Abstract
Severe postoperative wound infections caused by Nocardia farcinica were repeatedly observed in a German hospital surgical ward. A pulsed-field gel electrophoresis (PFGE) protocol was established to characterize the genetic relatedness of the bacterial isolates from these infections. All 18 isolates from postoperative infections that have occurred since 1985 belong to a common endemic genotype; organisms of this genotype were also detected in the air of two rooms of the department where these postoperative infections occurred. In contrast, two environmental isolates from another building on the same campus showed a distinct genotype. Three cases of pulmonary infections, at a department which is located in proximity to the surgical department, were also caused by the endemic type, which suggests aerogenic spread of the endemic strain to these patients. Controls consisting of epidemiologically unrelated isolates from sporadic infections in other towns belonged in each case to a different genotype. PFGE was well suited to differentiate various types of N. farcinica and revealed an endemic strain causing postoperative wound infections possibly after aerogenic transmission.
Members of the genus Nocardia are ubiquitous soil inhabitants which cause sporadic opportunistic human infections with clinical manifestations that vary widely (13). Besides the sporadic distribution of these infections, however, nocardiosis was also recently observed as an endemic nosocomial infection and Nocardia farcinica appears to be increasingly acquired by patients in European hospitals (1, 13). Fourteen cases of nocardial wound infections occurred in two German university hospitals between 1984 and 1991 (22). Infections by endemic N. farcinica manifested themselves not only as classical pulmonary or systemic disease but also as severe postoperative wound infections.
To investigate whether the organisms causing these infections were epidemiologically related and thus derived from a common source, further strain characterization was necessary. The genus Nocardia can be identified to the species level by a combination of chemotaxonomy (18, 19), hydrolysis, and other biochemical tests (2, 4, 10). Recently, PCR-based methods (24, 25) have been reported. Demonstration of epidemiological relationships between different clinical isolates, however, demands differentiation of these isolates below the species level. Pulsed-field gel electrophoresis (PFGE) is one of the strongest discriminatory DNA-based typing methods (12) and has gained broad application in characterizing epidemiologically related isolates. In this study, we established a PFGE protocol for the differentiation of N. farcinica isolates and here provide evidence that genetically related members of a strain of N. farcinica caused nosocomial infections in a surgical department of a German university hospital between 1985 and 1995.
MATERIALS AND METHODS
Bacterial isolates.
All 27 clinical N. farcinica isolates obtained from Würzburg, Germany, in the last 11 years were included in this study. As a control for epidemiologically unrelated strains, we also characterized seven clinical isolates from other towns in Germany and Austria. Air sampling was done with a slit sampler (Casella Ltd., London, United Kingdom). Samples of 150 liters of air were collected at a height of 1 m in various rooms of the Surgical Clinic [SC], as in other buildings on the hospital campus of the Universität Würzburg. In addition, isolation of nocardiae was also attempted by exposing open agar plates to air for 1 h. For isolating the nocardiae, we used two blood agar plates, one chocolate agar plate containing 10 mg of vancomycin per liter, and two selective agar plates containing 1 g of Na2HPO4 per liter, 0.5 g of MgSO4 per liter, 2.5 g of KNO3 per liter, 2.5 g of sodium-acetate per liter, 5.0 g of sodium propionate per liter, 10 g of Casitone (free of vitamins) per liter, 20 g of agar [pH 7.5] per liter, and 0.017% K+-tellurit (17). One blood agar plate, one chocolate agar plate, and one selective agar plate were incubated at 37°C for 48 h, and the other plates were incubated at 46°C for 48 h. Colonies grown on blood agar at 37°C were counted to determine the amounts of microorganisms in the air samples, and Nocardia-like colonies were subcultivated for species identification. Isolation from soil was done with paraffin baiting cultures (15), followed by subcultivation on blood agar or selective agar.
Species identification. (i) Physiological characteristics.
The ability of an organism to decompose adenine, guanine, hypoxanthine, xanthine, tyrosine, elastin, keratin, and testosterone was tested by the method of Gordon and Smith (7). The test to determine esculin decomposition was performed by the method of Gordon (6), and casein and gelatin hydrolysis tests were performed by the method of Gordon and Mihm (8). Tests to determine the use of various substrates as carbon sources or as simultaneous carbon and nitrogen sources were performed as previously described (27).
(ii) Mycolic acid analysis.
Mycolic acids were detected by using acid methanolysates of dry bacteria (30 mg) as described in reference 14. Analytical thin-layer chromatography (TLC) was performed with Merck 5554 silica gel 60F254 aluminum sheets. A triple development with petroleum ether (boiling point, 60 to 80°C)-acetone (95:5, vol/vol) was used. The presence of nocardomycolic acid was revealed by spraying with 10% ethanolic phosphomolybdic acid followed by heating at 150°C.
(iii) Pyrolysis gas chromatography.
Methylmycolates were prepared by acid methanolysis and were separated by preparative TLC. The purified mycolic acid methylesters were subjected to pyrolysis gas chromatography with a Shimadzu GC-14A gas chromatograph.
(iv) Antibiotic susceptibility tests.
The susceptibilities of the organisms to various antibiotics were studied by using the agar dilution technique. The results were determined microscopically as described in reference 21. The following antibiotics were tested: ampicillin, amoxicillin plus clavulanic acid, imipenem, cefotaxime, gentamicin, amikacin, cotrimoxazole, and erythromycin.
PFGE.
Freshly grown colonies were cultured in 10 ml of brain heart infusion liquid medium at 37°C under vigorous shaking until the medium showed homogeneous turbidity (2 to 14 days). A 4-h subculture of 2.5 ml of bacteria in 10 ml of brain heart infusion liquid medium containing 15% sucrose and 2% glycine followed to sensitize the bacteria against lysozyme (23). Bacteria were washed two times in 5 ml of TEN (0.1 M Tris-Cl [pH 8], 0.1 M EDTA, 0.15 M NaCl) and were subsequently resuspended in an appropriate amount of EC buffer (6 mM Tris-Cl [pH 7.6], 0.1 M EDTA [pH 7.6], 1 M NaCl, 1% sodium lauryl sarcosine, 0.2% sodium-deoxycholate) to yield a suspension of about 2 × 108 cells per ml. The suspension was mixed with an equal volume of molten 1% agarose (low-melting preparative-grade agarose; Bio-Rad, Munich, Germany), and 250-μl agarose blocks were cast. Blocks were incubated overnight at 37°C in 1 ml of EC buffer containing 20 mg of lysozyme (Boehringer, Mannheim, Germany) per ml. On the next day, the EC buffer was replaced with 1 ml of ESP buffer (0.5 M EDTA [pH 9.5], 1% sodium lauryl sarcosine, 1 mg of proteinase K) and the blocks were incubated at 50°C for 24 h. The ESP buffer was changed, and incubation was repeated for a further 24 h. Blocks were washed three times for 2 h in 15 ml of TE buffer (0.1 M Tris-Cl [pH 8], 1 mM EDTA) and stored until use. About a fifth of an agarose block was used for overnight restriction enzyme digestion with 20 U of restriction enzyme AseI (New England Biolabs, Bad Schwalbach, Germany) in 200 μl of enzyme buffer. Blocks were washed two times for 30 min in 1 ml of TE buffer and loaded onto a 1% agarose gel (Seakem LE Agarose; Biozym, Hameln, Germany). Lambda concatemers (lambda ladder; New England Biolabs) were used as size markers. PFGE was performed on a CHEF-DRII system (Bio-Rad), with pulse times increasing linearly from 5 to 50 s during the 26-h run. Voltage was constant at 200 V. Gels were stained with 0.4 μg of ethidium bromide per ml and photographed under UV light.
Interpretation of DNA fragment patterns.
The restriction fragment patterns of the PFGE analysis were grouped according to proposed guidelines (26) with a slight modification. Briefly, the predominant pattern was designated endemic type A, and isolates with one chromosomal band or up to three different chromosomal bands were included in this endemic strain. Patterns with four to six differing fragments were considered possibly related types (designated A1, A2, etc.). Isolates with patterns differing in more bands were considered unrelated (designated type B, type C, etc.).
RESULTS
Bacterial isolates.
Between 1984 and 1996, 27 clinical isolates of N. farcinica were obtained at the University Hospital of Würzburg, Germany. Twenty of these isolates were derived from patients with postoperative wound and other infections at the SC; the other seven isolates came from various other departments on the university hospital campus. As a further control for epidemiologically unrelated strains, seven additional isolates of N. farcinica were included in this study. These strains were derived from sporadic infections at other towns in Germany and Austria. All investigated strains, clinical diagnoses, sources from which the bacteria had been isolated, and departments where the patients were hospitalized are summarized in Table 1.
TABLE 1.
Epidemiology and characterization of N. farcinica isolates
| Straina | Year | Source | Diagnosisb | Locationc | Hospitald | Type | No. of plasmid bands in PFGE |
|---|---|---|---|---|---|---|---|
| D82 | 1984 | Blood | P.op. bronchial carcinoma | Würzburg | SC | B | 0 |
| D105 | 1984 | Blood | P.op. heart valve replacement | Würzburg | SC | B | 0 |
| D146 | 1985 | Abscess | P.op. splenectomy, lymphoma | Würzburg | SC | A | 2 |
| D158 | 1985 | Pus punctate | P.op. dermal dislocation | Würzburg | SC | A | 1 |
| D160 | 1986 | Pus punctate | P.op. heart valve replacement | Würzburg | SC | A | 1 |
| D161 | 1986 | Ascites fluid | P.op. splenectomy | Würzburg | SC | A | 1 |
| D173 | 1986 | Air of operation ward | Würzburg | SC | A | 1 | |
| D185 | 1986 | Renal abscess | P.op. renal transplant | Würzburg | SC | A | 1 |
| D201 | 1986 | Air of operation ward | Würzburg | SC | A | 2 | |
| D222 | 1986 | Pus | P.op. open fracture, osteitis | Würzburg | SC | A | 0 |
| D234 | 1987 | Pus near aorta ascendens | P.op. bypass | Würzburg | SC | A | 3 |
| D463 | 1989 | Hematoma | P.op. aortal aneurysm | Würzburg | SC | A | 2 |
| D549 | 1990 | Abdominal abscess | P.op. colectomy | Würzburg | SC | A | 2 |
| D621 | 1990 | Wound abscess | P.op. renal transplantation | Würzburg | SC | A | 3 |
| D631 | 1990 | Pulmonary abscess | P.op. renal transplantation | Würzburg | SC | A | 0 |
| D643 | 1990 | Wound abscess | P.op. esophagus resection | Würzburg | SC | A | 3 |
| D675 | 1990 | Wound abscess | P.op. polytrauma | Würzburg | SC | A | 2 |
| D691 | 1991 | Abdominal abscess | P.op. cystoplasty | Würzburg | SC | A | 3 |
| D745 | 1991 | Wound abscess | P.op. bypass | Würzburg | SC | A | 0 |
| D749 | 1991 | Abdominal abscess | P.op. rectum resection | Würzburg | SC | A | 2 |
| D1353 | 1995 | Wound abscess | P.op. renal transplant | Würzburg | SC | A | 1 |
| D1355 | 1995 | Wound abscess | P.op. laparotomy | Würzburg | SC | A | 2 |
| D729 | 1991 | Wound abscess | P.op. ovarial carcinoma | Würzburg | GC | A | 2 |
| D223 | 1986 | Air of corridor | Würzburg | IM | C | 0 | |
| D521 | 1990 | Soil surrounding IM | Würzburg | IM | C | 0 | |
| D110 | 1985 | Abscess | Pneumonia, myeloma | Würzburg | MC | A | 1 |
| D179 | 1986 | Pulmonary abscess | AIDS | Würzburg | MC | A | 1 |
| D1372 | 1995 | Bronchial secretion | Bronchial carcinoma | Würzburg | MC | A1 | 0 |
| D251 | 1987 | Brain abscess | NN | Würzburg | NC | D | 0 |
| D362 | 1988 | Pulmonary abscess | P.op. pneumonia, fibroblastoma | Würzburg | NC | A | 0 |
| D1408 | 1995 | Pericardial punctate | Leukemia | Würzburg | OC | A | 0 |
| D600 | 1990 | Peritoneal abscess | NN | Passau | E | 0 | |
| D692 | 1991 | Blood | Polyarthritis | Innsbruck | F | 0 | |
| D1132 | 1993 | BALe fluid | Pneumonia | Weingarten | G | 0 | |
| D1148 | 1993 | Punctate | Hematoma | Stuttgart | I | 5 | |
| D1170 | 1993 | Brain abscess | NN | Osnabrück | J | 0 | |
| D1146 | 1993 | Pleura punctate | NN | Kassel | H | 1 | |
| D1174 | 1993 | Swab | NN | Bochum | K | 2 |
Internal laboratory strain number.
P.op, postoperation; NN, not noted.
All towns are in Germany, except for Innsbruck, which is in Austria.
The hospital department is indicated for isolates from Würzburg. GC, Gynecological Clinic; IM, Institute for Medical Microbiology; NC, Neurological Clinic; OC, Outpatient Clinic.
BAL, broncho alveolar lavage.
Environmental isolates.
Studies of the air in various buildings of the hospital campus were done between 1986 and 1990. In the SC, two isolates of N. farcinica were obtained by air sampling. Isolate D173 was isolated from a technical storeroom of the operation suite, which is open to the room where the operations took place. Isolate D201 was from the dressing room of the operation suite at the same building. Air sampling at other hospitals did not yield N. farcinica isolates. The CFU in the air of the operation suite or intensive care units in the SC were two- to threefold higher (134 to 804 CFU/m3) than those of other operation suites or intensive care units on the hospital campus (20 to 402 CFU/m3). The highest concentration (1,300 CFU/m3) was measured in the dressing rooms of the operation suite of the SC. At the Institute for Medical Microbiology (IM), which is located on the same hospital campus, N. farcinica D223 was isolated by exposing a blood agar plate for 1 h to dusty air which was a result of renovation. Strain D521 was isolated from the soil surrounding the IM.
Species identification.
Isolates were identified as N. farcinica by physiological characterization, mycolic acid analysis, pyrolysis gas chromatography, and antibiotic susceptibility testing.
(i) Physiological characteristics.
The studied strains decomposed esculin, urea, and testosterone but not adenine, casein, elastin, guanine, hypoxanthine, tyrosine, xanthine, gelatin, and keratin. Glucose, rhamnose, acetate, paraffin, 2,3-butandiol, 1,2-propandiol, and amyl alcohol were utilized as sole sources of carbon and energy, but arabinose, galactose, lactose, benzoate, citrate, gluconate, adonitol, inositol, erythritol, mannitol, sorbitol, and xylose were not utilized. m-Hydroxybenzoate was used by some strains as a carbon source. All strains used acetamide as a simultaneous carbon and nitrogen source but not alanine, proline, and serine.
(ii) Lipid analysis.
By TLC, the strains studied showed a simple mycolic acid pattern consisting of only one spot, which corresponded to α-mycolate. By pyrolysis, these mycolates released three short-chain fatty acids, namely, C14:0, C16:0, and C18:0, with C16:0 being the major cleavage product. The natures of the pyrolysis products confirmed that the mycolate is of the nocardomycolic acid type.
(iii) Antibiotic susceptibility.
All studied strains were strongly inhibited by amikacin (MIC, 1.56 to 3.13 mg/liter) and weakly inhibited by imipenem (MIC, 3.13 to 6.26 mg/liter). They were resistant to ampicillin, amoxicillin plus clavulanic acid, cefotaxime, gentamicin, cotrimoxazole, and erythromycin. However, some strains were occasionally weakly inhibited by amoxicillin plus clavulanic acid.
PFGE.
The purpose of this study was to characterize the genetic relatedness of epidemiologically related isolates of Nocardia farcinica from patients with postoperative wound infections. Ideally for PFGE analysis, appropriate restriction enzyme digestion should result in 10 to 20 chromosomal fragments which are clearly resolved by PFGE. We tested several restriction enzymes with AT-rich recognition sequences (AseI, DraI, and SspI), as they were expected to cut rarely the nocardial genome because of its high C+G content. AseI gave the best results, producing 10 to 18 distinct chromosomal fragments which allowed us to group the isolates into distinct genotypes.
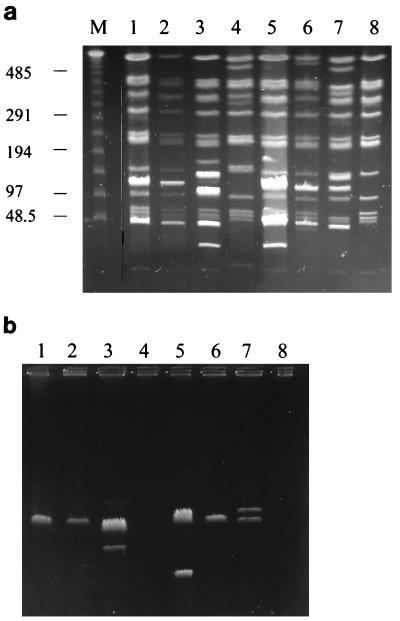
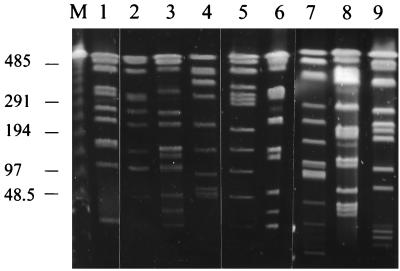
PFGE analysis was carried out with 38 isolates of N. farcinica: 31 from Würzburg, Germany, and 7 from other towns. Most of the isolates from Würzburg were from patients with postoperative wound infections. Table 1 summarizes the PFGE results of all investigated strains. Of the 31 Würzburgian isolates, 26 could be grouped with the predominant type A or related types (Table 1). Representative examples of type A restriction fragment patterns are shown in Fig. 1a. Sometimes, bright bands appeared in an otherwise uniform band pattern (Fig. 1a). In such cases, plasmids could be detected by subjecting the uncut DNA to PFGE (Fig. 1b). There was no correlation between clinical disease or location of an infected patient and plasmid pattern. Type A isolates showed considerable variation in plasmid patterns. The number of plasmid bands was determined for each isolate (Table 1) by subjecting the uncut DNA to PFGE, and bright bands, obviously produced by plasmids, were not considered for typing. Despite the problems in pattern recognition caused by various plasmids, we could clearly identify the predominant type A. This type represented all isolates from patients with postoperative wound infections at the SC in Würzburg since 1985 (Table 1). PFGE, however, was clearly suited to differentiate various genotypes of N. farcinica, since controls of epidemiologically unrelated isolates from other towns in Germany and Austria showed in each case a different genotype (Fig. 2).
FIG. 1.
PFGE analysis of representative N. farcinica type A isolates. DNA fragments were separated after digestion with AseI (a), or undigested DNA was run under the same PFGE conditions (b). Positions of DNA size markers are indicated on the left. Lanes: M, lambda DNA size markers; 1 to 8, D160, D161, and D234 from the SC, D362 from the Neurological Clinic, D729 from the Gynecological Clinic, D173 and D201 from the SC environment, and D1408 from the Outpatient Clinic, respectively. Sizes are given in kilobase pairs.
FIG. 2.
Representative AseI restriction patterns of distinct N. farcinica genotypes. Lanes: M, lambda DNA size markers; 1, D82 (type B); 2, D223 (type C); 3, D600 (type E); 4, D631 (type A); 5, D1132 (type G); 6, D1146 (type H); 7, D1148 (type I); 8, D1170 (type J); 9, D1174 (type K). Sizes are given in kilobase pairs.
Four environmental isolates of N. farcinica were obtained from air samples of various buildings of the hospital campus and from the soil surrounding these buildings. Two airborne isolates (D173 and D201) from rooms in the operation suite of the SC, where the type A infections occurred, were also type A (Fig. 1, lanes 6 and 7, respectively). In contrast, two environmental isolates from the IM (D223 and D521), which is located on the same campus, belonged to another genotype (type C) (Fig. 2, lane 2).
Analysis of seven isolates, collected from patients from other departments of the University Hospital at Würzburg, revealed in one case a distinct genotype (D251). Another was possibly related to type A (D1372), and five cases were also type A (D110, D179, D362, D729, and D1408). Three of these were pulmonary infections of immunocompromised patients who were hospitalized in a department located near the SC (D110, D179, D1372). The spread of type A to these patients is discussed below.
DISCUSSION
Hospital-acquired nocardiosis seems to be an emerging problem (13, 22). The goal of this study was to provide evidence that epidemiologically related isolates of N. farcinica from patients with postoperative wound infections are also genetically related and are therefore endemic strains. This information is helpful for understanding the spread of disease in both hospitals and communities. As nocardiosis is a rather rare disease and the isolates were collected over a period of 11 years, a strain of the predominant type, A, was considered an endemic strain rather than an outbreak strain on the basis of proposed guidelines (26). Endemic strains are temporally more distant than strains of a temporally limited outbreak of disease. As spontaneous mutations probably occur over a longer period in endemic strains than in limited outbreak strains and change the PFGE pattern of the same bacterial population over time, an endemic strain does not fulfill the strict criteria for identity that an outbreak strain is expected to meet. Therefore, we modified slightly the criteria proposed for isolates of short-term epidemics (26): isolates with one genetic change (two to three different bands) were grouped with the endemic strain (type A) and isolates with two genetic differences (four to six different bands) were considered related (type A1, type A2, etc.).
The bright bands sometimes overlaying the chromosomal fragments could be certainly identified as plasmids by running uncut DNA in PFGE. While the large uncut bacterial chromosome fails to migrate in PFGE and remains in the sample well of the gel under these conditions, small circular plasmids will form distinct bands (3, 11). Moreover, PFGE also reveals the large-sized (about several hundred kilobases) linear plasmids of actinomycetes (9), which migrate in PFGE according to the linear fragments of the size marker. However, PFGE is not useful for detection of circular plasmids larger than 40 kb (3, 11), as they do not migrate during PFGE. Migration of circular DNA is severely restricted in PFGE, and circular plasmids as small as 5 kb form bands at the range of the lambda ladder size marker, so size estimation of circular plasmids is hardly possible by PFGE. However, extensive characterization of plasmids was not the aim of this study. Plasmid characterization was the subject of a recent study (16) which showed that pathogenic strains of Nocardia do not necessarily contain plasmids. This finding suggests that plasmids are not directly involved in virulence. In our study, plasmid patterns frequently differed in isolates of the endemic type A, so we found no correlation between the plasmid profile and the type of disease or a certain infection route. Despite the various bands caused by plasmids, we were able to identify clearly an endemic strain among our isolates.
The isolates from Würzburg belonged exclusively to type A, while isolates from other towns revealed distinct genotypes. Only one isolate (D1170) (Fig. 2, lane 8) from another town showed a certain similarity to type A, but it did not meet the criteria for being a related strain. Computer-aided interpretation of fragment patterns should more precisely describe the relatedness between the various genotypes. All isolates from patients with postoperative wound infections at the SC between 1985 and 1995 were type A. Only two older isolates (D82 and D110) from this department belonged to another genotype (type B). Type B seems to have been replaced by type A, because type B was never isolated in subsequent years. The fact that nocardiae were isolated from operation wounds (e.g., abdominal abscesses after colectomy or renal abscesses after renal transplantation) makes it likely that the bacteria had entered the wound during the operation itself (or during postoperative treatments).
Environmental isolations were done in order to describe the source of infection. An airborne isolate (D173) from a storeroom in the operating suite of the SC was type A, and another airborne isolate (D521) from the dressing room of the same department also belonged to type A. The congruence of the environmental isolates from the SC with the isolates from the patients with postoperative wound infections, together with the long period of time over which these infections occurred, suggests that these infections had been acquired in this hospital setting. In contrast, an airborne isolate (D223) from a corridor at the IM, which is located on the same campus, and another isolate (D201) from the soil surrounding the IM revealed a common genotype distinct from type A. Therefore, type A is not the only type found on the university hospital campus and distribution of type A seems to be rather restricted to a certain building and its surrounding.
During the period 1984 to 1991, when most (18 of 20) of the postoperative infections occurred, extensive renovation work took place at the old surgical hospital (SC) constructed in 1912. Only very few sporadic Nocardia infections had been observed before the reconstruction work started. So, a general lack of hygienic conditions in the old rooms cannot explain the infections due to N. farcinica. Since the reconstruction work was completed in 1991, only two postoperative infections by N. farcinica have been detected in this hospital, though no specific interventions were made to prevent the surgical N. farcinica infections. So the reconstruction work may have led to conditions favoring the spread of N. farcinica. Interestingly, an increase of surgical wound infections due to other bacteria (e.g., staphylococci or pseudomonads) was not documented during this period, which indicates that the conditions were rather specific for N. farcinica. In the air of the operation ward at the SC, the CFU of microorganisms were two- to threefold higher than those of operation wards at other buildings. We isolated N. farcinica from dusty air during the renovation of a wall in the IM, simply by exposing a blood-agar plate for 1 h. So it seems possible that N. farcinica was liberated from its niche in the building during the renovation work and may have spread in the dusty air, directly or through contaminated materials (e.g., gloves, clothing, dressing material, or operation tools) to the patients. We recognize that we may not yet have identified the definite niche where N. farcinica mainly occurs in this hospital setting, because other surfaces or materials were not investigated at this time. In our and others’ experience, N. farcinica can be isolated mainly from dry soils (5) and N. farcinica could be detected at high counts in dust derived from the reconstruction of an old half-timber house (20).
Infection with type A was, however, not strictly restricted to the SC, because this type was also isolated from six of seven patients hospitalized in other departments in the hospital at Würzburg. Three of these type A infections occurred at the Medical Clinic (MC), which is located in the vicinity of the SC. As the nocardial infections at the MC were pulmonary diseases of immunocompromised patients, aerogenic spread of type A from the proximate SC or from bacteria living in the soil surrounding these two departments is a likely mode of infection. The three other type A-infected patients were hospitalized in clearly distant departments of the hospital at Würzburg. The following explanations for the type A infections of the patients from the more distant departments are possible. (i) Though the two environmental isolates from a location other than the SC were not of type A, type A may be rather widely distributed in the Würzburg hospital and may have colonized surrounding departments too. (ii) Endemic type A may have been transmitted by instruments or persons from the SC to the other departments. (iii) The patients themselves may have been at the SC to receive pre- or postoperative treatments which were not documented. (iv) N. farcinica may be present, either constantly or occasionally, in materials (e.g., disinfectant fluid) or systems distributed throughout the hospital campus. Further environmental investigations are necessary to determine the exact distribution of the endemic prototype A in the Würzburg hospital.
The PFGE method presented here will probably be a well-suited tool for identifying genetically related strains of N. farcinica in future nocardial endemics or epidemics. Reliable typing methods are a prerequisite for detecting the infectious source and the mode of transmission. Our data indicate that it might be possible to differentiate sporadic infections, acquired in other towns, from the endemic strain occurring in a certain hospital setting by PFGE. Though we could not definitely identify the source of the infectious agents, it appears that aerogenic transmission of N. farcinica during operative or postoperative treatments has to be considered as a possible mode of transmission. Aerogenic spread to immunocompromised patients resulting in pulmonary nocardiosis seems to be a further problem in hospitals. As we noted, because of the correlation between the reconstruction work and the surgical infections by N. farcinica, precautions against nocardial infections should be taken in hospitals where such work is planned.
ACKNOWLEDGMENTS
We thank G. Klemm for photographical work and D. Gierth and I. Lux for technical assistance.
REFERENCES
- 1.Beaman, B. L., P. Boiron, L. Beaman, G. H. Brownell, K. Schaal, and M. E. Gombert. 1992. Nocardia and nocardiosis. J. Med. Vet. Mycol. 30(Suppl. 1):317–331. [PubMed]
- 2.Beaman B L, Saubolle M A, Wallace R J. Nocardia, Rhodococcus, Streptomyces, Oerskovia, and other aerobic actinomycetes of medical importance. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: ASM Press; 1995. pp. 379–399. [Google Scholar]
- 3.Beverley S M. Characterization of the unusual mobility of large circular DNAs in pulsed field-gradient electrophoresis. Nucleic Acids Res. 1988;16:925–939. doi: 10.1093/nar/16.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biehle J R, Cavalieri S J, Felland T, Zimmer B L. Novel method for identification of Nocardia species by detection of preformed enzymes. J Clin Microbiol. 1996;34:103–107. doi: 10.1128/jcm.34.1.103-107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cross T, Rowbotham T J, Mishustin E N, Zepper E Z, Antoine-Portaels F, Schaal K P, Bickenbach H. The ecology of nocardioform actinomycetes. In: Goodfellow M, Brownell G H, Serrano J A, editors. The biology of the nocardiae. New York, N.Y: Academic Press; 1976. pp. 337–371. [Google Scholar]
- 6.Gordon R E. Some criteria for the recognition of Nocardia madurae (Vincent) Blanchard. J Gen Microbiol. 1966;45:355–364. doi: 10.1099/00221287-45-2-355. [DOI] [PubMed] [Google Scholar]
- 7.Gordon R E, Smith M M. Proposed group of characters for the separation of Streptomyces and Nocardia. J Bacteriol. 1955;69:147–150. doi: 10.1128/jb.69.2.147-150.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon R E, Mihm J M. A comparative study of some strains received as nocardiae. J Bacteriol. 1957;73:15–27. doi: 10.1128/jb.73.1.15-27.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalkus J, Reh M, Schlegel H G. Hydrogen autotrophy of Nocardia opaca strains is encoded by linear megaplasmids. J Gen Microbiol. 1990;136:1145–1151. doi: 10.1099/00221287-136-6-1145. [DOI] [PubMed] [Google Scholar]
- 10.Lechevalier H A, Goodfellow M. Nocardioform actinomycetes. In: Holt J G, Krieg N R, Sneath P H A, Staly J T, Williams S T, editors. Bergey’s manual of determinative bacteriology. 9th ed. Baltimore, Md: The Williams & Wilkins Co.; 1994. pp. 625–650. [Google Scholar]
- 11.Levene S D, Zim B D. Separation of open-circular DNA using pulsed-field electrophoresis. Proc Natl Acad Sci USA. 1987;84:4045–4047. doi: 10.1073/pnas.84.12.4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maslow J N, Mulligan M E, Arbeit R D. Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin Infect Dis. 1993;17:153–164. doi: 10.1093/clinids/17.2.153. [DOI] [PubMed] [Google Scholar]
- 13.McNeil M M, Brown J M. The medically important aerobic actinomycetes: epidemiology and microbiology. Clin Microbiol Rev. 1994;7:375–417. doi: 10.1128/cmr.7.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minnikin D E, Hutchinson I A, Caldicott A B, Goodfellow M. Thin-layer chromatography of methanolysates of mycolic acid-containing bacteria. J Chromatogr. 1980;188:221–233. [Google Scholar]
- 15.Mishra S K, Randhawa H S. Application of paraffin bait technique to the isolation of Nocardia asteroides from clinical specimens. Appl Microbiol. 1969;18:686–687. doi: 10.1128/am.18.4.686-687.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Provost F, Blanc M V, Beaman B L, Boiron P. Occurrence of plasmids in pathogenic strains of Nocardia. J Med Microbiol. 1996;45:344–348. doi: 10.1099/00222615-45-5-344. [DOI] [PubMed] [Google Scholar]
- 17.Schaal K P. Zur mikrobiologischen Diagnose der Nocardiose. Zentralbl Bakteriol Hyg 1 Abt Orig A. 1972;220:242–246. [PubMed] [Google Scholar]
- 18.Schaal K P. Laboratory diagnosis of actinomycete diseases. In: Goodfellow M, Mordarski M, Williams S T, editors. The biology of the actinomycetes. London, United Kingdom: Academic Press; 1984. pp. 425–456. [Google Scholar]
- 19.Schaal K P. Identification of clinically significant actinomycetes and related bacteria, using chemical techniques. In: Goodfellow M, Minmikin D E, editors. Chemical methods in bacterial systematics. Orlando, Fla: Academic Press; 1985. pp. 359–381. [Google Scholar]
- 20.Schaal, K. P. 1991. Medical and microbiological problems arising from airborne infection in hospitals. J. Hosp. Infect. 18(Suppl. A):451–459. [DOI] [PMC free article] [PubMed]
- 21.Schaal K P, Schütt-Gerowitt H, Goldmann A. In vitro and in vivo studies on the efficacy of various antimicrobial agents in the treatment of human nocardiosis. In: Szabó G, Biró S, Goodfellow M, editors. Biological, biochemical and biomedical aspects of actinomycetes, part B. Budapest, Hungary: Académiai Kiadó; 1986. pp. 619–633. [Google Scholar]
- 22.Schaal K P, Lee H-J. Actinomycete infections in humans—a review. Gene. 1992;115:201–211. doi: 10.1016/0378-1119(92)90560-c. [DOI] [PubMed] [Google Scholar]
- 23.Sensfuss C, Reh M, Schlegel H G. No correlation exists between the conjugative transfer of the autotrophic character and that of plasmids in Nocardia opaca strains. J Gen Microbiol. 1986;132:997–1007. doi: 10.1099/00221287-132-4-997. [DOI] [PubMed] [Google Scholar]
- 24.Steingrube V A, Brown B A, Gibson J L, Wilson R W, Brown J, Blacklock Z, Jost K, Locke S, Ulrich R F, Wallace R J., Jr DNA amplification and restriction endonuclease analysis for differentiation of 12 species and taxa of Nocardia, including recognition of four new taxa within the Nocardia asteroides complex. J Clin Microbiol. 1995;33:3096–3101. doi: 10.1128/jcm.33.12.3096-3101.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steingrube V A, Wilson R W, Brown B A, Jost K C, Jr, Blacklock Z, Gibson J L, Wallace R J., Jr Rapid identification of clinically significant species and taxa of aerobic actinomycetes, including Actinomadura, Gordona, Nocardia, Rhodococcus, Streptomyces, and Tsukamurella isolates, by DNA amplification and restriction endonuclease analysis. J Clin Microbiol. 1997;35:817–822. doi: 10.1128/jcm.35.4.817-822.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yassin A F, Rainey F A, Brzezinka H, Burghardt J, Lee H L, Schaal K P. Tsukamurella inchonensis sp. nov. Int J Syst Bacteriol. 1995;45:522–527. doi: 10.1099/00207713-45-3-522. [DOI] [PubMed] [Google Scholar]