Abstract
DNA fingerprinting techniques were used to type 273 isolates of Mycobacterium bovis from Australia, Canada, the Republic of Ireland, and Iran. The results of restriction fragment length polymorphism (RFLP) analysis with DNA probes from IS6110, the direct repeat (DR), and the polymorphic GC-rich sequence (PGRS) were compared with those of a new PCR-based method called spacer oligonucleotide typing (spoligotyping) developed for the rapid typing of Mycobacterium tuberculosis (J. Kamerbeek et al., J. Clin. Microbiol. 35:907–914, 1997). Eighty-five percent of the isolates harbored a single copy of IS6110, and 81.5% of these carried IS6110 on the characteristic 1.9-kb restriction fragment. RFLP analysis with IS6110 identified 23 different types, RFLP analysis with the DR probe identified 35 types, RFLP analysis with the PGRS probe identified 77 types, and the spoligotyping method identified 35 types. By combining all results, 99 different strains could be identified. Isolate clusters were frequently associated within herds or were found between herds when epidemiological evidence confirmed animal movements. RFLP analysis with IS6110 was sufficiently sensitive for the typing of isolates with more than three copies of IS6110, but RFLP analysis with the PGRS probe was the most sensitive typing technique for strains with only a single copy of IS6110. Spoligotyping may have advantages for the rapid typing of M. bovis, but it needs to be made more sensitive.
DNA typing techniques are now frequently used for epidemiological investigations of many infectious diseases. The insertion sequence (IS) IS6110 is accepted as an excellent tool for the identification of restriction fragment length polymorphisms (RFLPs) in Mycobacterium tuberculosis strains and is used in epidemiological studies worldwide (31). RFLP analysis with IS6110 has been used to define disease outbreaks and to trace the spread of multidrug-resistant strains of M. tuberculosis (2, 7, 16, 21, 34, 37). Other repetitive elements such as the direct repeat (DR) (13) and the GC-rich repetitive sequence (PGRS) (8, 25) now appear to be finding acceptance for use in the characterization of M. tuberculosis strains (10, 30, 33, 38).
Some of these methods have been applied to the typing of Mycobacterium bovis. In a previous study (6), a plasmid containing PGRS (pTBN12) was found to be more useful as a probe in RFLP analysis than the left-hand side (LHS) of the PvuII fragment in IS6110 (IS6110-L) in differentiating between Australian M. bovis isolates. However, a similar study found that pTBN12 was only marginally better than the entire IS6110 probe in the identification of polymorphisms in 109 isolates from cattle in Northern Ireland (27). Some workers have suggested that DNA fingerprinting with a sequence to the right-hand side (RHS) of the PvuII site of IS6110 (IS6110-R) as a probe may be useful in the study of M. bovis, but this appears to be confined to cases in which M. bovis isolates harbor multiple copies of IS6110 (12, 15). For example, in a study of 153 M. bovis isolates, van Soolingen et al. (32) reported the occurrence of strains of M. bovis with multiple copies of IS6110 in association with zoo and exotic animals, whereas M. bovis strains isolated from cattle in The Netherlands and Argentina generally harbored only a single copy of IS6110, usually carried in a characteristic 1.9-kb PvuII restriction fragment. When a small number of strains of M. bovis with single copies of IS6110 were analyzed by using the DR probe and pTBN12, further differentiation was achieved. In a comparison of RFLP techniques with IS6110, DR, and PGRS conducted with 85 M. bovis isolates from Argentina, DR and PGRS were recommended as the probes of choice for use in RFLP analysis (24). A comparison of the results of RFLP analyses with IS6110 and DR with 79 isolates of M. bovis from Texas and Mexico indicated that DR was a more sensitive probe than IS6110. Another comparison of the results of RFLP analyses with the entire IS6110, DR, and PGRS probes with 210 isolates of M. bovis, mostly from Northern Ireland, indicated that the effectiveness of RFLP analysis with each of these probes was almost the same, but when the results of RFLP analyses with all probes were used, the number of strains that were differentiated was almost doubled (28).
An innovative PCR-based typing method for the differentiation of M. tuberculosis strains has been reported recently (11). This method, termed spacer oligonucleotide typing (spoligotyping), relies on the in vitro amplification of DNA across the unique, highly polymorphic DR locus present in the M. tuberculosis complex chromosome. This region contains multiple short 36-bp DRs, and nonrepetitive spacers, which are 35 to 41 bp in length, are interspersed between the DRs. The PCR product from individual isolates is allowed to hybridize to 37 spacers identified in M. tuberculosis H37Rv and 6 spacers identified in M. bovis BCG P3 (14). Following hybridization and detection, the spacers that are common to the isolate being tested and the standard set of spacer oligonucleotides can be identified. Only one report on the use of spoligotyping compared with the use of RFLP analysis with IS6110 for the differentiation of M. bovis isolates has been published (1).
This study was undertaken to comprehensively compare the usefulness of the three most commonly used RFLP techniques (RFLP analyses with IS6110-R, DR, and PGRS) and the new spoligotyping method for the differentiation of M. bovis isolates from Australian sources. In addition, isolates from other countries were included to determine whether a geographic difference could be identified by any of the markers. The usefulness of these techniques in epidemiological studies of bovine tuberculosis in Australia and overseas was demonstrated.
MATERIALS AND METHODS
Source of M. bovis isolates.
Two hundred seventy-three M. bovis isolates were tested. Two hundred eleven animal isolates originated in Australia from the following states: Western Australia (n = 121), the Northern Territory (n = 46), Queensland (n = 35), Victoria (n = 8), and New South Wales (n = 1). The Western Australian isolates came from the agricultural area (n = 30), the northern pastoral area (n = 21), the Broome area (n = 5), the West Kimberley area (n = 57), and the East Kimberley area (n = 8). Sixty-one isolates of M. bovis obtained from overseas sources for comparison originated from Canada (n = 33), Iran (n = 10), the Republic of Ireland (n = 14), the United Kingdom (n = 3), and New Zealand (n = 1). In addition, the reference strain of M. bovis (strain AN5) was included.
Good epidemiological information was provided for 32 animal isolates and a single human isolate from Canada (4). The 33 Canadian isolates originated from 4 outbreaks (outbreaks A to D) involving 15 premises, 10 different animal species, and 1 human patient. Outbreak A involved four animal species and four related premises over the period from 1992 to 1994. Outbreak B was traced to four premises from 1990 to 1994 and involved four elk, a bison, and a veterinarian who was diagnosed with M. bovis infection after he treated one of the elk. Outbreak C involved a large collection of exotic species comprising seven animal species and four premises from 1989 to 1992. Epidemiological information suggested there was nose-to-nose contact between elk and a Sika deer, elk and a Pere David deer, and the Pere David deer and cattle. A cougar on one property was fed carcasses of other animals on the premises and chickens from a neighboring farm. Deer from New Zealand were imported to one of the previously infected premises after it had been depopulated, cleaned, disinfected, and released from quarantine. The deer were skin test negative before and after arrival in quarantine. Outbreak D in 1991 involved cattle on one property and an elk from a neighboring national park.
DNA probes.
PCR was used to amplify DNA from the right-hand side of the PvuII site in IS6110-R as described previously (31), and the IS6110-R probe was used to determine the number of IS6110 copies in each isolate. RFLPs were determined for all isolates by using probes prepared from amplified DNA of IS6110-R and from the oligonucleotides from the DR (5′ GTC GTC AGA CCC AAA ACC CCG AGA GGG GAC GGA AAC 3′) and PGRS (5′ CCG CCG TTG CCG CCG TTG CCG CCG TTG CCG CCG 3′).
RFLP methods.
Cells were grown and DNA was extracted as described previously (6, 31). RFLP analyses with IS6110-R and DR were performed basically by the standard method recommended for the DNA fingerprinting of M. tuberculosis (31), with the exception of the electrophoresis conditions and the probe labelling and detection methods. For RFLP analyses with IS6110-R and DR, electrophoresis was performed with 1 and 1.5% agarose gels, respectively, and gels were run in TAE (Tris, acetate, EDTA) buffer for 16 h at 45 V by using a refrigerated buffer recirculation pump at a constant temperature of 14°C. RFLP analysis with PGRS was performed as described previously (6), with the exception that the gels were run longer (45 V for 28 h) in an attempt to achieve better discrimination between strains by spreading the bands that hybridize with PGRS over a greater distance. All probes were labelled by using the nonradioactive digoxigenin system (Boehringer Mannheim), and hybridization and detection of bound probe with a chemiluminescent substrate were performed as described by the manufacturer.
DNAs from M. tuberculosis Mt 14323 and M. bovis BCG 3 were run on all gels in positions 1 and 30 as external standards. To allow for computer-assisted analysis of the IS6110-R, DR, and PGRS fingerprints, an internal marker (with high- and low-molecular-mass standards) was added to the loading buffer with each sample as recommended by van Embden and colleagues (31, 33) for their standardized method for RFLP analysis with IS6110-R for the differentiation of M. tuberculosis isolates.
Spoligotyping.
PCR of the DR locus was performed with extracted DNA (6, 31) or heat-treated cell suspensions, and the spoligotyping method was performed as described previously (1).
Analysis of results.
Analysis of the bands generated with the IS6110 and DR probes and the spoligotypes for the set of isolates tested was performed with the aid of a computer software program by using the Dice unweighted pair group method with arithmetic averages (UPGMA) (GelCompar, version 3.1; Applied Maths, Kortrijk, Belgium). The profiles obtained by RFLP analysis with the PGRS probe were analyzed by using the clustering correlation and UPGMA (GelCompar).
Type nomenclature.
No standard nomenclature for the naming of different M. tuberculosis complex DNA types has yet been agreed upon internationally. For the purposes of this study, different numbers were allocated to isolates when a genetic difference could be detected. These different DNA types were given a prefix to show the probe used; IS for IS6110-R types, DR for DR types, SP for spoligotypes, and PG for PGRS types. In addition, isolates that were identified as unique strains were given an overall DNA type based on the composite results of the four different typing methods. Strains that had the most common IS6110-R, DR, and SP types (IS01, DR01, and SP01) and differed only in PGRS type were called type A and allocated the number of the PGRS type (e.g., A061 is IS01, DR01, SP01, and PG61). The prefixes B and C were given to isolates that had the most common IS6110-R and SP types (IS01 and SP01) with DR type 02 and DR type 04 patterns, respectively. For these isolates the number following the prefix was the number of the PGRS type. When strains had a common prefix, they were considered to be genetically more closely related to each other than to other strains that did not have the same prefix. Other strains were allocated a number only. Numbers were allocated as the isolates were analyzed and did not necessarily bear any relationship to specific patterns. For example, strains 001, 002, and 003 may not be any more closely related to each other than they may be to strain 023 or 091.
RESULTS
RFLP analysis with IS6110-R.
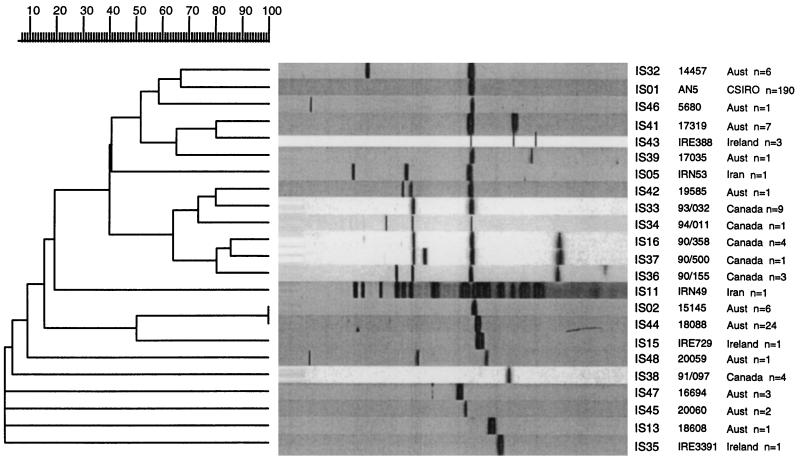
Two hundred thirty-three (85.4%) of the 273 isolates of M. bovis examined in this study harbored a single copy of IS6110. Of the 233 isolates with a single copy of IS6110, 190 (81.5%) carried the IS on a 1.9-kb restriction fragment, and this IS type was designated IS01. This means that almost 70% of all M. bovis isolates could not be differentiated by RFLP analysis with IS6110-R. Furthermore, 194 (91.9%) of the 211 Australian isolates had a single copy of IS6110, and 158 (81.4%) of these were type IS01. Figure 1 shows the 23 representative patterns (types) obtained by RFLP analysis with IS6110-R in a dendrogram indicating the relationships between the types. The Canadian isolates contained one (n = 15), two (n = 9), three (n = 5), or four (n = 4) copies of IS6110, and overall, nine different strains were identified among the Canadian isolates. Different strains were implicated in each of the four outbreaks (Table 1).
FIG. 1.
Dendrogram drawn by the GelCompar program showing the relationship of 23 representative fingerprints (IS types) for 273 isolates of M. bovis from Australia (n = 211) and overseas (n = 61) obtained by RFLP analysis with IS6110-R. Aust, Australia.
TABLE 1.
Results of DNA fingerprinting 61 M. bovis isolates from countries other than Australia and reference strain AN5 showing individual RFLP types with each of three probes, spoligotype, and overall RFLP type assigned to strains isolated from various animal hosts and geographical regions
| No. of isolates (no. of propertiesa) | RFLP type | No. of IS6110 copies | Type by RFLP analysis with the following probe:
|
Spoligotype | Host, no. of isolates (geographic originb) | ||
|---|---|---|---|---|---|---|---|
| IS6110 | DR | PGRS | |||||
| 3 (3) | A001 | 1 | 1 | 1 | 1 | 1 | Cattle, 2 (IRE, 1; IRN, 1); deer, 1 (IRE) |
| 1 (1) | A112 | 1 | 1 | 1 | 112 | 1 | Cattle (IRE) |
| 1 (1) | A128 | 1 | 1 | 1 | 128 | 1 | Deer (IRE) |
| 2 (2) | BCG1c | 1 | 1 | 24 | 120 | 7 | Human (IRN) |
| 1 (1) | 019 | 1 | 1 | 2 | 107 | 36 | Badger (IRE) |
| 1 (1) | 021 | 1 | 1 | 3 | 73 | 6 | Possum (NZ) |
| 1 (1) | 022 | 1 | 1 | 4 | 110 | 27 | Cattle (IRE) |
| 1 (1) | 024 | 1 | 1 | 6 | 129 | 6 | Human (IRE) |
| 3 (3) | 028 | 1 | 1 | 7 | 103 | 43 | Cattle (IRN) |
| 1 (1) | 029 | 1 | 1 | 7 | 106 | 43 | Cattle (IRN) |
| 1 (1) | 039 | 1 | 1 | 27 | 104 | 46 | Cattle (IRN) |
| 1 (1) | 040 | 1 | 1 | 29 | 74 | 42 | AN5 reference strain |
| 1 (1) | 045 | 1 | 1 | 47 | 113 | 37 | Cattle (UK) |
| 1 (1) | 046 | 1 | 1 | 47 | 114 | 37 | Cattle (UK) |
| 1 (1) | 049 | 1 | 1 | 57 | 109 | 47 | Cattle (IRE) |
| 2 (1) | 052 | 1 | 1 | 9 | 117 | 44 | Eland (CAN-A) |
| 10 (1) | 053 | 1 | 1 | 26 | 116 | 47 | Elk, 2; red deer, 2; cattle, 1; fallow deer, 1; Pere David deer, 1; cougar, 1; red deer crossbreed, 1; Sika deer, 1 (CAN-C) |
| 1 (1) | 059 | 3 | 5 | 2 | 7 | 1 | Cattle (IRN) |
| 1 (1) | 067 | 1 | 15 | 25 | 130 | 38 | Human (IRN) |
| 4 (1) | 068 | 3 | 16 | 23 | 115 | 43 | Elk, 3; human, 1 (CAN-B) |
| 9 (1) | 070 | 2 | 33 | 23 | 115 | 43 | Bison, 3; cattle, 2; elk, 1 (CAN-A) |
| 1 (1) | 071 | 3 | 34 | 23 | 115 | 43 | Yak (CAN-A) |
| 2 (1) | 072 | 1 | 35 | 28 | 108 | 53 | Badger (IRN) |
| 3 (1) | 073 | 4 | 36 | 23 | 115 | 43 | Elk (CAN-B) |
| 1 (1) | 074 | 4 | 37 | 23 | 115 | 43 | Bison (CAN-B) |
| 1 (1) | 075 | 1 | 38 | 53 | 111 | 50 | Cattle (UK) |
| 2 (1) | 076 | 1 | 38 | 30 | 118 | 51 | Cattle (CAN-D) |
| 1 (1) | 077 | 1 | 38 | 32 | 119 | 52 | Elk (CAN-D) |
| 3 (1) | 081 | 3 | 43 | 1 | 106 | 1 | Cattle, 2; badger, 1 (IRN) |
| 1 (1) | 087 | 19 | 11 | 22 | 29 | 56 | Cattle (IRN) |
The number of different properties from which isolates originated.
CAN, Canada; CAN-A, Canadian outbreak A; CAN-B, Canadian outbreak B; CAN-C, Canadian outbreak C; CAN-D, Canadian outbreak D; IRE, Republic of Ireland; IRN, Iran; NZ, New Zealand; UK, United Kingdom.
BCG1 isolates had RFLP patterns and spoligotypes typical of those of M. bovis BCG.
RFLP analysis with the DR probe.
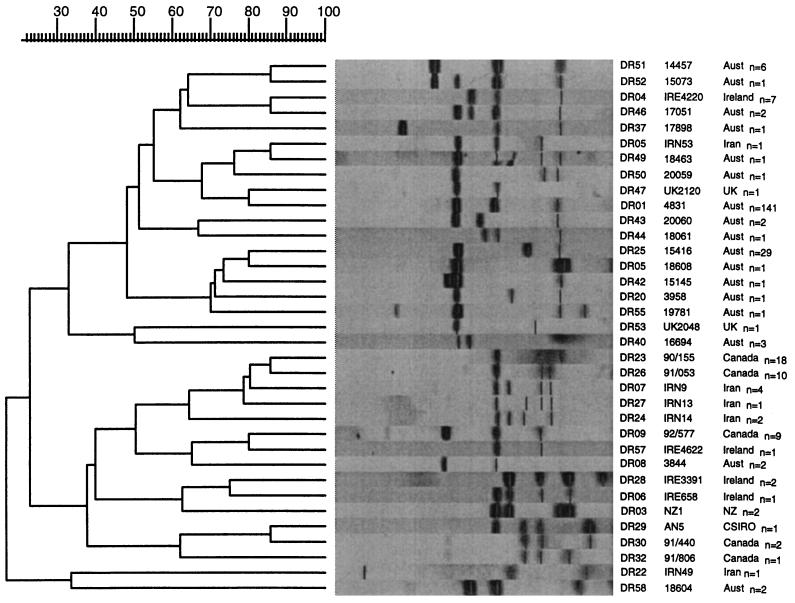
RFLP analysis with the DR probe identified 35 different types among the 273 M. bovis isolates. By RFLP analysis with DR, 141 (51.6%) of the isolates had a common fingerprint, designated DR01. Many of the Canadian and Iranian strains had DR patterns that appeared to be unique to the country of origin, and these were not identified among the Australian isolates. Figure 2 shows the 35 representative fingerprints obtained by RFLP analysis with the DR probe in a dendrogram showing the relationship between the types.
FIG. 2.
Dendrogram drawn by the GelCompar program showing the relationship of 35 representative fingerprints (DR types) for 273 isolates of M. bovis from Australia (n = 211) and overseas (n = 61) obtained by RFLP analysis with the DR probe. Aust., Australia, UK, United Kingdom; NZ, New Zealand.
Spoligotyping.
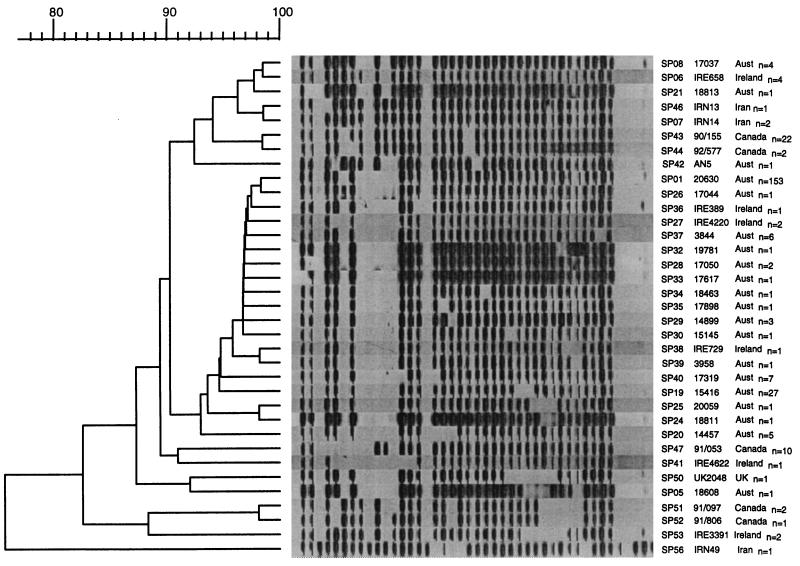
The spoligotyping method identified 35 different types among the 273 M. bovis isolates. One hundred fifty-three (56%) of the isolates had a common spoligotype (designated SP01) and therefore could not be differentiated by spoligotyping. Figure 3 shows the 35 representative spoligotypes in a dendrogram showing their relationships. With one exception (Iranian isolate IRN49), all of the isolates lacked the five spacers at the 3′ end of the DR locus. When the Iranian isolate was reexamined biochemically, its identity was more consistent with M. tuberculosis.
FIG. 3.
Dendrogram drawn by the GelCompar program showing the relationship of 35 representative SP types obtained for 273 isolates of M. bovis from Australia (n = 211) and overseas (n = 61) obtained by spoligotyping. Aust, Australia; UK, United Kingdom.
RFLP analysis with the PGRS probe.
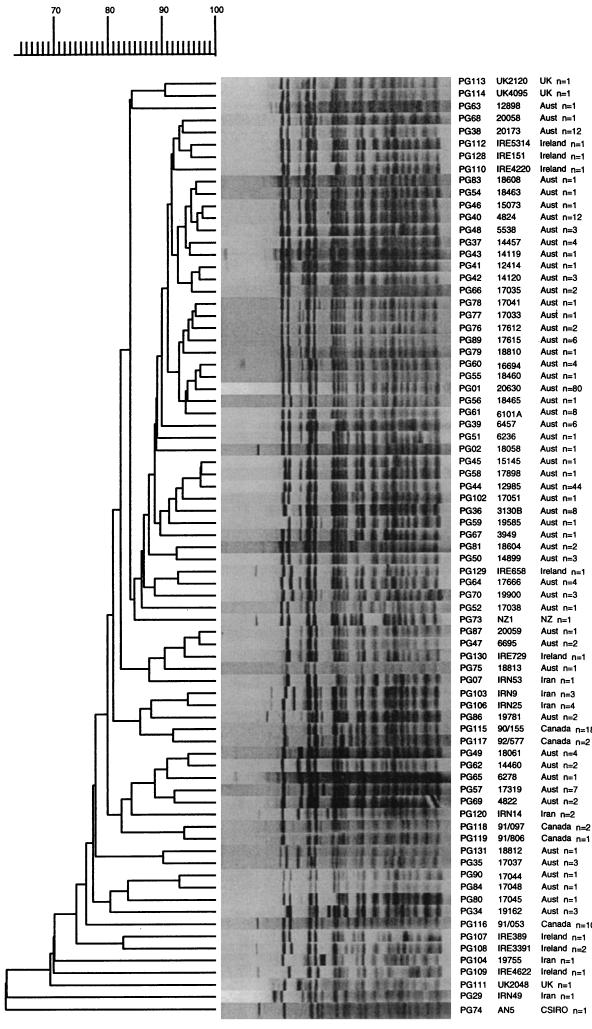
Use of the PGRS probe differentiated the M. bovis isolates into the most types, with 77 different types being identified among the 273 M. bovis isolates. Eighty (29.3%) of the isolates, including 77 (36.5%) of the Australian isolates, were identified as being of a common type, designated PG01. All of the isolates identified as PG01 harbored a single copy of IS6110, and 50 of the PG01 isolates, including 47 of the Australian isolates, carried a single copy of IS6110 on the 1.9-kb restriction fragment (type IS01). Figure 4 shows the 77 representative patterns obtained by RFLP analysis with the PGRS probe in a dendrogram showing their relationships. The Iranian M. tuberculosis-like isolate (IRN49) and the type strain, AN5, clustered separately from the other isolates of M. bovis.
FIG. 4.
Dendrogram drawn by the GelCompar program showing the relationship of 77 representative fingerprints (PG types) for 273 isolates of M. bovis from Australia (n = 211) and overseas (n = 61) obtained by RFLP analysis with the PGRS probe. UK, United Kingdom; Aust, Australia; NZ, New Zealand.
Composite typing based on four genetic markers for M. bovis.
When the results of the RFLP analyses with all three probes and the results of spoligotyping were considered, 99 different fingerprint combinations were identified among all of the M. bovis isolates. Forty-five (16.5%) of the isolates were of a common type, designated strain A001 (IS01, DR01, PG01, and SP01), and 42 (93.3%) of these originated from Australian animals (37 bovines, 1 buffalo, and 4 feral pigs).
Australian M. bovis isolates.
Most (n = 90) of the Australian isolates were type A strains, and they infected animals in all Australian states whose animals were tested and animals in the Northern Territory (Table 2). Many of these types were confined to particular properties or properties that were geographically or historically related. In Western Australia, type A strains were commonly found in cattle from all geographic regions (all 11 agricultural area properties, 10 of 13 northern pastoral properties, 12 of 15 West Kimberley properties, 5 of 6 East Kimberley properties, and both properties tested in the Broome area). Strain A001 was a relatively common cause of infection in Western Australia (14 of 47 properties), Queensland (8 of 18 properties), and the Northern Territory (9 of 25 properties). In Western Australia, strain A001 was found most commonly in cattle on properties in the agricultural (seven properties, including one outbreak involving five properties) and East Kimberley (four of five properties) areas. In one outbreak of tuberculosis in the agricultural area of Western Australia in 1989, M. bovis was isolated from 30 cattle and one goat from one property, and after tracing animal movements, a further four properties with infected animals and 55 infected cattle were detected. Representative isolates from animals on the five properties (10 bovine and 1 goat) were tested, and all of the bovine isolates were strain A001. The goat isolate was a different strain (A041) as a result of a unique type by RFLP analysis with PGRS (PG41); this type differed from type PG01 in one band in the high-molecular-mass region of the fingerprint (Fig. 4).
TABLE 2.
Results of DNA fingerprinting of 211 Australian M. bovis isolates showing individual RFLP types with each of three probes, spoligotype, and the overall RFLP type assigned to strains isolated from various animal hosts and geographical regions
| No. of isolates (no. of propertiesa) | RFLP type | No. of IS6110 copies | Type by RFLP analysis with the following probe:
|
Spoligotype | Host, no. of isolates (geographical originb) | ||
|---|---|---|---|---|---|---|---|
| IS6110 | DR | PGRS | |||||
| 42 (32) | A001 | 1 | 1 | 1 | 1 | 1 | Cattle, 27 (SW, 12; NT, 6; EK, 5; WK, 2; NSW, 1); buffalo, 1 (NT); feral pig, 4 (NT) |
| 8 (3) | A036 | 1 | 1 | 1 | 36 | 1 | Cattle (Vic) |
| 12 (1) | A038 | 1 | 1 | 1 | 38 | 1 | Cattle (SW) |
| 6 (3) | A039 | 1 | 1 | 1 | 39 | 1 | Cattle, 6 (PL, 5; SW, 1) |
| 11 (7) | A040 | 1 | 1 | 1 | 40 | 1 | Cattle, 11 (WK, 10; BM, 1) |
| 1 (1) | A041 | 1 | 1 | 1 | 41 | 1 | Goat (SW) |
| 3 (1) | A042 | 1 | 1 | 1 | 42 | 1 | Cattle (SW) |
| 1 (1) | A043 | 1 | 1 | 1 | 43 | 1 | Cattle (SW) |
| 1 (1) | A044 | 1 | 1 | 1 | 44 | 1 | Cattle (PL) |
| 1 (1) | A047 | 1 | 1 | 1 | 47 | 1 | Cattle (WK) |
| 3 (1) | A048 | 1 | 1 | 1 | 48 | 1 | Cattle, 3 (WK, 2; BM, 1) |
| 1 (1) | A051 | 1 | 1 | 1 | 51 | 1 | Cattle (BM) |
| 1 (1) | A052 | 1 | 1 | 1 | 52 | 1 | Cattle (NT) |
| 1 (1) | A055 | 1 | 1 | 1 | 55 | 1 | Cattle (QLD) |
| 1 (1) | A056 | 1 | 1 | 1 | 56 | 1 | Cattle (QLD) |
| 1 (1) | A060 | 1 | 1 | 1 | 60 | 1 | Cattle (WK) |
| 8 (5) | A061 | 1 | 1 | 1 | 61 | 1 | Cattle (PL) |
| 2 (1) | A062 | 1 | 1 | 1 | 62 | 1 | Cattle (WK) |
| 1 (1) | A063 | 1 | 1 | 1 | 63 | 1 | Cattle (PL) |
| 4 (3) | A064 | 1 | 1 | 1 | 64 | 1 | Cattle (WK) |
| 1 (1) | A065 | 1 | 1 | 1 | 65 | 1 | Cattle (BM) |
| 1 (1) | A066 | 1 | 1 | 1 | 66 | 1 | Cattle (NT) |
| 1 (1) | A069 | 1 | 1 | 1 | 69 | 1 | Cattle (WK) |
| 3 (1) | A070 | 1 | 1 | 1 | 70 | 1 | Cattle (QLD) |
| 2 (1) | A076 | 1 | 1 | 1 | 76 | 1 | Cattle (NT) |
| 1 (1) | A077 | 1 | 1 | 1 | 77 | 1 | Cattle (NT) |
| 1 (1) | A078 | 1 | 1 | 1 | 78 | 1 | Cattle (NT) |
| 1 (1) | A080 | 1 | 1 | 1 | 80 | 1 | Cattle (NT) |
| 1 (1) | A084 | 1 | 1 | 1 | 84 | 1 | Cattle (NT) |
| 6 (1) | A089 | 1 | 1 | 1 | 89 | 1 | Cattle (NT) |
| 3 (1) | B034 | 1 | 1 | 2 | 34 | 1 | Cattle (QLD) |
| 2 (2) | C001 | 1 | 1 | 4 | 1 | 1 | Cattle, 2 (NT, 1; QLD, 1) |
| 1 (1) | C079 | 1 | 1 | 4 | 79 | 1 | Cattle (NT) |
| 5 (1) | 002 | 1 | 2 | 25 | 1 | 19 | Cattle (WK) |
| 21 (1) | 003 | 1 | 44 | 25 | 1 | 19 | Cattle (WK) |
| 1 (1) | 004 | 1 | 44 | 25 | 68 | 19 | Cattle (WK) |
| 4 (2) | 005 | 2 | 32 | 51 | 37 | 20 | Cattle (PL) |
| 1 (1) | 006 | 2 | 32 | 51 | 67 | 20 | Cattle (WK) |
| 2 (1) | 007 | 1 | 1 | 4 | 50 | 29 | Cattle (WK) |
| 1 (1) | 008 | 1 | 1 | 1 | 40 | 6 | Cattle (WK) |
| 1 (1) | 009 | 2 | 32 | 52 | 46 | 1 | Cattle (WK) |
| 1 (1) | 010 | 1 | 1 | 51 | 1 | 28 | Cattle (WK) |
| 1 (1) | 011 | 1 | 2 | 42 | 45 | 30 | Cattle (WK) |
| 1 (1) | 012 | 1 | 44 | 20 | 1 | 30 | Cattle (PL) |
| 2 (1) | 013 | 1 | 1 | 8 | 49 | 37 | Cattle (EK) |
| 1 (1) | 014 | 2 | 46 | 1 | 47 | 1 | Cattle (WK) |
| 1 (1) | 016 | 1 | 1 | 1 | 1 | 28 | Buffalo (NT) |
| 1 (1) | 017 | 1 | 1 | 1 | 1 | 33 | Cattle (NT) |
| 1 (1) | 018 | 1 | 1 | 1 | 50 | 29 | Cattle (BM) |
| 1 (1) | 020 | 1 | 1 | 3 | 2 | 6 | Cattle (QLD) |
| 1 (1) | 023 | 1 | 1 | 4 | 90 | 26 | Cattle (NT) |
| 1 (1) | 032 | 1 | 1 | 9 | 131 | 8 | Buffalo (NT) |
| 3 (1) | 033 | 1 | 1 | 9 | 35 | 8 | Buffalo (NT) |
| 1 (1) | 034 | 1 | 1 | 9 | 75 | 21 | Buffalo (NT) |
| 1 (1) | 035 | 1 | 1 | 9 | 86 | 21 | Cattle (NT) |
| 1 (1) | 041 | 1 | 1 | 37 | 58 | 35 | Cattle (QLD) |
| 1 (1) | 042 | 1 | 1 | 44 | 49 | 1 | Cattle (QLD) |
| 1 (1) | 043 | 1 | 1 | 44 | 49 | 27 | Cattle (QLD) |
| 1 (1) | 044 | 1 | 1 | 46 | 102 | 1 | Buffalo (NT) |
| 1 (1) | 047 | 1 | 1 | 49 | 54 | 34 | Cattle (QLD) |
| 1 (1) | 048 | 1 | 1 | 55 | 86 | 32 | Cattle (NT) |
| 2 (1) | 050 | 1 | 1 | 58 | 81 | 37 | Buffalo (NT) |
| 1 (1) | 065 | 1 | 13 | 5 | 83 | 5 | Cattle (NT) |
| 1 (1) | 078 | 2 | 39 | 1 | 66 | 1 | Cattle (NT) |
| 7 (1) | 079 | 2 | 41 | 44 | 57 | 40 | Cattle (QLD) |
| 1 (1) | 080 | 3 | 42 | 44 | 59 | 1 | Cattle (QLD) |
| 1 (1) | 082 | 1 | 44 | 25 | 1 | 24 | Buffalo (NT) |
| 2 (2) | 083 | 1 | 45 | 43 | 1 | 1 | Cattle (NT) |
| 3 (2) | 084 | 1 | 47 | 40 | 60 | 1 | Cattle (QLD) |
| 1 (1) | 085 | 3 | 38 | 50 | 87 | 25 | Buffalo (NT) |
The number of different properties from which isolates originated.
WK, West Kimberley, Western Australia; EK, East Kimberley, Western Australia; SW, southwest agricultural area, Western Australia; PL, Pilbara area, Western Australia; BM, Broome area, Western Australia; WA, Western Australia; NT, Northern Territory; QLD, Queensland; NSW, New South Wales; Vic, Victoria.
Canadian M. bovis isolates.
Outbreaks A and B involved multicopy IS6110 strains that had common bands (outbreak A, two common bands; outbreak B, three common bands), but single band changes were detected after infection in a yak (outbreak A) and a bison (outbreak B) (Fig. 1; Table 1). On the original premises in outbreak A, isolates from two elands obtained in 1992 and 1993 were identified as strain 052, whereas an elk, two cattle, and two bison were infected with strain 070 and the yak was infected with strain 071. Isolates from another four bison from three premises were also strain 070. In outbreak B, all isolates from four elk and a bison had identical DR, SP, and PG types, but differences were detected in the number of IS6110 copies and the banding patterns, despite three shared IS6110-R fragments. Two different types by RFLP analysis with IS6110-R, IS36 and IS16 (strains 073 and 068), were identified in four infected elk, and a third type by RFLP analysis with IS6110-R, IS37 (strain 074), was identified in an infected bison. Strain 068, isolated from the veterinarian, was different from the strain isolated from the elk that he treated (strain 073), but it was isolated from another elk that had been in contact with the elk that he treated. All animals involved in outbreak C, including the deer imported from New Zealand, were infected with the same strain of M. bovis, identified as strain 053. The cougar was infected with M. bovis and Mycobacterium avium. In outbreak D, the strain infecting cattle on one property (strain 076) had DR, PG, and SP types different from those of the strain isolated from an elk (strain 077) from the national park on the border of the premises from which strain 076 was isolated, suggesting that infection had originated from different sources.
Effectiveness of GelCompar in the analysis of genetic types.
GelCompar was a very effective means of analyzing the different fingerprints obtained by the RFLP analyses with IS6110-R and DR and by the spoligotyping method. However, in analyzing the fingerprints obtained by RFLP analysis with PGRS, which were much more complex because of the number of bands that hybridized with the PGRS probe, the program was less effective. Because of this, all isolates that had similar patterns by RFLP analysis with PGRS had to be checked manually.
DISCUSSION
The M. bovis isolates examined in this study could be characterized to various degrees by up to four genotyping procedures. Each of the procedures was able to differentiate strains of M. bovis and could be used for epidemiological studies of M. bovis isolates from animals. However, the combined use of the four procedures resulted in superior differentiation of strains for a detailed epidemiological investigation. The finding of a genetic difference between two strains by the use of any particular marker would imply infection with different strains, presumably originating from different sources. Confidence in this assumption would be increased if more than one typing system detected a difference, since three of the genetic markers detect changes or mutations within different parts of the genome. One example of this was a case in which strain 008 (IS42, DR44, PG59, and SP01) was identified from an animal in 1984 and strain 009 (IS32, DR52, PG46, and SP01) was isolated from animals on the same property in 1991. Apart from the spoligotyping method, each of the markers identified a difference between the two isolates, suggesting that they were not closely related genetically and that infection was introduced from different sources. It must be remembered, however, that some genetic change or drift must occur to account for the development of different strains, and one method might identify an initial change earlier than another technique. The DNA polymorphism driven by insertion elements, such as IS6110, is due to their inherent capacity to move about the genome with little target specificity. Studies on the nature of genetic rearrangements within the DR region suggested that homologous recombination between the small DRs was the predominant kind of rearrangement for this repetitive element (11), and it seems likely that the same mechanism may contribute to polymorphisms with PGRS (35). Certainly, there was evidence of stability with each of the markers used in this study, providing confidence in the ability of these techniques to provide useful information for epidemiological studies. In several cases in Western Australia, where isolates from cattle from the same property were available for study over periods of up to 7 years, studies with each of the markers gave consistent results.
Just how frequently a strain of M. bovis will alter its genetic makeup and how often these differences will be detectable are unknown. The fact that the PGRS probe detected the greatest number of types is best explained by the fact that these repetitive elements are found at various locations around the genome of M. bovis, whereas the DR probe and the spacers used in spoligotyping target the same region. RFLP analysis with the DR probe targets the DRs around the point of the IS6110 insertion (13), and spoligotyping identifies the presence or absence of specific spacers between the DRs (11).
In strains with a single copy of IS6110, the IS is inserted between two DRs and has been found to transpose relatively infrequently (13). In this study, 85.4% of the M. bovis isolates examined had a single copy of IS6110, and the percentage of Australian isolates that had a single copy was slightly higher (91.9%). In addition, the majority of these single-copy strains carried the IS on the 1.9-kb fragment considered characteristic for M. bovis isolates from cattle (32), thereby minimizing the effectiveness of IS6110-R for use in the characterization of M. bovis in this study. These results are consistent with the results of RFLP analysis with IS6110 for isolates of M. bovis from New Zealand (3) and bovine and human isolates of M. bovis from Argentina (32). In a study which examined Argentinian isolates (32), it was suggested that strains with multiple copies of IS6110 were more likely to have originated from wild or zoo animals other than cattle or from humans from The Netherlands. Six to eight copies of IS6110 were present in all 23 goat strains examined in Spain, yet 16 of 17 cattle were infected with strains with a single copy of IS6110 (12), suggesting that different reservoirs for caprine and bovine tuberculosis existed in Spain. A more comprehensive study involving 129 isolates of M. bovis from cattle, goats, cats, and a sheep in Spain demonstrated that almost 50% of the bovine strains examined harbored multiple IS6110 copies, but found that bovine and caprine isolates still clustered separately (15). The results of those studies and the results reported in this paper indicate that different countries or animal hosts may harbor unique clonal populations of M. bovis.
The Canadian isolates examined in this study were quite distinct from the Australian M. bovis isolates that were examined. Many of the Canadian isolates had multiple copies of IS6110 (outbreaks A and B), and their DR, PG, and SP patterns were also different from those commonly found in Australian strains. RFLP analysis with IS6110 is considered to be a sensitive method for the detection of genetic changes in isolates of M. tuberculosis or M. bovis that have multiple copies of this IS (5, 32, 33). This was well demonstrated in isolates from Canadian outbreaks A and B in which genetic change was evident in the slight rearrangement of IS6110 fragments, without alteration in the patterns obtained by RFLP analysis with the PGRS and DR probes. In outbreaks A and B, two and three common IS6110 bands were present, respectively, and single band changes were detected after infection in a yak (outbreak A) and a bison (outbreak B). The rate of transposition of the IS element is considered to be a factor of time (35); however, there was some evidence from the fingerprints for the Canadian isolates obtained by RFLP analysis with IS6110-R suggesting that the M. bovis strains may have altered in different hosts during the course of the infection in outbreaks A and B. Similar band changes were identified in an outbreak of M. bovis in Swedish deer (29) and in cattle in Burundi (23). It is not known whether the isolation of strains, from different animal species, with minor differences in profiles by RFLP analysis with IS6110 (insertions or deletions), such as those seen in outbreaks A and B, was related to host factors or represented natural evolution of the bacterium over time. In contrast, isolates recovered from two elands in outbreak A were clearly different from those recovered from other animals in 1993 and 1994 by each of the fingerprinting methods used, suggesting a clear genetic difference in the strain infecting the two elands and confirming an alternate source of infection for these animals.
PGRS was by far the best single genetic marker for RFLP analysis with which to characterize the majority of the M. bovis isolates encountered in Australia and, with the exception of isolates from Canada, the M. bovis isolates from other countries examined in this study. In fact, RFLP analysis with PGRS identified twice as many types as RFLP analysis with DR and spoligotyping and more than three times as many types as RFLP analysis with IS6110-R. This is consistent with previous findings (6) but differs from those of a study in Northern Ireland in which the results of RFLP analyses with PGRS and IS6110 were found to be virtually equivalent (27). Skuce et al. (27) used the entire IS6110 probe, which is known to identify slightly more polymorphisms than the RHS IS6110 probe used in this study. In a direct comparison of LHS and RHS IS6110 probes, the LHS probe was able to identify three different types among a group of 15 isolates that were a common IS6110 type (single band at 1.9 kb) (15). The digests used for our studies were run longer than those of Skuce et al. (27) and those reported previously (6) in an attempt to improve the resolution of bands. In addition, we included RFLP bands of >1.3 kb in the analysis. In Northern Ireland, more than 40% of 109 isolates were identified as having a common pattern by using a combination of PGRS, IS6110, and IS1081 probes, and almost 50% of the isolates had a common pattern when the PGRS probe was used (27). By comparison, 29.3% of the isolates were identified in this study as a common type by RFLP analysis with PGRS, PG01, and 36.5% of Australian isolates were identified as PG01. The use of other genetic markers resulted in further characterization of 45% of these isolates with the common PG01 type, so that 19.9% of Australian isolates were identified as the most common strain, designated A001. In a more recent study in Northern Ireland (28), bands greater than 2.26 kb were included in the analysis, but the sensitivity of RFLP analysis with PGRS was not markedly improved. Additional studies may determine whether the common strain found in Northern Ireland can be further differentiated and whether it is the same as any of the more common strains found in Australia.
The finding of a common strain distributed throughout Australia suggests that animals on many properties were infected from the same source. This may have occurred over time as infected cattle moved from one area to another throughout the country and spread infection caused by a single clone. It is generally accepted that bovine tuberculosis was introduced into Australia with the introduction of infected cattle during the early settlement period (26). The number of introductions would have been limited, and this may account for some of the genetic similarity of strains seen in Australia. The finding of a further 22.7% of isolates with common IS, DR, and SP types but different PG types (type A strains other than type A001) suggests the clonal expansion of strain A001. The question of host influence versus genetic drift over time was also raised with the finding of type A041 in a single goat in a mixed cattle and goat herd in Western Australia in which cattle were infected with strain A001. Four other cattle herds that were epidemiologically linked to this herd were also infected with strain A001. The goats on the affected property were in poor condition and were considered to be highly stressed because of the excessively high stocking rate. It is possible that the host response to infection in this goat may have caused the genetic makeup of the organism to alter.
RFLP analysis with the DR probe produced results similar to those obtained by the spoligotyping method, a finding that is not surprising, considering that both target the same chromosomal locus. In one previous small-scale comparison of IS01 isolates, analysis with PGRS allowed for a somewhat better differentiation than analysis with DR (32), but another study suggested that the results of analyses with the two markers were equivalent (12).
The advantages of spoligotyping lie in the speed and low cost of the technique and in the ease of analysis. The spoligotyping method was originally designed for epidemiological studies of M. tuberculosis, and as such, 37 of the 43 spacers presently used on the membranes originate from M. tuberculosis H37Rv and 6 spacers were derived from M. bovis BCG (11). Better discrimination of M. bovis strains may be achieved by this technique in the future by the identification and use of spacer sequences that are specific for M. bovis strains. The spoligotyping technique has the added advantage that it can be used directly with isolates and tissue specimens for the rapid screening of M. bovis isolates. It would appear to be sensible to further characterize isolates with common spoligotypes at least with the PGRS probe. RFLP analyses with IS6110-R and DR could be used if full characterization was needed. The lack of the five spacers at the 3′ end of the DR locus detected by spoligotyping is considered to be consistent with M. bovis (14). All but 1 of the 273 isolates of M. bovis tested in this study lacked these spacers. The one isolate (IRN49) that was found to contain four of the five 3′ end spacers was biochemically more consistent with M. tuberculosis, and use of each of the probe methods produced an unusual pattern for M. bovis. These findings confirmed the lack of the 3′ DR spacers as being consistent with the identification of M. bovis isolates.
Spread of infection with M. bovis between animals is generally considered to be via aerosols, although ingestion of infectious materials is also accepted as a route of transmission (20, 22). Contact with infected nasal mucus was identified as an important source of infection in a series of studies with naturally and experimentally infected cattle in Northern Ireland (17–19). In the present study, DNA fingerprinting results for isolates from Canadian outbreak C were able to support the proposal that nose-to-nose contact between elk and deer and between deer and cattle on opposite sides of a fence had spread the infection. In addition, DNA fingerprinting provided confirmatory evidence that a cougar on one property had become infected with M. bovis and M. avium after being fed tuberculosis-affected carcasses of other animals on the premises and chickens from a neighboring farm, respectively.
Although early studies suggested that M. bovis could survive in cow feces for between 2 and 5 months, depending on the season, and in soil for 2 years, more recent evidence suggests that survival in soil or feces exposed to natural conditions of sunlight is limited (36). Under Australian conditions, M. bovis survived for 4 weeks in soil in 80% shade or more, but no isolation from dry or moist soils exposed to sunlight or from feces held in any conditions was made at 4 weeks (9). In Australia, when infected properties are destocked, the owner is advised to wait a minimum of 30 days before restocking. This precaution is taken to minimize the risk of reinfection when new stock are brought in. In the case of outbreak C in Canada, one property that had undergone cleaning and disinfection and that had been released from quarantine was restocked with deer from New Zealand that were skin test negative. The finding in three imported New Zealand deer of a strain of M. bovis identical to the strain previously identified on the property confirmed infection with the same strain in the imported animals. It is possible that the M. bovis organisms on the property remained infectious, despite thorough decontamination procedures. Another possible explanation is that the New Zealand animals, which had passed pre- and postquarantine testing and isolation, were infected, coincidentally, with an identical strain before they arrived at the premises. Alternatively, a worker in contact with both groups of animals may have acted as a vector in passing on infection to the newly arrived animals. Typing of strains from the herd or region of origin in New Zealand may assist in clarifying this situation.
It was expected that some similarities would be found between isolates from Australia and those from the United Kingdom and the Republic of Ireland since M. bovis was believed to have been originally imported with infected cattle from these countries. In fact, only a few similarities were seen between some of the Australian strains and a small number of isolates from Ireland and one isolate from Iran. This similarity was seen by the identification of some type A strains from Ireland, suggesting a common clonal origin for these strains in Australia and Ireland. The majority of isolates from other countries appeared to be unique. Certainly, the isolates from Canada were clearly different from the “Australian” strains, and this is consistent with observations that geographically distinct strains exist in The Netherlands and Argentina (32), regions where clones of organisms may have spread within countries with little opportunity for genetic exchange. The finding of geographically distinct populations of M. bovis may be useful for confirming the source of infection in any imported animals that are subsequently diagnosed with bovine tuberculosis.
ACKNOWLEDGMENTS
This work was supported by the Australian Brucellosis and Tuberculosis Eradication Campaign.
We gratefully acknowledge the many collaborators who provided isolates or reference strains for study, in particular, Elizabeth Rohonczy and Claude Turcotte, Animal Disease Research Institute, Nepean, Ontario, Canada; Anne Fanning, TB Services, Alberta Health, Edmonton, Alberta, Canada; Louis O’Reilly, Veterinary Research Laboratory, Abbottstown, Castleknock, Ireland; Mohammad Feizabadi, courtesy of the Razi Institute, Tehran, Iran; and Jeremy Dale, Surrey University, Guildford, United Kingdom. In addition, we acknowledge Barry Francis for excellent technical assistance and colleagues in Australian State, Northern Territory, and CSIRO Animal Health laboratories for supplying Australian isolates for typing.
REFERENCES
- 1.Aranaz A, Liébana E, Mateos A, Dominguez L, Vidal D, Domingo M, Gonzáles O, Rodrigues-Ferri E F, Bunshoten A, van Embden J, Cousins D. Spoligotyping of Mycobacterium bovis strains from cattle and other animals: a tool for epidemiology of tuberculosis. J Clin Microbiol. 1996;34:2734–2740. doi: 10.1128/jcm.34.11.2734-2740.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck-Sagué C, Dooley S W, Hutton M D, Otten J, Breeden A, Crawford J T, Pitchenik A E, Woodley C, Cauthen G, Jarvis W R. Hospital outbreak of multidrug-resistant Mycobacterium tuberculosis infections. JAMA. 1992;268:1280–1286. doi: 10.1001/jama.1992.03490100078031. [DOI] [PubMed] [Google Scholar]
- 3.Collins D M, Erasmuson S K, Stephens D M, Yates G F, de Lisle G W. DNA fingerprinting of Mycobacterium bovis strains by restriction fragment analysis and hybridization with insertion elements IS1081 and IS6110. J Clin Microbiol. 1993;31:1143–1147. doi: 10.1128/jcm.31.5.1143-1147.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cousins D V. Molecular epidemiology and diagnosis of Mycobacterium bovis and M. bovis-like organisms causing tuberculosis. Ph.D. thesis. Perth, Australia: University of Western Australia; 1996. [Google Scholar]
- 5.Cousins, D. V., R. A. Skuce, R. R. Kazwala, and J. D. A. van Embden. Towards a standardized approach to DNA fingerprinting of Mycobacterium bovis. Int. J. Tuberc. Lung Dis., in press. [PubMed]
- 6.Cousins D V, Williams S N, Ross B C, Ellis T M. Use of a repetitive element isolated from Mycobacterium tuberculosis in hybridization studies with Mycobacterium bovis: a new tool for epidemiological studies of bovine tuberculosis. Vet Microbiol. 1993;37:1–17. doi: 10.1016/0378-1135(93)90178-a. [DOI] [PubMed] [Google Scholar]
- 7.Daley C L, Small P M, Schecter G F, Schoolnik G K, McAdam R A, Jacobs W R, Jr, Hopewell P C. An outbreak of tuberculosis with accelerated progression among persons infected with human immunodeficiency virus: an analysis using restriction fragment length polymorphisms. N Engl J Med. 1992;326:231–235. doi: 10.1056/NEJM199201233260404. [DOI] [PubMed] [Google Scholar]
- 8.Doran T J, Hodgson A L M, Davies J K, Radford A J. Characterisation of a highly repeated DNA sequence from Mycobacterium bovis. FEMS Microbiol Lett. 1993;111:147–152. doi: 10.1111/j.1574-6968.1993.tb06377.x. [DOI] [PubMed] [Google Scholar]
- 9.Duffield B J, Young D A. Survival of Mycobacterium bovis in defined environmental conditions. Vet Microbiol. 1985;10:193–197. doi: 10.1016/0378-1135(85)90021-5. [DOI] [PubMed] [Google Scholar]
- 10.Dwyer B, Jackson K, Raios K, Sievers A, Wilshire E, Ross B. DNA restriction fragment analysis to define an extended cluster of tuberculosis in homeless men and their associates. J Infect Dis. 1993;167:490–494. doi: 10.1093/infdis/167.2.490. [DOI] [PubMed] [Google Scholar]
- 11.Groenen P M A, Bunschoten A E, van Soolingen D, van Embden J D A. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application of strain differentiation by a novel typing method. Mol Microbiol. 1993;10:1057–1065. doi: 10.1111/j.1365-2958.1993.tb00976.x. [DOI] [PubMed] [Google Scholar]
- 12.Gutiérrez M, Samper S, Gavigan J A, García-Marín J F, Martín C. Diffentiation by molecular typing of Mycobacterium bovis strains causing tuberculosis in cattle and goats. J Clin Microbiol. 1995;33:2953–2956. doi: 10.1128/jcm.33.11.2953-2956.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hermans P W M, van Soolingen D, Bik E M, de Haas P E W, Dale J W, van Embden J D A. The insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect Immun. 1991;59:2695–2705. doi: 10.1128/iai.59.8.2695-2705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamerbeek J, Schouls L, van Agterveld M, Kolk A, Kuijper S, van Soolingen D, de Haas P, Bunschoten A, van Embden J. Simultaneous strain detection and differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liébana E, Aranaz A, Dominguez L, Mateos A, Gonzáles-Llamazares O, Rodrigues-Ferri E F, Domingo M, Vidal D, Cousins D. The insertion element IS6110 is a useful tool for DNA fingerprinting of Mycobacterium bovis isolates from cattle and goats in Spain. Vet Microbiol. 1997;54:223–233. doi: 10.1016/s0378-1135(96)01282-5. [DOI] [PubMed] [Google Scholar]
- 16.Mazurek G H, Cave M D, Eisenach K D, Wallace R J J, Bates J H, Crawford J T. Chromosomal DNA fingerprint patterns produced with IS6110 as strain-specific markers for epidemiologic study of tuberculosis. J Clin Microbiol. 1991;29:2030–2033. doi: 10.1128/jcm.29.9.2030-2033.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McIlroy S G, Neill S D, McCracken R M. Pulmonary lesions and Mycobacterium bovis excretion from the respiratory tract of tuberculin reacting cattle. Vet Rec. 1986;118:718–721. doi: 10.1136/vr.118.26.718. [DOI] [PubMed] [Google Scholar]
- 18.Neill S D, Hanna J, Mackie D P, Bryson T G D. Isolation of Mycobacterium bovis from the respiratory tract of skin test-negative cattle. Vet Rec. 1992;131:45–47. doi: 10.1136/vr.131.3.45. [DOI] [PubMed] [Google Scholar]
- 19.Neill S D, Hanna J, O’Brien J J, McCracken R M. Transmission of tuberculosis from experimentally infected cattle to in-contact calves. Vet Rec. 1989;124:269–271. doi: 10.1136/vr.124.11.269. [DOI] [PubMed] [Google Scholar]
- 20.Neill S D, Pollock J M, Bryson D B, Hanna J. Pathogenesis of Mycobacterium bovis infection in cattle. Vet Microbiol. 1994;40:41–52. doi: 10.1016/0378-1135(94)90045-0. [DOI] [PubMed] [Google Scholar]
- 21.Otal I, Martín C, Vincent-Levy-Frébault V, Thierry D, Gicquel B. Restriction fragment length polymorphism analysis using IS6110 as an epidemiological marker in tuberculosis. J Clin Microbiol. 1991;29:1252–1254. doi: 10.1128/jcm.29.6.1252-1254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pritchard D G. A century of bovine tuberculosis 1888–1988: conquest and controversy. J Comp Pathol. 1988;99:357–399. doi: 10.1016/0021-9975(88)90058-8. [DOI] [PubMed] [Google Scholar]
- 23.Rigouts L, Maregeya B, Traore H, Collart J P, Fissette K, Portaels F. Use of DNA restriction fragment typing in the differentiation of Mycobacterium tuberculosis complex isolates from animals and humans in Burundi. Tubercle Lung Dis. 1996;77:264–268. doi: 10.1016/s0962-8479(96)90011-8. [DOI] [PubMed] [Google Scholar]
- 24.Romano M I, Alito A, Fisanotti J C, Bigi F, Kantor I, Cicuta M E, Cataldi A. Comparison of different genetic markers for molecular epidemiology of bovine tuberculosis. Vet Microbiol. 1996;50:59–71. doi: 10.1016/0378-1135(95)00197-2. [DOI] [PubMed] [Google Scholar]
- 25.Ross B C, Raios K, Jackson K, Dwyer B. Molecular cloning of a highly repeated DNA element from Mycobacterium tuberculosis and its use as an epidemiologic tool. J Clin Microbiol. 1992;30:942–946. doi: 10.1128/jcm.30.4.942-946.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seddon H R, Albiston H E. Bacterial diseases. Vol. 1. Canberra, Australia: Commonwealth Department of Health; 1965. [Google Scholar]
- 27.Skuce R A, Brittain D, Hughes M S, Beck L-A, Neill S D. Genomic fingerprinting of Mycobacterium bovis from cattle by restriction fragment length analysis. J Clin Microbiol. 1994;32:2387–2392. doi: 10.1128/jcm.32.10.2387-2392.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skuce R A, Brittain D, Hughes M S, Neill S D. Differentiation of Mycobacterium bovis isolates from animals by DNA typing. J Clin Microbiol. 1996;34:2469–2474. doi: 10.1128/jcm.34.10.2469-2474.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szewzyk R, Svenson S B, Hoffner S E, Bölske G, Wahlström H, Englund L, Engvall A, Källenius G. Molecular epidemiological studies of Mycobacterium bovis infections in humans and animals in Sweden. J Clin Microbiol. 1995;33:3183–3185. doi: 10.1128/jcm.33.12.3183-3185.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torrea G, Levee G, Grimont P, Martin C, Chanteau S, Gicquel B. Chromosomal DNA fingerprinting analysis using the insertion sequence IS6110 and the repetitive element DR as strain-specific markers for epidemiolical study of tuberculosis in French polynesia. J Clin Microbiol. 1995;33:1890–1895. doi: 10.1128/jcm.33.7.1899-1904.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Embden J D A, Cave D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Soolingen D, de Haas P E W, Haagsma J, Eger T, Hermans P W M, Ritacco V, Alito A, van Embden J D A. Use of various genetic markers in differentiation of Mycobacterium bovis strains from animals and humans and for studying epidemiology of bovine tuberculosis. J Clin Microbiol. 1994;32:2425–2433. doi: 10.1128/jcm.32.10.2425-2433.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Soolingen D, de Haas P E W, Hermans P W M, Groenen P M A, van Embden J D A. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J Clin Microbiol. 1993;31:1987–1995. doi: 10.1128/jcm.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Soolingen D, Hermans P W M, de Haas P E W, Soll D R, van Emden J D A. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Soolingen D, Qian L, de Haas P E W, Douglas J T, Traore H, Portaels F, Xi Qing H, Enkhsaikan D, Nymadawa P, van Embden J D A. Predominance of a single genotype of Mycobacterium tuberculosis in countries of Asia. J Clin Microbiol. 1995;33:3234–3238. doi: 10.1128/jcm.33.12.3234-3238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wray C. Survival and spread of pathogenic bacteria of veterinary importance within the environment. Vet Bull. 1975;45:543–550. [Google Scholar]
- 37.Yang Z H, de Haas P E W, van Soolingen D, van Embden J D A, Anderson Å B. Restriction fragment length polymorphism of Mycobacterium tuberculosis strains isolated from Greenland during 1992: evidence of tuberculosis transmission between Greenland and Denmark. J Clin Microbiol. 1994;32:3018–3025. doi: 10.1128/jcm.32.12.3018-3025.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuen L K W, Ross B C, Jackson K M, Dwyer B. Characterization of Mycobacterium tuberculosis strains from Vietnamese patients by Southern blot hybridization. J Clin Microbiol. 1993;31:1615–1618. doi: 10.1128/jcm.31.6.1615-1618.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]