| 42°C | + | + | − |
| α-Hemolysis | + | + | + |
| Growth on chocolate agar | (+)a | (+) | (+) |
| Growth on Schaedler agar | + | + | + |
| Facultative anaerobe | + | + | + |
| Growth on TSA plates supplemented withb: | |||
| Pyridoxal hydrochloride | + | + | − |
| l-Cysteine hydrochloride | + | + | (+) |
| d-Alanine | − | − | − |
| l-Alanine | − | − | − |
| Growth in THB supplemented withc: | |||
| Pyridoxal hydrochloride | + | + | − |
| l-Cysteine hydrochloride | + | + | + |
| d-Alanine | + | − | − |
| l-Alanine | − | − | − |
| Growth in complex casein-soy peptone medium supplemented with: | |||
| Pyridoxal hydrochlorided | + | + | − |
| l-Cysteine hydrochloridee | + | + | + |
(+), strongly reduced growth.
Growth surrounding filter discs containing 5 μl of 5 mM amino acid solution or 0.1% pyridoxal hydrochloride after 48 h.
Final concentrations: amino acids, 0.01%; pyridoxal hydrochloride, 0.001%. After incubation at 37°C for 24 h a bacterial density with an OD600 of >0.15 indicated growth.
BACTEC blood culture medium.
Oxoid blood culture medium.
We tested whether B1333 also grows in the BACTEC blood culture system (Becton Dickinson, Cockeysville, Md.) containing pyridoxal hydrochloride (vitamin B6) as the growth factor. Aerobic and anaerobic BACTEC-Plus blood culture bottles were inoculated with strain B1333, A. adiacens, and A. defectiva, respectively (100 μl; OD600, 1.0). While the BACTEC blood culture system detected the growth of A. adiacens and A. defectiva after 1 or 2 days, strain B1333 was not detected in this system (period of incubation, 7 days). Gram staining of a sample taken from the blood culture inoculated with strain B1333 revealed no visible bacteria. However, at day 7 strain B1333 from the bottles could be reisolated on Schaedler blood agar plates, suggesting that B1333 did not multiply in BACTEC blood culture medium but persisted in this medium for at least 7 days. In contrast, strain B1333 grew well in Oxoid blood culture bottles, which are supplemented with l-cysteine.
DISCUSSION
The growth behavior, microscopic appearance, and negative catalase reaction of unidentified clinical isolate B1333 resembled those of bacteria of the genus Abiotrophia, formerly known as NVS. Sequencing of the 16S rRNA gene revealed that this pathogen is most closely related to A. adiacens.
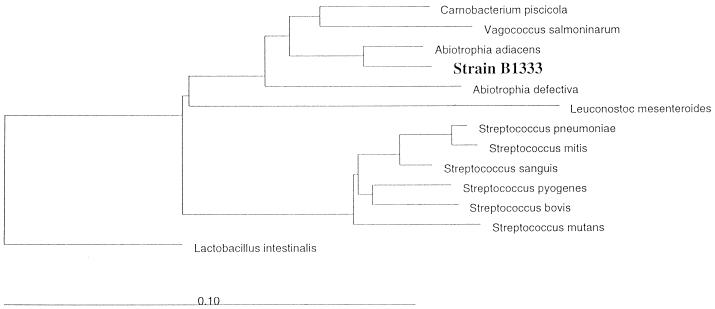
The genus Abiotrophia represents a phylogenetically distinct cluster and was only remotely related to the Aerococcus and Carnobacterium clusters (11). The relationship between Abiotrophia and other phylogenetic groups containing medically relevant bacteria was weak (16S rRNA homology to Streptococcus, 85%) (Fig. 2). Hitherto, the 16S rRNA sequences of two species of the genus Abiotrophia have been determined. The low degree of homology between these two species (93%) might be a hint that the genus comprises more than these two species. The phylogenetic distance between strain B1333 and A. adiacens is greater than 3%, which is indicative of a separate species (16). Consistent with this assumption, the biochemical properties of strain B1333 differed from those known for Abiotrophia spp. and had a restricted growth temperature range compared with those of Abiotrophia spp., a characteristic whole-cell lysate polypeptide profile, and unusual nutritional requirements compared with those of Abiotrophia spp. In biochemical tests members of the genus Abiotrophia often showed different enzyme profiles (2, 5). In the most comprehensive study Bouvet et al. (5) characterized 60 strains of NVS and divided them into three biotypes. Strain B1333 fits none of them. A characteristic enzymatic reaction might be the hippurate hydrolysis of B1333 which was reported by Bouvet et al. (5) for 2 of the 60 strains.
FIG. 2.
Unrooted tree showing the phylogenetic relationships of strain B1333 to Abiotrophia spp. and related taxa. Distances were calculated by the neighbor-joining method. The evolutionary distance between any two taxa is the sum of the lengths of the horizontal lines between them. The bar indicates substitution rate per nucleotide.
It is generally accepted that pyridoxal hydrochloride at a concentration of 0.001% will support the growth of Abiotrophia spp. (14). This was not the case for strain B1333, which did not grow in pyridoxal hydrochloride-supplemented THB or complex casein-soy peptone medium, but the type strains of Abiotrophia did. This deficiency has consequences for the cultivation of B1333-like strains in commercially available blood culture systems. As a possible causative agent of endocarditis, it will rarely be isolated in laboratories which use a pyridoxal hydrochloride-supplemented blood culture system. Therefore, it might be a cause of culture-negative endocarditis.
l-Cysteine works quite well as a growth factor for B1333, but the exact enzymatic defect that is responsible for the nutritional requirement must be elucidated. Beighton et al. (2) also suggested that NVS may have different nutritional requirements. Although it is speculative, we assume that the genus Abiotrophia contains more than the three species discussed herein, and these additional species may possibly have other, yet unknown growth requirements. In general, it is problematic to describe a new species on the basis of the characterization of a single strain. The successful isolation and identification of fastidious pathogens, however, will be possible only if clinical microbiologists are aware of the unique growth requirements of the organisms.
Description of Abiotrophia elegans sp. nov.
Abiotrophia elegans (e′le.gans. L. adj. elegans, fastidious, referring to the fastidious growth requirements) is a gram-positive microorganism. The shape is dependent on the nutritional state. In sufficiently supplemented nutritional broth the bacterium appears coccoid in short chains. Lack of appropriate growth factors results in pleomorphism and the appearance of elongated, swollen forms. The microorganism is nonmotile, nonsporulating, and catalase and oxidase negative and grows as a facultative anaerobe with complex growth requirements. It grows as satellite colonies adjacent to S. epidermidis on trypticase-soy-sheep blood agar plates with α-hemolysis. Tiny colonies up to 0.2 mm in diameter are formed on Schaedler-sheep blood agar plates after 48 h, but only minimal growth is visible on chocolate agar plates. Growth occurs at 27 and 37°C but not at 20 or 42°C. It grows in THB or casein-soy peptone bouillon supplemented with 0.01% l-cysteine hydrochloride. It does not grow in THB or casein-soy peptone bouillon supplemented with 0.001% pyridoxal hydrochloride. It produces a red chromophore visualized by boiling the microorganism at pH 2 for 5 min. It produces pyrrolidonyl-arylamidase and leucine aminopeptidase but not alkaline phosphatase, α- or β-galactosidase, β-glucuronidase, or β-glucosidase. It hydrolyzes hippurate. It ferments raffinose but not trehalose, inulin, lactose, starch, or glycogen.
Strain B1333 has been deposited in the German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany, as strain DSM 11693.
ACKNOWLEDGMENT
We are thankful to Helmut Walter for accurate sequencing of the 16S rRNA gene.
REFERENCES
- 1.Anonymous. Culture-negative endocarditis. Lancet. 1977;ii:1164–1165. . (Editorial.) [PubMed] [Google Scholar]
- 2.Beighton D, Homer K A, Bouvet A, Storey A R. Analysis of enzymatic activities for differentiation of two species of nutritionally variant streptococci, Streptococcus defectivus and Streptococcus adjacens. J Clin Microbiol. 1995;33:1584–1587. doi: 10.1128/jcm.33.6.1584-1587.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouvet A, Grimont F, Grimont P A. Streptococcus defectivus sp. nov. and Streptococcus adjacens sp. nov., nutritionally variant streptococci from human clinical specimens. Int J Syst Bacteriol. 1989;39:290–294. doi: 10.1099/00207713-41-4-483. [DOI] [PubMed] [Google Scholar]
- 4.Bouvet A, van de Rijn I, McCarty M. Nutritionally variant streptococci from patients with endocarditis: growth parameters in a semisynthetic medium and demonstration of a chromophore. J Bacteriol. 1981;146:1075–1082. doi: 10.1128/jb.146.3.1075-1082.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouvet A, Villeroy F, Cheng F, Lamesch C, Williamson R, Gutmann L. Characterization of nutritionally variant streptococci by biochemical tests and penicillin-binding proteins. J Clin Microbiol. 1985;22:1030–1034. doi: 10.1128/jcm.22.6.1030-1034.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carey R B, Gross K C, Roberts R B. Vitamin B6-dependent Streptococcus mitior (mitis) isolated from patients with systemic infections. J Infect Dis. 1975;131:722–726. doi: 10.1093/infdis/131.6.722. [DOI] [PubMed] [Google Scholar]
- 7.Edwards U, Rogall T, Blöcker H, Emde M, Böttger E C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S rRNA. Nucleic Acids Res. 1989;17:7843–7852. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Facklam R R. Physiological differentiation of viridans streptococci. J Clin Microbiol. 1977;5:184–201. doi: 10.1128/jcm.5.2.184-201.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frenkel A, Hirsch W. Spontaneous development of L forms of streptococci requiring secretions of other bacteria or sulphydryl compounds for normal growth. Nature. 1961;191:728–730. doi: 10.1038/191728a0. [DOI] [PubMed] [Google Scholar]
- 10.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. Vol. 3. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 11.Kawamura Y, Hou X G, Sultana F, Liu S, Yamamoto H, Ezaki T. Transfer of Streptococcus adjacens and Streptococcus defectivus to Abiotrophia gen. nov. as Abiotrophia adiacens comb. nov. and Abiotrophia defectiva comb. nov., respectively. Int J Syst Bacteriol. 1995;45:798–803. doi: 10.1099/00207713-45-4-798. [DOI] [PubMed] [Google Scholar]
- 12.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.Roberts R B, Krieger A G, Schiller N L, Gross K C. Viridans streptococcal endocarditis: the role of various species, including pyridoxal-dependent streptococci. Rev Infect Dis. 1979;1:955–966. doi: 10.1093/clinids/1.6.955. [DOI] [PubMed] [Google Scholar]
- 14.Ruoff K L. Nutritionally variant streptococci. Clin Microbiol Rev. 1991;4:184–190. doi: 10.1128/cmr.4.2.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 16.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 17.Studier J A, Keppler K J. A note on the neighbor-joining algorithm of Saitou and Nei. Mol Biol Evol. 1988;5:729–731. doi: 10.1093/oxfordjournals.molbev.a040527. [DOI] [PubMed] [Google Scholar]
- 18.van de Rijn I, Bouvet A. Characterization of a pH-dependent chromophore from nutritionally variant streptococci. Infect Immun. 1984;43:28–31. doi: 10.1128/iai.43.1.28-31.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]