Abstract
Simple sample-processing methods for PCR detection of Porphyromonas gingivalis, a major pathogen causing adult periodontitis, from saliva were studied. The ability to detect P. gingivalis from 118 salivary samples by PCR after boiling and Chelex 100 processing was compared with bacterial culture. P. gingivalis was detected three times more often by PCR than by culture. Chelex 100 processing of saliva proved to be effective in preventing PCR inhibition and was applied to determine the occurrence of P. gingivalis in saliva samples from 263 Finnish subjects between 5 and 80 years of age. The occurrence of P. gingivalis increased with age, and it was detected by PCR in the saliva of 5.0% of subjects between 5 and 10 years of age, 13.8% of subjects between 11 and 20 years of age, 13.4% of subjects between 21 and 30 years of age, and 63.3% of subjects between 31 and 80 years of age. The results indicate that P. gingivalis is a rare finding in saliva from periodontally healthy children and young adults but a frequent one in saliva from adult periodontitis patients.
Porphyromonas gingivalis, a black-pigmented gram-negative anaerobic rod, is a major pathogen causing adult periodontitis (18). P. gingivalis is frequently isolated from subgingival plaque of periodontitis patients, whereas it can be cultured only occasionally from periodontally healthy adults and is usually not isolated from children (4, 8, 9, 13, 15, 25).
Saliva is the most probable vehicle for person-to-person transmission of oral bacteria. Thus, it is likely that the presence of P. gingivalis in saliva is a prerequisite for its transmission. Saliva represents an easily and noninvasively obtainable sample containing bacteria from all oral sites, e.g., the mucosa and supra- and subgingival plaque. Furthermore, it is also rather easily obtainable from the oral cavities of young children. However, the proportion of shed periodontal bacteria in the salivary microbiota is relatively low, a fact that makes bacterial culture an insensitive detection method. Selective media, such as kanamycin vancomycin laked blood agar, which are useful for isolation of other oral black-pigmented gram-negative anaerobes from polymicrobial sources, cannot be used for culturing P. gingivalis, since Porphyromonas spp. isolates are usually susceptible to vancomycin (11).
PCR allows the specific amplification of target bacterial DNA in samples for which the background caused by other species is high. Thus, PCR could be applicable for the detection of P. gingivalis from saliva. However, biological samples may contain compounds that are inhibitory to PCR amplification, leading to false-negative results (2, 10). Aside from that, PCR-based detection methods require proper validation and quality control to avoid false-positive results. PCR-based methods for detection of P. gingivalis from subgingival samples have been described earlier (3, 14, 21, 22, 26), whereas no reports on PCR detection of periodontal bacteria from salivary samples are available.
This paper describes a simple sample-processing method which can be used to prevent PCR inhibition by saliva. The efficacy of the present PCR method for detection of P. gingivalis from salivary samples was compared with that of bacterial culture. Additionally, the present PCR method was applied to determine the occurrence of P. gingivalis in saliva specimens from Finnish subjects 5 to 80 years of age.
MATERIALS AND METHODS
Subjects, sampling, and bacterial culture.
The material used in this study consisted of salivary samples from 263 periodontally healthy or diseased subjects (age range, 5 to 80 years). The periodontal status of a subject was defined as healthy when no signs of periodontal breakdown were found. Periodontitis was diagnosed when the presence of periodontal breakdown was verified in a clinical and/or radiological examination. Periodontitis was further classified according to guidelines of the American Academy of Periodontology (1).
A subset of 118 samples was used for evaluation of the PCR method. The occurrence of P. gingivalis in Finnish subjects as determined by PCR was studied using all 263 saliva samples. The samples were collected during several previous studies, between 1985 and 1996, by using paraffin chewing stimulation and preserved at −70°C until used in the present study. For the detection of P. gingivalis by bacterial culture, 142 of the 263 saliva samples had been serially diluted immediately after sampling and cultured on Brucella agar (BBL Microbiology Systems, Cockeysville, Md.) supplemented with lysed horse blood (5%), hemin (5 μg/ml), and vitamin K1 (10 μg/ml). The plates were incubated anaerobically in jars filled by the evacuation-replacement method with a mixture of gases (85% N2, 10% H2, 5% CO2). The isolates were identified as P. gingivalis on the basis of having the typical colony color and morphology, lacking colony autofluorescence, having positive trypsin-like enzyme activity (19), and having a positive indole reaction.
Sample processing for PCR.
Two rapid methods of sample processing for PCR were tested.
(i) Boiling.
A 30-μl aliquot of each of the 118 saliva specimens was boiled for 10 min and then centrifuged at 10,000 × g for 5 min, and 5 μl (or 0.5 μl) of the supernatant was used as a template for PCR.
(ii) Chelex 100 treatment.
A 50-μl aliquot of each of the 263 samples of saliva was incubated with 12.5 μl of 25% (wt/vol) Chelex 100 (Bio-Rad Laboratories, Hercules, Calif.), a cation-chelating resin, at 56°C for 30 min before being boiled and centrifuged as described above. A 6-μl aliquot of the supernatant was then used as a template for PCR.
PCR amplification.
Two P. gingivalis-specific primers described by Slots et al. (21) were used to amplify a 404-bp fragment of the 16S rRNA gene: primer 1 (5′-AGG CAG CTT GCC ATA CTG CG-3′) and primer 2 (5′-ACT GTT AGC AAC TAC CGA TGT-3′). The specificity of the PCR method was investigated by using purified DNA from 26 clinical P. gingivalis isolates from unrelated subjects and that from 34 isolates of 30 species/genera other than P. gingivalis as templates for PCR. The latter included Porphyromonas asaccharolytica ATCC 25260, four clinical isolates of P. asaccharolytica (AHN 1728, AHN 10916, AHN 10803, and AHN 10927), Porphyromonas endodontalis ATCC 35406, Porphyromonas macacae ATCC 33141, Porphyromonas salivosa NCTC 11632 (reclassified as P. macacae), Porphyromonas levii ATCC 29147, Porphyromonas canoris NCTC 12835, Porphyromonas cangingivalis AHN 4138, Porphyromonas cansulci AHN 4364, Prevotella intermedia ATCC 25611, Prevotella nigrescens ATCC 33563, Prevotella corporis ATCC 33547, Prevotella denticola AHN 9656, Prevotella loescheii AHN 9806, Prevotella melaninogenica 25AA, Prevotella oris ATCC 33573, Fusobacterium nucleatum ATCC 25586, Fusobacterium naviforme AHN 9610, Selenomonas sp. AHN 9988, Capnocytophaga sp. AHN 10373, Bacteroides forsythus R878D, Bacteroides gracilis AHN 9641, Campylobacter rectus ATCC 33238, Campylobacter concisus ATCC 33237, Eikenella corrodens AHN 9363, Veillonella sp. AHN 5836, Actinobacillus actinomycetemcomitans ATCC 29523, Haemophilus aphrophilus 1659, Peptostreptococcus micros D3Ja, Streptococcus mutans 75.3, and Streptococcus sobrinus 475.4.
Amplification reactions were performed in a total volume of 50 μl consisting of 0.2 mM each deoxynucleoside triphosphate (dATP, dTTP, dCTP, and dGTP; Pharmacia LKB, Piscataway, N.J.), 1.5 mM MgCl2, 10× Taq buffer (Perkin-Elmer), 1 μM each primer, 2.5 U of AmpliTaq polymerase (Perkin-Elmer Cetus, Norwalk, Conn.), and 0.5 to 6 μl of template overlaid with mineral oil. PCR amplification was performed in a thermocycler (Perkin-Elmer Cetus). Cycling parameters were as follows: an initial denaturation at 95°C for 1 min; 36 cycles consisting of 95°C for 30 s, 65°C for 1 min, and 72°C for 1 min; and a final extension at 72°C for 2 min.
To avoid contamination during PCR amplification, the reagents were premixed and sterile tips with aerosol barriers were used. P. gingivalis ATCC 33277 cells (50 cells per PCR) were used as a positive control for the PCR, and 5 μl of water constituted the negative control. Positive and negative controls were included in each PCR set and in all sample processings.
The amplification products were subjected to electrophoresis in a 1% agarose gel containing ethidium bromide (0.5 μg/ml) and photographed under UV illumination by using the Polaroid MP4 system. A 1-kb DNA ladder (Gibco BRL Life Technologies, Inc., Gaithersburg, Md.) was used as a molecular size standard.
PCR detection limits and inhibitory effect of saliva on PCR.
The detection limits of PCR after boiling and after Chelex 100 processing were determined by using known numbers of P. gingivalis ATCC 33277 cells (0, 1, 2, 5, 50, and 500 cells per PCR as determined by viable-cell counts) suspended in sterile distilled water or in saliva with no cultivable P. gingivalis.
To investigate the possible inhibition of PCR amplification by saliva, P. gingivalis ATCC 33277 cells (50 cells per PCR) were added to 118 saliva samples processed by the boiling method. Inhibition was recorded when no or a very weak amplification signal was detected. The samples that showed inhibition when processed by boiling were analyzed additionally by spiking a 10-fold dilution of saliva processed by boiling, as well as by Chelex 100 processing of spiked saliva.
RESULTS
Specificity of PCR.
Both the specificity and sensitivity of the PCR primers were 100%. No amplification was detected for any of the 34 isolates that were not P. gingivalis, whereas all 26 clinical P. gingivalis isolates gave an amplification product of the expected size.
Comparison of bacterial culture and PCR techniques.
PCR, after both boiling and Chelex 100 processing, was used on 118 salivary samples that were also cultured for the presence of P. gingivalis (Table 1). P. gingivalis-positive samples were detected three times more often by PCR than by bacterial culture: 11 of 118 (9.3%) samples were P. gingivalis-positive by culture, whereas 40 of 118 (33.9%) samples were P. gingivalis-positive by PCR after being boiled and 37 of 118 (31.4%) were positive by PCR after being subjected to Chelex 100 processing. The PCR results obtained after Chelex 100 processing of the sample correlated better with bacterial culture than did those obtained after boiling, since all 11 culture-positive samples were also positive by PCR after Chelex 100 processing whereas 4 culture-positive samples failed to give amplification products after being processed by boiling (Table 1).
TABLE 1.
Detection of P. gingivalis in saliva samples from 118 subjects by bacterial culture and PCR
| PCR result | Culture result
|
Total | |
|---|---|---|---|
| No. positive (%) | No. negative (%) | ||
| After boiling | |||
| Positive | 7 (5.9) | 33 (28.0) | 40 |
| Negative | 4 (3.4) | 74 (63.8) | 78 |
| Total | 11 | 107 | 118 |
| After Chelex 100 treatment | |||
| Positive | 11 (9.3) | 26 (22.0) | 37 |
| Negative | 0 | 81 (68.6) | 81 |
| Total | 11 | 107 | 118 |
There was some discrepancy between the results obtained after sample boiling and those obtained after Chelex 100 processing (Table 2). Of the 118 samples, 28 (23.7%) were P. gingivalis-positive and 69 (58.5%) were P. gingivalis-negative by both methods, whereas 21 (17.8%) samples repeatedly gave discrepant results.
TABLE 2.
Comparison of two PCR sample processing methods used for the detection of P. gingivalis in saliva samples from 118 subjects
| PCR result after Chelex 100 treatment | PCR result after boiling
|
Total | |
|---|---|---|---|
| No. positive (%) | No. negative (%) | ||
| Positive | 28 (23.7) | 9 (7.6) | 37 |
| Negative | 12 (10.2) | 69 (58.5) | 81 |
| Total | 40 | 78 | 118 |
Inhibitory effect of saliva on PCR.
Of the 118 saliva samples processed by the boiling method, 23 (19.5%) showed a distinct inhibition of PCR amplification when 50 P. gingivalis cells were added to each 50-μl PCR mixture. Dilution of the saliva decreased the inhibitory effect, since all but 3 of the 23 inhibitory samples showed good amplification when 0.5 μl instead of 5 μl of saliva (in both cases with 50 P. gingivalis cells) was used as a template for PCR. Although dilution resulted in amplification of most of the 23 salivary samples, the simultaneously occurring 10-fold rise in the detection level is not desirable. When Chelex 100 was applied to process the 23 samples that showed inhibition after boiling, 18 samples gave distinct amplicons and 5 samples gave weak amplicons.
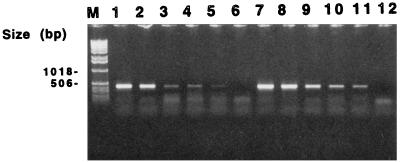
The detection limit of the PCR after boiling and after Chelex 100 processing of the sample was one P. gingivalis cell per PCR in water. The detection limit in saliva showing no PCR inhibition was also one cell per PCR. However, the amplification signal obtained with a few cells (1 to 5 cells per PCR) was constantly weaker in saliva than in water (Fig. 1).
FIG. 1.
Detection limit of PCR after Chelex 100 processing. Lanes 1 to 6, 500, 50, 5, 2, 1, and 0 P. gingivalis ATCC 33277 cells in saliva per PCR, respectively; lanes 7 to 12, 500, 50, 5, 2, 1, and 0 P. gingivalis cells in water per PCR, respectively; M, 1-kb DNA ladder.
Occurrence of P. gingivalis.
Since Chelex 100 processing of the saliva samples decreased PCR inhibition most effectively, it was used to investigate the occurrence of P. gingivalis in 263 Finnish subjects from 5 to 80 years of age (Table 3). The saliva samples from 142 of these 263 subjects were also cultured for the presence of P. gingivalis (Table 3). The occurrence of P. gingivalis increased with age, since the organism was detected by PCR in 3 (5.0%) of the 60 subjects between 5 and 10 years of age, in 12 (13.8%) of the 87 subjects between 11 and 20 years of age, in 9 (13.4%) of the 67 subjects between 21 and 30 years of age, and in 31 (63.3%) of the 49 subjects between 31 and 80 years of age. P. gingivalis was not found by bacterial culture in any of the 9 subjects between 5 and 10 years of age, whereas it was isolated from 3 (6.1%) of the 49 subjects between 11 and 20 years of age, from 1 (2.3%) of the 44 subjects between 21 and 30 years of age, and from 7 (17.5%) of the 40 subjects between 31 and 80 years of age (Table 3).
TABLE 3.
Occurrence of P. gingivalis in Finnish subjects with different periodontal statuses as determined by PCR after Chelex 100 processing and by bacterial culture
| Mean age (range), in yr | Periodontal status | Detection by:
|
|||
|---|---|---|---|---|---|
| PCR
|
Culture
|
||||
| n | No. positive (%) | n | No. positive (%) | ||
| 5.6 (5–10) | Healthy | 59 | 3 (5.1) | 8 | 0 |
| Prepubertal periodontitis | 1 | 0 | 1 | 0 | |
| 15.2 (11–20) | Healthy | 83 | 11 (13.3) | 45 | 2 (4.4) |
| Localized juvenile periodontitis | 3 | 0 | 3 | 0 | |
| Diabetes-associated periodontitis | 1 | 1 | 1 | 1 (100) | |
| 24.5 (21–30) | Healthy | 34 | 2 (5.9) | 16 | 0 |
| Localized juvenile periodontitis | 16 | 2 (12.5) | 14 | 0 | |
| Early-onset periodontitis | 8 | 4 (50.0) | 8 | 1 (12.5) | |
| No data | 9 | 1 (11.1) | 6 | 0 | |
| 42.8 (31–80) | Healthy | 9 | 3 (33.3) | 9 | 0 |
| Adult periodontitis | 40 | 28 (70) | 31 | 7 (22.6) | |
| 22.5 (5–80) | 263 | 55 (20.9) | 142 | 11 (7.7) | |
Data on periodontal status were available for 254 subjects. P. gingivalis was detected by PCR in 19 (10.3%) of the 185 periodontally healthy subjects; in 6 (21%) of the 28 subjects with prepubertal, localized juvenile periodontitis or some other type of early-onset periodontitis; and in 28 (70%) of the 40 subjects with adult periodontitis. The corresponding figures obtained by bacterial culture were 2 (2.6%) of 78, 1 (3.8%) of 26, and 7 (22.6%) of 31 subjects (Table 3).
DISCUSSION
In the present study, 20% of the saliva samples showed PCR inhibition when the boiling method was used for sample processing. PCR inhibition of other biological samples (e.g., sputum, stool, and genital ulcer specimens) is well known, and different methods (e.g., immunomagnetic separation, phenol-chloroform extraction, and use of capture resins) have been suggested for the inactivation of PCR inhibitors (2, 10, 12). However, many of these other methods are laborious and expensive. In the present study, a simple sample-processing technique involving the use of Chelex 100 resin prior to PCR amplification proved to be very applicable for the detection of P. gingivalis in salivary samples. In the Chelex 100 processing method, only one reagent was added to the sample and the processing was performed in a single tube, which minimizes the number of steps involving handling and hence the risk of contamination. The applicability of Chelex 100 for inactivating PCR inhibitors in saliva probably stems from the ability of Chelex 100 to chelate divalent ions. Chelex 100 has previously been shown to enhance the efficiency of DNA extraction, especially from gram-positive and acid-fast bacteria, and to protect DNA at high temperatures (6).
In the present study, some discrepancies in PCR detection of P. gingivalis from saliva were observed when the two sample preparation methods were compared. Why some samples were P. gingivalis-positive by PCR after being subjected to Chelex 100 processing but P. gingivalis-negative by PCR after being boiled can be explained by the better ability of the Chelex 100 method to abolish PCR inhibition by saliva. The existence of samples that were P. gingivalis-positive by bacterial culture but P. gingivalis-negative by PCR after being boiled supports this suggestion. The reason why some samples were P. gingivalis-negative by PCR after being subjected to Chelex 100 processing but P. gingivalis-positive by PCR after being boiled remained unknown. The results were reproducible, and the finding can be explained neither by differences in the sensitivities of the two methods nor by the uneven distribution of very low numbers of P. gingivalis cells in the samples.
In the present study, P. gingivalis was detected in saliva samples three times more often by PCR than by bacterial culture, which is well in accordance with data from earlier PCR studies using the same primers for the detection of P. gingivalis in subgingival plaque samples (3, 21). Also, in other studies using primers targeted to different regions of the 16S rRNA gene or to the collagenase gene, P. gingivalis has been detected in subgingival samples more frequently by PCR than by bacterial culture (21, 26). The higher rate of detection by PCR is most likely due to the higher sensitivity of the PCR technique, which is especially important in studies on mixed bacterial flora. In addition, in contrast to bacterial culture, PCR amplification also detects nonviable bacterial cells present in the sample. In all earlier studies in which detection of P. gingivalis by PCR and detection of this bacterium by bacterial culture have been compared, the sample material has been subgingival plaque. P. gingivalis has been cultured more often and in higher proportions from subgingival plaque than from saliva (24, 25). However, due to the high detection limit for P. gingivalis in saliva, culture studies may underestimate its prevalence in saliva samples.
In this study, the occurrence of P. gingivalis as determined by PCR detection seemed to increase with age of the Finnish subjects. P. gingivalis was infrequently detected in samples from children under 10 years of age (5%), and it was only rarely detected in specimens from teenagers and young adults (13.4 to 13.8%), whereas most adults (63.3%) over 30 years of age harbored this bacterium. The low rate of detection of P. gingivalis in children is in accordance with a study by Ashimoto et al. (3) in which they detected P. gingivalis by PCR in 14% of children aged around 7 years. However, in a study by McClellan et al. (14), P. gingivalis was detected by PCR in 37% of subjects under 18 years of age, with similar frequencies irrespective of age. Differences in the PCR methodologies are a likely cause for the discrepant results, since the nested-PCR method used by McClellan et al. (14) is more sensitive than the PCR methods used in other studies. However, McClellan et al. did not report the specificity of the nested-PCR method. In culture studies, P. gingivalis has not been isolated (7–9, 13), or has been isolated only extremely rarely (4, 16, 17), from the oral cavities of children and young adults, which coincides well with the low isolation frequency of the present study. Similar to PCR studies, hybridizations with DNA probes have revealed higher rates of detection of P. gingivalis in the older age groups as well as a positive correlation between detection of P. gingivalis and increasing age (20). However, the higher frequency of detection of P. gingivalis with total chromosomal DNA probes compared with that of bacterial culture may be explained in part by the hybridization of these probes to other species, leading to false-positive results (20, 23).
While P. gingivalis was rarely detected by PCR in saliva from periodontally healthy subjects or from subjects with localized juvenile periodontitis in the present study, it was common in adult periodontitis patients. Since the prevalence of periodontitis is very low in children and adolescents but increases with age, being almost ubiquitous in middle-aged individuals (5), it is difficult to find a representative study population in which subjects of various ages would have similar periodontal statuses. In the present study, most of the subjects under 30 years of age were periodontally healthy whereas older subjects commonly exhibited adult periodontitis. The increased rates of detection of P. gingivalis in the older age groups may be related to the differences in the periodontal statuses of the subjects. Also, in previous PCR studies, P. gingivalis has rarely been detected in subjects without periodontitis (3, 22) but has frequently been found in adult periodontitis patients (3).
In conclusion, the present study shows the applicability of Chelex 100 processing of salivary samples for PCR amplification. P. gingivalis was rarely detected in saliva from periodontally healthy Finnish children and young adults. The prevalence of P. gingivalis increased with age, suggesting that oral colonization takes place mainly during adulthood.
ACKNOWLEDGMENTS
This work was supported by the Academy of Finland and by the Emil Aaltonen Foundation.
REFERENCES
- 1.American Academy of Periodontology. Proceedings of the world workshop on clinical periodontics. 1989. pp. I-23–I-24. , I-32. The American Academy of Periodontology, Chicago, Ill. [Google Scholar]
- 2.Amicosante M, Richeldi L, Trenti G, Paone G, Campa M, Bisetti A, Saltini C. Inactivation of polymerase inhibitors for Mycobacterium tuberculosis DNA amplification in sputum by using capture resin. J Clin Microbiol. 1995;33:629–630. doi: 10.1128/jcm.33.3.629-630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashimoto A, Chen C, Bakker I, Slots J. Polymerase chain reaction detection of 8 putative periodontal pathogens in subgingival plaque of gingivitis and advanced periodontitis lesions. Oral Microbiol Immunol. 1996;11:266–273. doi: 10.1111/j.1399-302x.1996.tb00180.x. [DOI] [PubMed] [Google Scholar]
- 4.Ashley F P, Gallagher J, Wilson R F. The occurrence of Actinobacillus actinomycetemcomitans, Bacteroides gingivalis, Bacteroides intermedius and spirochaetes in the subgingival microflora of adolescents and their relationship with the amount of supragingival plaque and gingivitis. Oral Microbiol Immunol. 1988;3:77–82. doi: 10.1111/j.1399-302x.1988.tb00086.x. [DOI] [PubMed] [Google Scholar]
- 5.Brown L J, Brunelle J A, Kingman A. Periodontal status in the United States, 1988–91: prevalence, extent, and demographic variation. J Dent Res. 1996;75:672–683. doi: 10.1177/002203459607502S07. . (Special issue.) [DOI] [PubMed] [Google Scholar]
- 6.De Lamballerie X, Zandotti C, Vignoli C, Bollet C, de Micco P. A one-step microbial DNA extraction method using “Chelex 100” suitable for gene amplification. Res Microbiol. 1992;143:785–790. doi: 10.1016/0923-2508(92)90107-y. [DOI] [PubMed] [Google Scholar]
- 7.Delaney J E, Ratzan S K, Kornman K S. Subgingival microbiota associated with puberty: studies of pre-, circum-, and postpubertal human females. Pediatr Dent. 1986;8:268–275. [PubMed] [Google Scholar]
- 8.Frisken K W, Higgins T, Palmer J M. The incidence of periodontopathic microorganisms in young children. Oral Microbiol Immunol. 1990;5:43–45. doi: 10.1111/j.1399-302x.1990.tb00225.x. [DOI] [PubMed] [Google Scholar]
- 9.Gusberti F A, Mombelli A, Lang N P, Minder C E. Changes in subgingival microbiota during puberty: a 4-year longitudinal study. J Clin Periodontol. 1990;17:685–692. doi: 10.1111/j.1600-051x.1990.tb01054.x. [DOI] [PubMed] [Google Scholar]
- 10.Johnson S R, Martin D H, Cammarata C, Morse S A. Alterations in sample preparation increase sensitivity of PCR assay for diagnosis of chancroid. J Clin Microbiol. 1995;33:1036–1038. doi: 10.1128/jcm.33.4.1036-1038.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jousimies-Somer H R, Summanen P H, Finegold S M. Bacteroides, Porphyromonas, Prevotella, Fusobacterium, and other anaerobic gram-negative bacteria. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: ASM Press; 1995. pp. 603–620. [Google Scholar]
- 12.Kongmuang U, Luk J M C, Lindberg A A. Comparison of three stool-processing methods for detection of Salmonella serogroups B, C2, and D by PCR. J Clin Microbiol. 1994;32:3072–3074. doi: 10.1128/jcm.32.12.3072-3074.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Könönen E, Jousimies-Somer H, Asikainen S. Relationship between oral gram-negative anaerobic bacteria in saliva of the mother and the colonization of her edentulous infant. Oral Microbiol Immunol. 1992;7:273–276. doi: 10.1111/j.1399-302x.1992.tb00587.x. [DOI] [PubMed] [Google Scholar]
- 14.McClellan D L, Griffen A L, Leys E J. Age and prevalence of Porphyromonas gingivalis in children. J Clin Microbiol. 1996;34:2017–2019. doi: 10.1128/jcm.34.8.2017-2019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore L V H, Moore W E C, Cato E P, Smibert R M, Burmeister J A, Best A M, Ranney R R. Bacteriology of human gingivitis. J Dent Res. 1987;66:989–995. doi: 10.1177/00220345870660052401. [DOI] [PubMed] [Google Scholar]
- 16.Petit M D A, van Steenbergen T J M, Timmerman M F, de Graaff J, van der Velden U. Prevalence of periodontitis and suspected periodontal pathogens in families of adult periodontitis patients. J Clin Periodontol. 1994;21:76–85. doi: 10.1111/j.1600-051x.1994.tb00283.x. [DOI] [PubMed] [Google Scholar]
- 17.Sixou J-L, Bonnaure-Mallet M, Mouton C. Serum antibodies to Porphyromonas gingivalis in children. J Periodontol. 1995;66:369–376. doi: 10.1902/jop.1995.66.5.369. [DOI] [PubMed] [Google Scholar]
- 18.Slots J, Bragd L, Wikström M, Dahlen G. The occurrence of Actinobacillus actinomycetemcomitans, Bacteroides gingivalis and Bacteroides intermedius in destructive periodontal disease in adults. J Clin Periodontol. 1986;13:570–577. doi: 10.1111/j.1600-051x.1986.tb00849.x. [DOI] [PubMed] [Google Scholar]
- 19.Slots J. Detection of colonies of Bacteroides gingivalis by a rapid fluorescence assay for trypsin-like activity. Oral Microbiol Immunol. 1987;2:139–141. doi: 10.1111/j.1399-302x.1987.tb00279.x. [DOI] [PubMed] [Google Scholar]
- 20.Slots J, Chen C. Detection of Porphyromonas gingivalis associated with human periodontitis by DNA methods. Clin Infect Dis. 1993;16:S317–S318. doi: 10.1093/clinids/16.supplement_4.s317. [DOI] [PubMed] [Google Scholar]
- 21.Slots J, Ashimoto A, Flynn M J, Li G, Chen C. Detection of putative periodontal pathogens in subgingival specimens by 16S ribosomal DNA amplification with the polymerase chain reaction. Clin Infect Dis. 1995;20:S304–S307. doi: 10.1093/clinids/20.supplement_2.s304. [DOI] [PubMed] [Google Scholar]
- 22.Tran S D, Rudney J D. Multiplex PCR using conserved and species-specific 16S rRNA gene primers for simultaneous detection of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. J Clin Microbiol. 1996;34:2674–2678. doi: 10.1128/jcm.34.11.2674-2678.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Steenbergen T J M, Timmerman M F, Mikx F H M, de Quincey G, van der Weijden G A, van der Velden U, de Graaff J. Discrepancy between culture and DNA probe analysis for the detection of periodontal bacteria. J Clin Periodontol. 1996;23:955–959. doi: 10.1111/j.1600-051x.1996.tb00518.x. [DOI] [PubMed] [Google Scholar]
- 24.van Winkelhoff A J, van der Velden U, Clement M, de Graaff J. Intra-oral distribution of black-pigmented Bacteroides species in periodontitis patients. Oral Microbiol Immunol. 1988;3:83–85. doi: 10.1111/j.1399-302x.1988.tb00087.x. [DOI] [PubMed] [Google Scholar]
- 25.Von Troil-Lindén B, Torkko H, Alaluusua S, Wolf J, Jousimies-Somer H, Asikainen S. Periodontal findings in spouses: a clinical, radiographic and microbiological study. J Clin Periodontol. 1995;22:93–99. [PubMed] [Google Scholar]
- 26.Wahlfors J, Meurman J H, Väisänen P, Alakuijala P, Korhonen A, Torkko H, Jänne J. Simultaneous detection of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis by a rapid PCR method. J Dent Res. 1995;74:1796–1801. doi: 10.1177/00220345950740111301. [DOI] [PubMed] [Google Scholar]