Abstract
Two methods were compared for the analysis of 48 unrelated and epidemiologically related Legionella pneumophila serogroup 1 isolates. These are the infrequent-restriction-site PCR (IRS-PCR) assay with adapters designed for XbaI and PstI restriction sites and the pulsed-field gel electrophoresis (PFGE) analysis determined after DNA restriction with SfiI. Both methods demonstrated a high level of discrimination with a similar capacity for differentiating 23 of the 24 unrelated isolates. PFGE analysis and IRS-PCR assay were both able to identify epidemiologically related isolates of L. pneumophila from three outbreaks. Hence, IRS-PCR assay appears to be a reproducible (intergel reproducibility, 100%) and discriminative (discriminatory index, ≥0.996) tool for typing of Legionella. Compared to PFGE, however, IRS-PCR presented an advantage through ease of performance and with attributes of rapidity and sensitivity of target DNA.
The family Legionellaceae is represented to date by 42 species (3, 11), most of which are potentially pathogenic for humans. Legionella pneumophila serogroup 1 is the major causative agent associated with legionellosis (2). L. pneumophila is widely present in the environment, especially in water distribution systems; thus, the source of a given human infection cannot be presumed on the single basis of an isolation of L. pneumophila from an environmental source. Hence, epidemiological tools are needed to determine the clonal relatedness between isolates of human and environmental origins. A variety of molecular typing techniques have been developed to compare L. pneumophila serogroup 1 strains, including analysis by monoclonal antibodies, arbitrarily primed and repetitive element PCR assays, plasmid analysis, multilocus enzyme analysis, restriction fragment length polymorphism, ribotyping, and pulsed-field gel electrophoresis (PFGE) (1, 4, 6, 7, 14–16, 19, 20–23). Most of the recent findings suggest that restriction enzyme analysis by PFGE is the most discriminative epidemiological marker for subtyping L. pneumophila strains (15, 20, 22). Nevertheless, PFGE is a time-consuming method which requires expensive and specialized equipment.
A recently described method referred to as infrequent-restriction-site PCR (IRS-PCR) assay (17) consists of double digestion of genomic DNA with a restriction enzyme that infrequently cuts the chromosome and a second enzyme that frequently cuts it, followed by amplification of DNA with primers and adapters targeting the extremities of the restricted fragments. This technique has the advantage of using minute quantities of target DNA, and the separation of amplified fragments can be achieved by conventional agarose gel electrophoresis. The method has not previously been applied to Legionella.
In this study, we have adapted the IRS-PCR method to analyze 48 unrelated and epidemiologically related L. pneumophila serogroup 1 isolates and compared the results with those obtained by PFGE. The IRS-PCR technique appeared to be rapid, versatile, reproducible, and useful for discrimination of L. pneumophila serogroup 1 isolates.
MATERIALS AND METHODS
Bacterial strains.
Twenty-five environmental and 23 clinical isolates of L. pneumophila serogroup 1 were obtained from the National Reference Center (France) for Legionella (Table 1). Among these, 6 human and 18 environmental isolates were associated with three outbreaks, the others being epidemiologically unrelated. Strains were cultured on buffered charcoal yeast extractα agar and were biochemically characterized according to standard methods (3). Strains were identified by direct immunofluorescence assays with a commercial monoclonal antibody (Monofluo Kit Legionella pneumophila; Diagnostics Pasteur, Paris, France) (8) and with specific antisera prepared by rabbit immunization at the Reference Center.
TABLE 1.
Clinical and environmental isolates of L. pneumophila used for PFGE and IRS-PCR analysis
| Strain | Origin of strain (city or country)b | PFGE pattern | IRS-PCR pattern
|
|
|---|---|---|---|---|
| (1-band difference) | (3-band difference) | |||
| Unrelated strains | ||||
| Human strains | ||||
| L27 | Reims | A | A | A1 |
| L47 | St. Omer | B | B | A2 |
| L12 | Pau | C | C | B |
| L54 | Marseille | D | D | C |
| L3 | Nantes (hospital A) | E | E | D |
| L13 | Toulouse | F | F | E |
| L23 | Colmar | G1 | G | F |
| L48 | Neufchateau | G2 | H | G |
| L51 | St. Brieuc | H | I | H |
| L52 | Créteil | I | J | I |
| L392 | Nancy | J | K | J |
| L403 | Nice | K | L | K |
| L827 | Poitiers | L | M | L |
| Bellingham-1 | USA | M | N | M |
| Detroit-1 | USA | N | O | N |
| Knoxville-1 | USA | O | P | O |
| OLDA | USA | P | Q | P |
| Environmental strains (water) | ||||
| L489 | Lyon | Q | R | Q |
| Pontiac-1 | USA | R | S | R |
| L900 | Nevers | S | T | S |
| L524 | Strasbourg | T | U | T |
| L401 | Poitiers | U | V | U |
| L387 | Golfech | V | W | V |
| L211 | Nantes (hospital B) | W | X | W |
| Epidemiologically related strains | ||||
| Outbreak 1 | ||||
| Human isolatea L31 | Morsbronn | a | a | a |
| Environmental strains (water) | ||||
| L60 | Morsbronn | a | a | a |
| L61 | Morsbronn | a | a | a |
| L62 | Morsbronn | a | a | a |
| L64 | Morsbronn | a | a | a |
| L65 | Morsbronn | a | a | a |
| L66 | Morsbronn | a | a | a |
| L67 | Morsbronn | a | a | a |
| L68 | Morsbronn | a | a | a |
| L71 | Morsbronn | a | a | a |
| Outbreak 2 | ||||
| Human strains | ||||
| L37 | St. Etienne | b | b | b1 |
| L35 | St. Etienne | b | b | b1 |
| L33 | St. Etienne | b | d | b2 |
| L36 | St. Etienne | b | d | b2 |
| Environmental strains (water) | ||||
| L32 | St. Etienne | b | b | b1 |
| L38 | St. Etienne | b | b | b1 |
| L39 | St. Etienne | b | b | b1 |
| L40 | St. Etienne | b | b | b1 |
| L41 | St. Etienne | b | b | b1 |
| L45 | St. Etienne | b | b | b1 |
| L34 | St. Etienne | b | b | b1 |
| Outbreak 3 | ||||
| Human straina L215 | Nantes (hospital B) | c | c | c |
| Environmental strains (water) | ||||
| L214 | Nantes (hospital B) | c | c | c |
| L212 | Nantes (hospital B) | c | c | c |
Other cases were serological.
Cities without countries listed are in France. USA, United States.
IRS-PCR. (i) Adapters and primers.
Adapters were constructed as previously described by Mazurek et al. (17) with oligonucleotides purchased from Eurogentec SA (Seraing, Belgium). These were AX1, AX2, and PX-G, as described elsewhere (17), and PS1 (5′-GAC TCG ACT CGC ATG CA-3′) and PS2 (5′-TGC GAG T-3′), which were specifically designed in this study to generate PstI adapters. Adapters were also designed to ligate specifically to the cohesive ends of the corresponding restricted fragments. To prepare the adapters, oligonucleotides PS1 and PS2 or AX1 and AX2 were mixed in equal molar amounts in 1× PCR buffer (Perkin-Elmer Cetus, Branchburg, N.J.) and were allowed to anneal as the mixture cooled from 80 to 4°C over 1 h in a thermocycler. Oligonucleotides PS1 and PX-G were used as primers in PCR.
(ii) Preparation of template DNA.
Bacterial cultures were harvested and resuspended in 500 μl of STE buffer (100 mM NaCl, 50 mM Tris-HCl, 10 mM sodium EDTA [pH 7.5]) and incubated at room temperature with 100 μl of lysozyme (10 mg/ml) for 1 h. Cells were lysed with 20 μl of sodium dodecyl sulfate (25 mg/ml) at 37°C and digested for 1 h with 100 μl of proteinase K (25 mg/ml)–5 μl of RNase (10 mg/ml) at 37°C. DNA was purified with phenol-chloroform-isoamyl alcohol (50:48:2) (13) and chloroform-isoamyl alcohol (24:1) and precipitated by the addition of absolute ethanol. The pellet was air dried, resuspended in 100 μl of sterile distilled water, and stored at −20°C until used. A portion of the extracted DNA was digested with 40 U of PstI–40 U of XbaI in 1× buffer for 90 min at 37°C. T4 DNA ligase (15 U), ATP (12.6 pmol), 10× ligase buffer (0.75 μl), the XbaI adapter (20 pmol), the PstI adapter (20 pmol), and sterile distilled water were added to 12.5 μl of extract for a total volume of 20 μl. The mixture was incubated at 16°C for 1 h and then at 65°C for 20 min to inactivate T4 DNA ligase. The sample was digested with 10 U of XbaI–10 U of PstI at 37°C for 15 min to cleave any restriction sites re-formed by ligation and then submitted to amplification. All enzymes were obtained from Boehringer Mannheim (Meylan, France).
(iii) Amplification.
Each PCR mixture included 10 μl of template DNA, 0.5 U of Taq DNA polymerase (Perkin-Elmer Cetus), deoxynucleoside triphosphates (200 μM each) (Pharmacia Biotech, Uppsala, Sweden), and the oligonucleotide primers in 1× PCR buffer. Typically, the oligonucleotides PS1 and PX-G were used together as primers. Amplification was performed in a PHC-3 Dri-Block cycler (Techne Ltd., Cambridge, United Kingdom) with an amplification profile that consisted of an initial denaturation step at 94°C for 5 min and then 30 cycles of denaturation at 94°C for 30 s, primer annealing at 60°C for 30 s, and extension at 72°C for 90 s. All experiments included negative controls which were processed with the samples.
Electrophoretic patterns.
Gel electrophoresis was performed for 4 h at 100 V on the PCR products loaded into wells of 1.5% agarose prepared in 0.5× Tris-borate-EDTA (SeaKem GTG; FMC Bioproducts, Rockland, Maine). DNA VI molecular weight markers (Boehringer Mannheim) were used.
PFGE typing.
PFGE patterns were obtained by the modified technique of Grothues and Tümmler (10). Briefly, agarose blocks were digested with SfiI overnight at 50°C followed by electrophoresis with the contour-clamped homogeneous electric field DRII system (Bio-Rad Laboratories, Hercules, Calif.). Separations were accomplished at constant pulse times (25 s) for 11 h and increasing pulse times (35 to 60 s) for 11 h. Lambda concatemers (PFGE marker I; Boehringer Mannheim) were used as size markers.
Gel staining and data processing.
Gels were stained with ethidium bromide (0.5 mg/ml) (Bioprobe Systems, Paris, France) for 10 min and photographed (Polaroid, Cambridge, Mass.) with UV illumination. Pattern clustering on a matrix of Dice coefficient (5) was based on the unweighted pair group method with averages (UPGMA), and dendrograms were constructed with Taxotron software (Institut Pasteur, Paris, France). Interpretations for PFGE and IRS-PCR were based on differences of banding patterns as suggested by Tenover et al. (24). Strains differing in up to three fragments only were deemed clonally related, and these strains were described as subtypes of a given clonal type. In the case of no differences between banding patterns, strains were considered identical. When differing in four or more fragments, strains were considered separate types. Major genotypes were labeled by letters, and each of their variant subtypes was indicated by a numeral suffix.
Determination of reproducibility and discriminatory ability.
To assess the reproducibility of IRS-PCR typing, 24 different pairs of isolates were analyzed in two different runs. Reproducibility was defined as the percentage of pairs with concordant types. The index of discriminatory ability was calculated as described by Hunter and Gaston (12) on the basis of the type distribution among the 24 epidemiologically unrelated L. pneumophila serogroup 1 isolates.
RESULTS
IRS-PCR analysis.
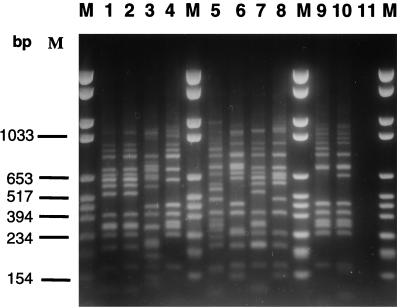
The patterns generated with PX-G and PS1 primers were composed of 12 to 15 bands ranging in size between 100 and 1,100 bp (Fig. 1). The intergel reproducibility of banding patterns was 100% for 24 duplicate pairs. When a three-band difference was used to distinguish IRS-PCR types, a total of 26 types were recognized and labeled A to c (Table 1). For the 24 isolates tested which were not epidemologically related, 23 distinctive patterns were obtained and the discriminatory index was 0.996. One of these patterns (A) was divided into two subtypes (A1 and A2) which differed by no more than two fragments (Table 1). For the outbreak-associated isolates, IRS-PCR analysis yielded three different patterns (types a, b, and c). Isolates from outbreaks 1 and 3 each presented consistent types; this contrasted with those of outbreak 2, manifesting two subtypes (b1 and b2), based on the presence of an additional 0.6-kb fragment in b2.
FIG. 1.
IRS-PCR electrophoretic patterns of L. pneumophila serogroup 1 isolates from patients or environmental samples in different geographic locations. Lanes M, VI molecular weight markers (Boehringer Mannheim); lane 1, isolate L32; lane 2, isolate L38; lane 3, isolate L3; lane 4, isolate L524; lane 5, isolate L387; lane 6, isolate L215; lane 7, isolate L41; lane 8, isolate L54; lane 9, isolate L401; lane 10, isolate L392; lane 11, negative control.
When a one-band difference was used to distinguish IRS-PCR types, a total of 28 types were recognized and termed A to d. For the 24 isolates tested which were not epidemiologically related, 24 distinctive patterns were obtained, giving a discriminatory index of 1. For the isolates associated with outbreaks, IRS-PCR analysis yielded four different patterns (types a, b, c, and d). For outbreak 2, when a one-band difference was used to distinguish IRS-PCR types, two types (b and d) of isolates were recognized.
Macrorestriction analysis of genomic DNAs with PFGE.
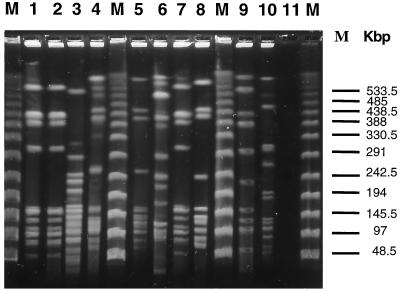
Macrorestriction profiles generated by SfiI cleavage consisted of 9 to 15 fragments varying in size between 100 and 1,000 kb (Fig. 2). A total of 26 types were recognized (A to c). Twenty-four isolates which were not epidemiologically related showed 23 distinctive patterns, and the discriminatory index was 0.996. Two isolates (L23 and L48) were recognized as subclonal types by comparing their PFGE patterns (G1 and G2), based on the presence of an additional 100-kb fragment in G2. For the outbreak-associated isolates, three major groups of macrorestriction patterns (a, b, and c) were recognized. Each isolate comprising a given outbreak was identical in pattern.
FIG. 2.
PFGE patterns of L. pneumophila serogroup 1 isolates from patients or environmental samples in different geographic locations. Lanes M, PFGE I markers (Boehringer Mannheim); lane 1, isolate L32; lane 2, isolate L38; lane 3, isolate L3; lane 4, isolate L524; lane 5, isolate L387; lane 6, isolate L215; lane 7, isolate L41; lane 8, isolate L54; lane 9, isolate L401; lane 10, isolate L392; lane 11, negative control.
DISCUSSION
PFGE analysis is a highly efficient means of distinguishing between strains of Legionella during outbreaks (20, 27). Together with SfiI-digested DNA, this very sensitive typing system is labor-intensive and expensive compared to the PCR-based methodology. PCR-based typing techniques used for analysis of L. pneumophila strains during outbreaks include the arbitrarily primed PCR assay, randomly amplified polymorphic DNA analysis (9, 26), and amplified fragment length polymorphism (AFLP) analysis (25). The reproducibility of arbitrarily primed PCR and randomly amplified polymorphic DNA methods is affected by various parameters including (i) the method of DNA extraction (9, 26), (ii) the purity of the oligonucleotide primers (26), (iii) the quality of materials (thermocyclers, Taq polymerase origin, and electrophoresis apparatus) (18), and (iv) the low-stringency hybridization conditions (7). AFLP and IRS-PCR overcome all these disadvantages since these methods depend on double digests of genomic DNAs, which are specifically amplified under stringent conditions with selective primers extending beyond the adapters. In the AFLP method, the two adapters consist of 18- to 22-bp oligonucleotides whereas in the IRS-PCR method, one of the two oligonucleotides is short enough to allow efficient ligation of the double-stranded adapter at 16°C but cannot form stable hybrids at higher temperatures nor compete for primer during subsequent PCR steps. Moreover, this short oligonucleotide (PS2) is not phosphorylated and therefore not ligated (it does not interfere with the PCR). Indeed, due to the presence of an excess of adapter after ligation in the AFLP method, an ethanol purification step would be recommended to avoid interference in the subsequent PCR amplification (25). The application of AFLP is also restricted by a patent registered in 1992 (European patent application 054858A1).
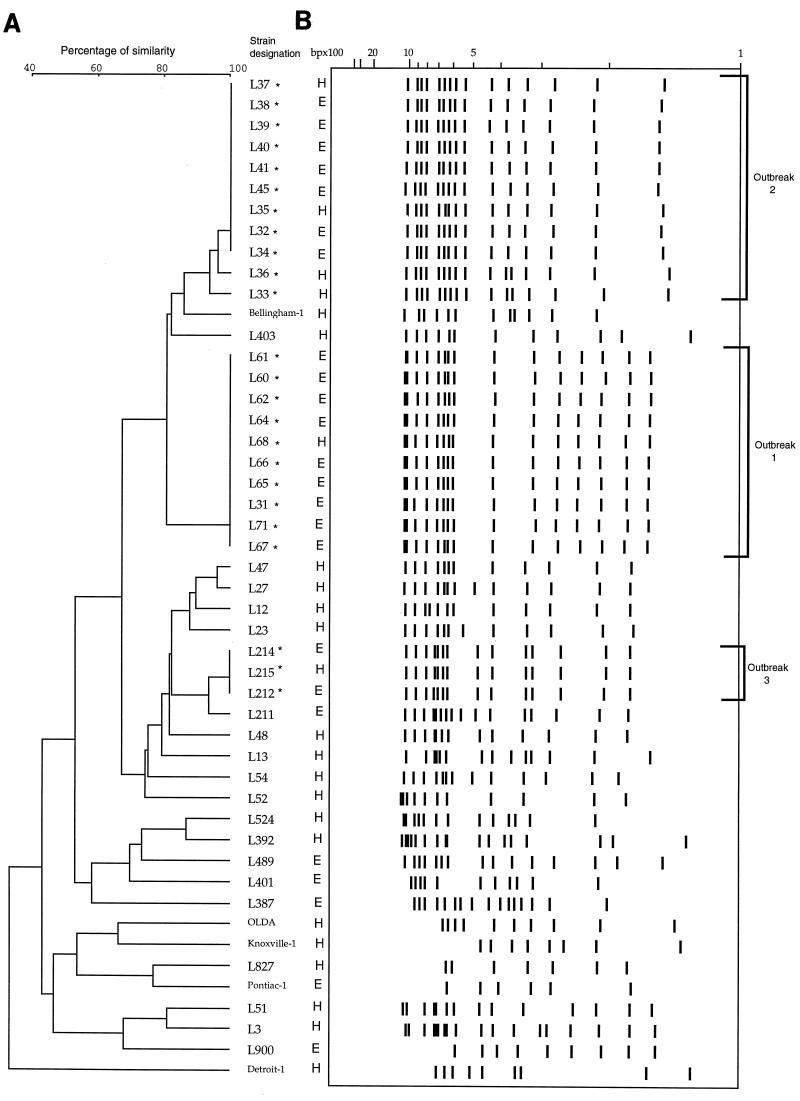
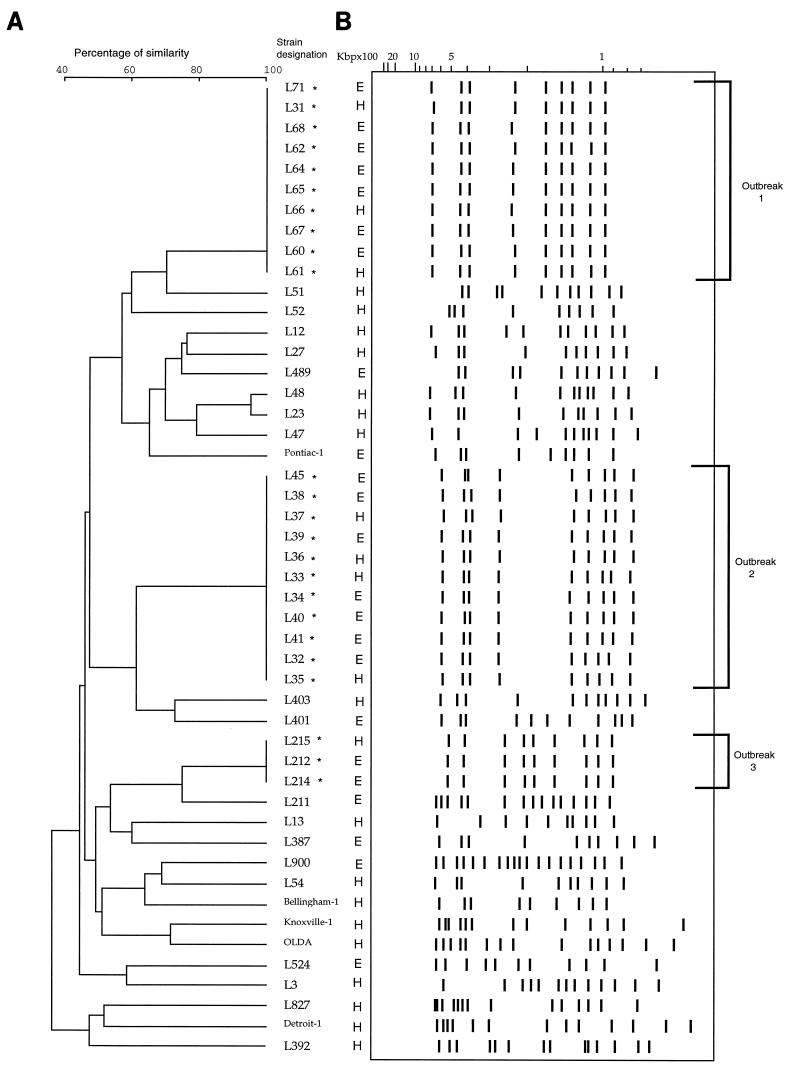
The results of this study show that PFGE analysis with SfiI and IRS-PCR assay with PstI and XbaI were both able to identify epidemiologically related isolates of L. pneumophila when applied to three outbreaks (Fig. 3 and 4). For each outbreak, both PFGE and IRS-PCR produced isolate profiles that were concordant between those from the patients and the sources of transmission. Rules on interpretation of PFGE and IRS-PCR were based principally on the guidelines described by Tenover et al. for interpreting macrorestriction patterns (24); these regard differences in three bands or less in defining members of a subtype. Hence, when applied to unrelated isolates, PFGE presented discriminatory power (D = 0.996) identical to that of IRS-PCR. PFGE considered IRS-PCR subtypes A1 and A2 of unrelated strains to be two different types (A and B); conversely, two PFGE subtypes (G1 and G2) were considered to be two distinct IRS-PCR types (F and G). If IRS-PCR types were defined by a difference of one band only, the discriminatory power of IRS-PCR for differentiating unrelated strains was equal to 1 and two types (b and d) of isolates associated with outbreak 2 were recognized.
FIG. 3.
Clustering of L. pneumophila serogroup 1 isolates by analysis of IRS-PCR patterns. (B) Schematic representation of IRS-PCR patterns. Lanes H, patient isolates; lanes E, environmental isolates. Names of epidemiologically related isolates are followed by an asterisk. (A) Dendrogram corresponding to panel B in accordance with UPGMA clustering (error, 3.5 to 4.5%) on a matrix based on the Dice coefficient (Taxotron software analysis; Institut Pasteur).
FIG. 4.
Clustering of L. pneumophila serogroup 1 isolates by analysis of PFGE patterns. (B) Schematic representation of PFGE patterns. Lanes H, patient isolates; lanes E, environmental isolates. Names of epidemiologically related isolates are followed by an asterisk. (A) Dendrogram corresponding to panel B in accordance with UPGMA clustering (error, 3.5 to 5%) on a matrix based on the Dice coefficient (Taxotron software analysis; Institut Pasteur).
Both techniques used in this study yielded well-resolved, easily compared restriction fragment patterns, but PFGE was time-consuming (at least 3 to 4 days to complete) and labor-intensive. IRS-PCR possesses the attributes of ease of performance, reproducibility (intergel reproducibility, 100%), and much less time consumption (from time of receipt of an isolate, less than 2 days to complete without intensive labor). Also, minute quantities of target DNA are sufficient for PCR amplification. The fragments amplified by IRS-PCR have small molecular sizes (less than 1,100 bp), facilitating separation in 3 to 4 h by standard agarose gel electrophoresis. Moreover, IRS-PCR is less expensive to operate than PFGE in terms of both equipment and consumables.
The IRS-PCR method appears to be a potentially useful epidemiologic tool for the early investigations of L. pneumophila serogroup 1 isolates in hospital and regional laboratories. The results based on epidemiological markers could then be confirmed in the reference laboratory.
ACKNOWLEDGMENTS
This work was supported by a grant from the Bureau des Ressources Génétiques.
We thank M. Goldner for editorial assistance. We are grateful to H. Lelièvre for photography and C. Bouveyron, D. de Longevialle, P. Lefevre, M. Siffert, and C. Vallier for technical support.
REFERENCES
- 1.Bangsborg J M, Gerner-Smidt P, Colding H, Fiehn N E, Bruun B, Hoiby N. Restriction fragment length polymorphism of rRNA genes for molecular typing of members of the family Legionellaceae. J Clin Microbiol. 1995;33:402–406. doi: 10.1128/jcm.33.2.402-406.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartlett C L R, Macrae A D, Macfarlane J T. Clinical aspects and diagnosis of Legionella infection. In: Bartlett C L R, Macrae A D, Macfarlane J T, editors. Legionella infections—1986. London, United Kingdom: Edward Arnold; 1986. pp. 37–55. [Google Scholar]
- 3.Benson R F, Thacker W L, Daneshvar M I, Brenner D J. Legionella waltersii sp. nov. and an unnamed Legionella genomospecies isolated from water in Australia. Int J Syst Bacteriol. 1996;46:631–634. doi: 10.1099/00207713-46-3-631. [DOI] [PubMed] [Google Scholar]
- 4.Bingen E H, Denamur E, Elion J. Use of ribotyping in epidemiological surveillance of nosocomial outbreaks. Clin Microbiol Rev. 1994;7:311–327. doi: 10.1128/cmr.7.3.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dice L R. Measures of the amount of ecological association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 6.Georghiou P R, Doggett A M, Kielhofner M A, Stout J E, Watson D A, Lupski J R, Hamill R J. Molecular fingerprinting of Legionella species by repetitive element PCR. J Clin Microbiol. 1994;32:2989–2994. doi: 10.1128/jcm.32.12.2989-2994.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomez-Lus P, Fields B S, Benson R F, Martin W T, O’Connor S P, Black C M. Comparison of arbitrarily primed polymerase chain reaction ribotyping and monoclonal antibody analysis for subtyping Legionella pneumophila serogroup 1. J Clin Microbiol. 1993;31:1940–1942. doi: 10.1128/jcm.31.7.1940-1942.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gosting L H, Cabrian K, Sturge J C, Goldstein L C. Monoclonal antibody to a species-specific antigen of Legionella pneumophila. J Clin Microbiol. 1984;20:1031–1035. doi: 10.1128/jcm.20.6.1031-1035.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grattard F, Berthelot P, Reyrolle M, Ros A, Etienne J, Pozzetto B. Molecular typing of nosocomial strains of Legionella pneumophila by arbitrarily primed PCR. J Clin Microbiol. 1996;34:1595–1598. doi: 10.1128/jcm.34.6.1595-1598.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grothues D, Tümmler B. Genome analysis of Pseudomonas aeruginosa by field inversion gel electrophoresis. FEMS Microbiol Lett. 1987;48:419–422. [Google Scholar]
- 11.Hookey J V, Saunders N A, Fry N K, Birtles R J, Harrison T G. Phylogeny of Legionellaceae based on small-subunit ribosomal DNA sequences and proposal of Legionella lytica comb. nov. for Legionella-like amoebal pathogens. Int J Syst Bacteriol. 1996;46:526–531. [Google Scholar]
- 12.Hunter P R, Gaston M A. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 14.Marques M T, Bornstein N, Fleurette J. Combined monoclonal antibody typing, multilocus enzyme electrophoresis, soluble protein profiles and plasmid analysis of clinical and environmental Legionella pneumophila serogroup 1 isolated in a Portuguese hospital. J Hosp Infect. 1995;30:103–110. doi: 10.1016/0195-6701(95)90150-7. [DOI] [PubMed] [Google Scholar]
- 15.Marrie T J, Johnson W, Tyler S, Bezanson G, Haldane D, Burbridge S, Joly J. Potable water and nosocomial Legionnaires’ disease—check water from all rooms in which patient has stayed. Epidemiol Infect. 1995;114:267–276. doi: 10.1017/s0950268800057939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marrie T J, Johnson W M, Tyler S D, Bezanson G S, Burbridge S. Genomic stability of Legionella pneumophila isolates recovered from two cardiac transplant patients with nosocomial Legionnaires’ disease. J Clin Microbiol. 1994;32:3085–3087. doi: 10.1128/jcm.32.12.3085-3087.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazurek G H, Reddy V, Marston B J, Haas W H, Crawford J T. DNA fingerprinting by infrequent-restriction-site amplification. J Clin Microbiol. 1996;34:2386–2390. doi: 10.1128/jcm.34.10.2386-2390.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meunier J-R, Grimont P A D. Factors affecting reproducibility of random amplified polymorphic DNA fingerprinting. Res Microbiol. 1993;144:373–379. doi: 10.1016/0923-2508(93)90194-7. [DOI] [PubMed] [Google Scholar]
- 19.Ott M, Bender L, Marre R, Hacker J. Pulsed field electrophoresis of genomic restriction fragments for the detection of nosocomial Legionella pneumophila in hospital water supplies. J Clin Microbiol. 1991;29:813–815. doi: 10.1128/jcm.29.4.813-815.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pruckler J M, Mermel L A, Benson R F, Giorgio C, Cassiday P K, Breiman R F, Whitney C G, Fields B S. Comparison of Legionella pneumophila isolates by arbitrarily primed PCR and pulsed-field gel electrophoresis: analysis from seven epidemic investigations. J Clin Microbiol. 1995;33:2872–2875. doi: 10.1128/jcm.33.11.2872-2875.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saunders N A, Harrison T G, Haththotuwa A, Kachwalla N, Taylor A G. A method for typing strains of Legionella pneumophila serogroup 1 by analysis of restriction fragment length polymorphisms. J Med Microbiol. 1990;31:45–55. doi: 10.1099/00222615-31-1-45. [DOI] [PubMed] [Google Scholar]
- 22.Schoonmaker D, Heimberg T, Birkhead G. Comparison of ribotyping and restriction enzyme analysis using pulsed-field gel electrophoresis for distinguishing Legionella pneumophila isolates obtained during a nosocomial outbreak. J Clin Microbiol. 1992;30:1491–1498. doi: 10.1128/jcm.30.6.1491-1498.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Struelens M J, Maes N, Rost F, Deplano A, Jacobs F, Liesnard C, Bornstein N, Grimont F, Lauwers S, McIntyre M P, Serruys E. Genotypic and phenotypic methods for the investigation of a nosocomial Legionella pneumophila outbreak and efficacy of control measures. J Infect Dis. 1992;166:22–30. doi: 10.1093/infdis/166.1.22. [DOI] [PubMed] [Google Scholar]
- 24.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valsangiacomo C, Baggi F, Gaia V, Balmelli T, Peduzzi R, Piffaretti J-C. Use of amplified fragment length polymorphism in molecular typing of Legionella pneumophila and application to epidemiological studies. J Clin Microbiol. 1995;33:1716–1719. doi: 10.1128/jcm.33.7.1716-1719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Belkum A. Molecular typing methods for epidemiological studies in medical microbiology. Med Microbiol Lett. 1996;5:271–283. [Google Scholar]
- 27.Van Belkum A, Struelens M, Quint W. Typing of Legionella pneumophila strains by polymerase chain reaction-mediated DNA fingerprinting. J Clin Microbiol. 1993;31:2198–2200. doi: 10.1128/jcm.31.8.2198-2200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]