Abstract
We constructed internal controls (ICs) to provide assurance that clinical specimens are successfully amplified and detected. The IC nucleic acids contain primer binding regions identical to those of the target sequence and contain a unique probe binding region that differentiates the IC from amplified target nucleic acid. Because only 20 copies of the IC are introduced into each test sample, a positive IC signal indicates that amplification was sufficient to generate a positive signal from targets present at the limit of test sensitivity. The COBAS AMPLICOR Chlamydia trachomatis, Neisseria gonorrhoeae, Mycobacterium tuberculosis, and human hepatitis C virus tests exhibited inhibition rates ranging from 5 to 9%. Approximately 64% of these inhibitory specimens were not inhibitory when a second aliquot was tested. Because repeatedly inhibitory specimens were not reported as false negative and because additional infected specimens were detected during retesting, test sensitivities were 1 to 6% greater than they would have been if the IC had not been used.
Because of its ability to specifically amplify minute quantities of nucleic acid, PCR has been applied with great success in clinical diagnostics (7, 8, 29). Relatively simple procedures for extracting nucleic acids from clinical specimens provide samples of reasonable purity without requiring hazardous chemicals and extensive manipulation (10). Nevertheless, extracted clinical specimens may contain impurities that inhibit enzyme-based nucleic acid amplification processes. Numerous studies have demonstrated that a small, but significant, proportion of clinical specimens contain substances that inhibit PCR, the ligase chain reaction (LCR), and transcription-mediated amplification (TMA) (1–6, 11, 14, 21, 22, 24–26, 30).
Unless inhibitory specimens are identified, negative amplification test results do not necessarily indicate absence of infection. Inhibitory specimens can be identified by monitoring amplification of a second target nucleic acid, which serves as an internal control (IC). Obtaining a positive signal from the second target demonstrates successful amplification, thereby validating a negative result for the primary target.
A normal cellular gene sequence, which is expected to be present in all specimens, can be used as an IC (13, 20). This approach has the advantage of monitoring the integrity of the nucleic acid target; in improperly collected, stored, or processed specimens, the endogenous target will be absent (or degraded) and fail to yield a positive result. A disadvantage is that endogenous sequences may not accurately reflect amplification of the primary target due to differences in the primer sequences, size of the amplified product, and the relative amounts of the two targets.
There is another approach to monitoring amplification, using a synthetic IC as a proxy for the primary target, that overcomes the inherent limitations of an endogenous IC. One design of a synthetic IC is a plasmid DNA or an in vitro RNA transcript with primer binding regions identical to those of the target sequence, a randomized internal sequence similar to the target sequence in length and base composition, and a unique probe binding region that differentiates the IC from amplified target nucleic acid (27). These features ensure equivalent amplification of the IC and the target nucleic acid. A limited number of IC molecules are added to each test sample and coamplified with target nucleic acid; thus, a positive IC signal assures amplification sufficient to generate a positive signal from very small quantities of target. When introduced into the amplification reaction mixture, the IC can monitor amplification and detection. When introduced into the unprocessed specimen, the IC can also monitor nucleic acid recovery during specimen preparation. Unlike a housekeeping gene, a synthetic IC cannot be used to monitor the integrity of the target nucleic acid in the specimen.
Here, we describe the properties of the IC used in COBAS AMPLICOR and AMPLICOR tests and explain how to use and interpret the IC results during routine clinical testing. We also demonstrate the clinical utility of using an IC: increased sensitivity is achieved because reporting false-negative results is avoided and because additional positive results are detected by retesting inhibitory specimens.
MATERIALS AND METHODS
Construction of ICs.
A unique IC was constructed for each test by inserting the test-specific IC DNA sequence into a DNA plasmid. All ICs contain the same probe binding sequence, which makes it possible to detect all ICs with a single probe. The probe binding sequence was designed to minimize homology with primers and target sequences.
Several pairs of partially overlapping oligonucleotides that collectively contained the entire IC sequence were first annealed and extended with Escherichia coli DNA polymerase I Klenow fragment. The double-stranded DNA fragments were joined in PCR mixtures to form the full-length insert DNA, which was then cloned into plasmid DNA [pUC18 for the Chlamydia trachomatis, Neisseria gonorrhoeae, and Mycobacterium tuberculosis tests and pSP64 [poly(A)] for the human hepatitis C virus test]. The resulting recombinant plasmid was transformed into an E. coli (DG101) host.
For the C. trachomatis, N. gonorrhoeae, and M. tuberculosis tests, the linearized, recombinant plasmid DNA served as IC. For the human hepatitis C virus (HCV) test, runoff RNA transcripts synthesized from the linearized, recombinant plasmid DNA served as the IC.
The C. trachomatis-N. gonorrhoeae test is a multiplex test that uses two pairs of primer oligonucleotides to simultaneously amplify C. trachomatis and N. gonorrhoeae target DNAs in a single reaction mixture. The IC for this test contains the primer sequences specific for C. trachomatis. We demonstrated that amplification of this IC can serve as an indicator for amplification of both the C. trachomatis and N. gonorrhoeae target DNAs. C. trachomatis or N. gonorrhoeae DNA was added to urogenital swab and urine specimens obtained from patients who were previously determined to be negative for both C. trachomatis and N. gonorrhoeae by PCR. In a selected subset of inhibitory and noninhibitory specimens, 13 of 33 yielded positive signals for the IC and both targets, 15 yielded negative signals for the IC and both targets, 1 was positive for C. trachomatis but negative for N. gonorrhoeae and IC, and 4 were positive for N. gonorrhoeae but negative for C. trachomatis and IC. Thus, the IC detected amplification failure in all specimens that gave false-negative results for at least one of the two targets.
Preparation of ICs.
Cells from cultures of plasmid-containing bacteria grown overnight were harvested and extracted by a standard alkaline lysis method (23). DNA was recovered from the lysate with isopropanol (23) and purified by CsCl2 density gradient centrifugation. The purified plasmid DNA was then linearized with an appropriate restriction endonuclease and purified by phenol-chloroform extraction.
(i) ICs for C. trachomatis-N. gonorrhoeae and M. tuberculosis tests.
The C. trachomatis-N. gonorrhoeae and M. tuberculosis ICs are supplied as stock solutions. Prior to use, 0.1 ml of IC stock solution is added to 1.7 ml of master mix to yield an amplification-ready working master mix containing 20 copies of IC plasmid per 50 μl. These IC stock solutions were prepared by serial dilution of purified plasmid DNA in a Tris-buffered solution containing EDTA, poly(A) RNA, sodium azide, and the dye amaranth (Sigma); amaranth was included to enable visual confirmation that IC was added to master mix.
(ii) IC for HCV test.
The HCV IC was generated by transcribing HCV plasmid DNA, using the MEGAscript (Ambion, Inc.) in vitro transcription kit. Purified plasmid DNA was linearized with EcoRI restriction endonuclease and transcribed to yield 352-nucleotide-long RNA transcripts. The transcripts contained the 229-nucleotide IC sequence, 6 upstream nucleotides (the transcription start site), 14 downstream nucleotides, 30 adenylate nucleotides, and 3 nucleotides of the EcoRI recognition sequence.
The transcripts were purified from plasmid DNA by digesting the transcription reaction mixture with RNase-free DNase, followed by extraction with phenol-chloroform. The full RNA transcripts were then separated from unincorporated ribonucleotides and truncated transcripts by oligo(dT) cellulose affinity chromatography and were serially diluted in a buffered solution containing EDTA, poly(A) RNA, and sodium azide to yield an IC stock solution. Prior to use, 50 μl of the stock solution is added to 14 ml of HCV specimen diluent. Diluting processed specimens with specimen diluent (see “Specimen collection and processing” below) results in 20 copies of IC being delivered to each test sample.
Poisson statistical analysis to determine IC copy number.
The concentrations of the IC stock solutions were quantitated by serially diluting the stocks and performing multiple amplifications (28). At a particular dilution, the set of replicate amplifications generated a mixture of positive and negative results. At this low concentration, IC molecules are distributed among replicate amplifications according to Poisson’s law, with some replicates having one or a few IC molecules and others having none. The average number of molecules in a given volume of solution (C) and the probability that no molecule exists in a particular sample of the given volume (Pn) are given by the formula C = −ln(Pn) where Pn was determined by counting the number of negative replicates and used to calculate C, the number of IC molecules in the volume of solution added to each replicate. [C is actually the number of signal-generating units (SGU), which is defined as the smallest unit in the given solution that will generate a PCR-positive signal. The physical meaning of a SGU is a particle consisting of one or more PCR-amplifiable molecule(s). Under ideal conditions, a SGU is equal to a single copy of the target molecule.] The IC concentration was calculated by multiplying C by the appropriate dilution factor.
Specimen collection and processing. (i) C. trachomatis and N. gonorrhoeae.
Endocervical and male urethral swabs were collected by standard procedures and inoculated into chlamydial culture transport medium (CTM; either 2SP or MicroTest M4; MicroTest, Inc., Snellville, Ga.). A second swab was collected from each patient and cultured for N. gonorrhoeae. Specimens in CTM were transported to the lab at 2 to 8°C where an aliquot was used for Chlamydia culture. A second aliquot was shipped on wet ice to Roche Molecular Systems, where it was processed and tested on the COBAS AMPLICOR system. A 100-μl sample of specimen was mixed with 100 μl of C. trachomatis-N. gonorrhoeae lysis buffer and incubated for 10 min at room temperature. The resulting mixture was combined with 200 μl of C. trachomatis-N. gonorrhoeae specimen diluent and incubated for an additional 10 min at room temperature.
Ten to 50 ml of first-catch urine was also collected from each subject. The urine was shipped on wet ice to Roche Molecular Systems, where it was processed and tested on the COBAS AMPLICOR system. A 500-μl sample of urine was combined with 500 μl of C. trachomatis-N. gonorrhoeae urine wash buffer and incubated at 37°C for 15 min. The mixture was then centrifuged at 12,500 × g for 5 minutes. The supernatant was discarded, and the pellet was resuspended in 250 μl of C. trachomatis-N. gonorrhoeae lysis buffer. After a 15-min incubation at room temperature, 250 μl of C. trachomatis-N. gonorrhoeae specimen diluent was added to the lysate. The specimens were centrifuged at 12,500 × g for 10 min, and the resulting supernatant was tested.
(ii) M. tuberculosis.
Sputum specimens were collected, liquified, decontaminated, and concentrated by standard procedures. Liquified, decontaminated, and concentrated specimens were shipped to the laboratory at 2 to 25°C, where aliquots were tested for the presence of mycobacteria by culture and by acid-fast smear. An additional aliquot was processed and tested for M. tuberculosis on the COBAS AMPLICOR system. A 100-μl sample of liquified, decontaminated, and concentrated sputum was mixed with 500 μl of respiratory specimen wash solution and centrifuged at 12,500 × g for 10 min. The supernatant was discarded, and 100 μl of respiratory specimen lysis reagent was added to the resulting pellet. The resuspended specimen was incubated at 60°C for 45 min and then combined with 100 μl of respiratory specimen neutralization reagent.
(iii) HCV.
Blood samples were collected in VACUTAINER serum separator tubes or in VACUTAINER blood collection tubes containing either ACD or EDTA as an anticoagulant. Collection tubes were stored at room temperature for up to 6 h until serum or plasma was prepared. Plasma and serum samples were shipped to the laboratory at 2 to 8°C, where aliquots were processed and tested for HCV on the COBAS AMPLICOR system. A 100-μl sample of serum or plasma was combined with 400 μl of HCV lysis reagent and incubated at 60°C for 10 min. The lysate was combined with 500 μl of isopropyl alcohol, incubated at room temperature for 2 min, and centrifuged at 14,000 × g for 15 min. The supernatant was discarded, and the resulting pellet was washed with 1 ml of 70% ethanol. The washed pellet was resuspended in 1 ml of HCV specimen diluent containing HCV IC. The serological status of patients was determined by enzyme immunoassay (EIA).
Amplification and detection.
A 50-μl sample of processed specimen was added to 50 μl of master mix containing IC and amplified by using the thermal cycler onboard the COBAS AMPLICOR system. Each test used a unique set of thermal cycling conditions, which were automatically performed by COBAS AMPLICOR (8, 12). Upon completing amplification, the COBAS AMPLICOR system automatically denatured the amplification reaction mixtures, hybridized the amplicon to target-specific oligonucleotides bound to magnetic microparticles, and colorimetrically detected the captured amplicon by using an avidin-horseradish peroxidase complex (8, 12). The specific target and the IC were detected in separate reactions using separate, target- and IC-specific, oligonucleotide capture probes.
Where indicated, certain samples being tested for C. trachomatis and N. gonorrhoeae were amplified on a GeneAmp PCR System 9600 thermal cycler (Perkin-Elmer Corporation, Norwalk, Conn.) using the following thermal cycling parameters: 2 min at 50°C, 5 min at 95°C, and then 35 cycles, with 1 cycle consisting of 10 s at 91°C, 50 s at 62°C, and 35 s at 72°C. Amplification products were detected colorimetrically after hybridization to microwell plates coated with oligonucleotide probes specific for C. trachomatis, N. gonorrhoeae, or IC (15).
Interpretation of results.
Specimens yielding target signals above the test cutoff were interpreted as positive, regardless of the IC result. Specimens yielding target signals below the test cutoff were interpreted as negative, provided that the IC signal was above the assigned cutoff. Specimens yielding below cutoff signals for both the target and IC were interpreted as inhibitory. Inhibitory specimens were retested by processing another aliquot of the original specimen. The repeat test results were classified with the above criteria.
For HCV, sensitivity was calculated by comparing PCR results to EIA results. For the C. trachomatis, N. gonorrhoeae, and M. tuberculosis tests, sensitivity was calculated by comparing PCR results to resolved results. Because culture is not 100% sensitive, these tests yielded specimens that were positive by PCR but negative by culture. For C. trachomatis and N. gonorrhoeae, these discrepant results were resolved by performing PCR for an alternative target DNA sequence: a portion of the major outer membrane protein gene was used for C. trachomatis (9), and a portion of the 16S rRNA gene was used for N. gonorrhoeae (16). Specimens were considered positive for infection if the culture was positive or if the specimen was PCR positive for both the primary and alternative targets. For M. tuberculosis, discrepancies between PCR and culture results were resolved based on patient chart review; specimens were considered positive if there was clinical evidence of M. tuberculosis infection.
RESULTS
Effect of primary target on IC signal.
A single set of primer oligonucleotides is used to amplify both the IC and target DNA. Both targets also draw from a common pool of nucleotides and polymerase. Thus, IC amplification may be suppressed due to competition in samples containing large amounts of target DNA. We determined the amount of target DNA required to suppress the IC signal by testing a set of samples that contained increasing amounts of C. trachomatis target DNA. These samples also contained a constant low concentration (25 copies per test sample) of N. gonorrhoeae target DNA to determine whether it was similarly sensitive to competition.
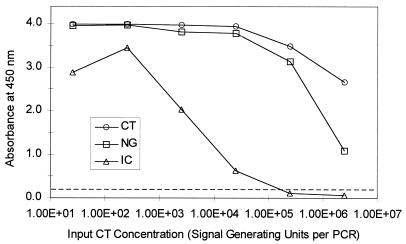
The IC signal progressively decreased as the amount of C. trachomatis target DNA increased (Fig. 1). Between 250 and 2,500 copies per test sample (corresponding to 5,000 and 50,000 copies per ml of specimen) of C. trachomatis DNA was required to produce a noticeable decrease in the IC signal. The IC signal was reduced to background by 2.5 × 105 copies per test sample (5 × 106 copies per ml of specimen) of C. trachomatis target DNA. Virtually identical IC signals were observed when the N. gonorrhoeae target DNA concentration was varied and the C. trachomatis target concentration was maintained at 25 copies per test sample (data not shown).
FIG. 1.
Effect of increasing concentration of target nucleic acid on IC signal. Samples containing increasing amounts of C. trachomatis (CT) target DNA, 25 copies of N. gonorrhoeae (NG) target DNA, and 20 copies of IC DNA per test sample were amplified in a Perkin-Elmer 9600 thermal cycler. C. trachomatis, N. gonorrhoeae, and IC amplification products were hybridized to separate microwell plates coated with target-specific oligonucleotide probes. The hybridized amplification products were detected colorimetrically with an avidin-horseradish peroxidase complex. The concentration of C. trachomatis target DNA is shown on the x axis (1.00E+01 is 101, 1.00E+02 is 102 etc.) and the absorbance at 450 nm is shown on the y axis. The broken line indicates the assay cutoff value.
Compared to the IC, C. trachomatis and N. gonorrhoeae target DNAs were less sensitive to competition. Approximately 2.5 × 105 copies per test sample of C. trachomatis (Fig. 1) or N. gonorrhoeae (data not shown) DNA were required to produce a small decrease in the signal generated by 25 copies of N. gonorrhoeae or C. trachomatis DNA. The N. gonorrhoeae (or C. trachomatis) signal was greatly reduced but was still well above background in the presence of 2 × 106 copies of C. trachomatis (or N. gonorrhoeae) target DNA. Because high levels of C. trachomatis DNA can suppress N. gonorrhoeae amplification and vice versa, competition for reagents other than primers is responsible for reduced signals.
Determination of IC cutoff.
The purpose of the IC is to maximize test sensitivity by identifying inhibitory (i.e., nonamplifiable) specimens that have the potential to generate false-negative results. The IC signal that identifies all such inhibitory specimens is chosen as the cutoff. As a first step in determining the IC cutoff, we analyzed the distribution of IC signals in a set of specimens that were negative for the target DNA. Specimens that were positive for target DNA (both true and false positives) were not included in this analysis because the depressed IC signals in these specimens did not necessarily serve as an indicator of inhibition.
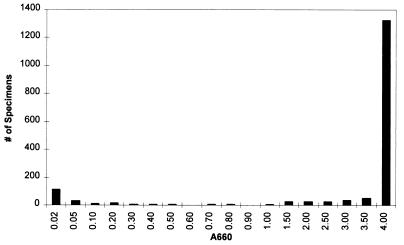
Of 1,915 urogenital specimens tested by the COBAS AMPLICOR C. trachomatis test, 1,688 gave negative signals for C. trachomatis. The distribution of IC signals in these 1,688 specimens was clearly bimodal, with most specimens having an IC signal of ≥1.0 A660 and a smaller set of specimens having an IC signal of <0.2 A660 (Fig. 2). IC signals between 0.2 and 0.5 A660 were observed in 0.8% (13 of 1,688) of the specimens; IC signals between 0.5 and 1.0 A660 were observed in 1.0% (17 of 1,688) of the specimens. When amplification was performed in the absence of clinical material, the IC signal was always ≥2.0 A660 (data not shown). These data suggest that specimens having IC signals below 0.2 A660 are inhibitory. Specimens having IC signals between 0.2 and 1.0 A660 probably contained weak inhibitors that partially suppressed amplification. To select the optimal IC cutoff, we determined whether test sensitivity was enhanced by classifying such partially suppressed specimens as inhibitory.
FIG. 2.
Distribution of IC signals in a set of clinical specimens. Specimens were tested by the COBAS AMPLICOR C. trachomatis and N. gonorrhoeae tests. The IC signal was evaluated in specimens that were negative for both C. trachomatis and N. gonorrhoeae. Each bar shows the number of specimens (on the y axis) having an IC absorbance less than the value the value shown directly below the bar (on the x axis) but greater than or equal the value to the immediate left (on the x axis). The bar over the 4.00 value includes specimens having signals equal to 4.00.
Test sensitivity is calculated by the following formula: sensitivity = number of PCR-positive specimens/total number of amplifiable, infected specimens. When no IC is used, all PCR-negative, infected specimens are considered amplifiable and are included in the denominator of the above equation, which minimizes test sensitivity. When the IC is used, PCR-negative, infected specimens that give negative IC signals are classified as inhibitory and excluded from the denominator, thereby maximizing sensitivity. Figure 3 illustrates the relationship between IC cutoff, test sensitivity, and number of inhibitory specimens for urogenital specimens tested by the COBAS AMPLICOR C. trachomatis test. Of 1,915 specimens tested, 226 were infected with C. trachomatis. Maximum sensitivity was achieved for IC cutoffs above 0.04 A660 (Fig. 2). Similarly, the number of inhibitory, infected specimens reached a plateau at the cutoff of 0.04 A660. At very high IC cutoffs, we obtained a small increase in sensitivity and in the number of inhibitory specimens because several COBAS AMPLICOR-negative, infected specimens had strong IC signals. These signals represent actual false-negative results that were being eliminated from sensitivity calculations by an artificially high IC cutoff.
FIG. 3.
Effect of IC cutoff on assay sensitivity. A set of clinical specimens was tested with the COBAS AMPLICOR C. trachomatis test. Different IC cutoff values were used to distinguish between inhibitory and noninhibitory specimens. Clinical sensitivity and the number of inhibitory, infection-positive specimens were calculated for each cutoff value. The IC cutoff is shown on the x axis, clinical sensitivity is shown on the left y axis, and the number of inhibitory specimens is shown on the right y axis.
Based on the above analyses, 0.2 A660 was selected as the IC cutoff for the COBAS AMPLICOR C. trachomatis test to provide a wide margin over the value required to achieve maximum sensitivity; this value also matches the cutoff for the primary target. Similar analyses were performed to determine the IC cutoffs for the other COBAS AMPLICOR tests (data not shown).
Effect of IC on assay performance.
The utility of the IC was demonstrated by testing clinical specimens obtained from patients suspected of having C. trachomatis, N. gonorrhoeae, M. tuberculosis, or HCV infection. Specimens were tested as described and classified as positive or negative for infection (see Materials and Methods). Any specimen that was inhibitory (i.e., negative for both IC and target DNA) was retested by processing and testing another aliquot of the original specimen.
The fraction of specimens that were inhibitory in the test ranged from 0 to 11.5% (Table 1). Inhibition was somewhat more frequent in uninfected specimens than in infected ones. This is expected because even minimal inhibition will suppress amplification below the threshold required to generate a positive IC result. In contrast, infected specimens that suppress amplification to a similar extent will not appear inhibitory, provided that the target DNA concentration is high enough.
TABLE 1.
Frequency of inhibition in infection-positive and -negative specimens
| COBAS AMPLICOR test and specimen | Infection-positive specimens
|
Infection-negative specimens
|
||||||
|---|---|---|---|---|---|---|---|---|
| Total no. | No. inhibitory (%) | Retest
|
Total no. | No. inhibitory (%) | Retest
|
|||
| No. positive | No. inhibitory | No. negative | No. inhibitory | |||||
| C. trachomatis | ||||||||
| Swab | 117 | 4 (3.4) | 1 | 3 | 839 | 82a (9.8) | 22 | 46 |
| Urine | 118 | 7 (5.9) | 6 | 1 | 847 | 79b (9.3) | 68 | 9 |
| N. gonorrhoeae | ||||||||
| Swab | 115 | 1 (0.9) | 1 | 0 | 831 | 63c (7.6) | 24 | 38 |
| Urine | 118 | 8 (6.8) | 6 | 2 | 836 | 70d (8.4) | 62 | 6 |
| M. tuberculosis | ||||||||
| Smear positive | 154 | 1 (0.6) | 1 | 0 | 29 | 0 (0.0) | 0 | 0 |
| Smear negative | 125 | 3 (2.4) | 1 | 2 | 4,068 | 217 (5.3) | 96 | 121 |
| HCV | 1,036 | 61 (5.9) | 50 | 11 | 929 | 107 (11.5) | 98 | 9 |
Fourteen of the inhibitory C. trachomatis swab specimens were not available for retesting.
Two of the inhibitory C. trachomatis urine specimens were not available for retesting.
One of the inhibitory N. gonorrhoeae swab specimens was not available for retesting.
Two of the inhibitory N. gonorrhoeae urine specimens were not available for retesting.
Approximately 64% (436 of 684) of inhibitory specimens were not inhibitory when another aliquot of the specimen was processed and tested (Table 1). This could indicate that some inhibitors were labile. Alternatively, a low level and/or nonuniform distribution of inhibitors could account for this interaliquot variation. For HCV, variation in carryover of extraction reagents could be the reason why some inhibitory HCV specimens were not inhibitory when tested after reextraction. Reagent carryover is not a concern for C. trachomatis, N. gonorrhoeae, and M. tuberculosis, because the PCR-compatible extraction reagents are not removed during processing. Regardless of the mechanism, the absence of inhibition during retesting enables additional infections to be detected by PCR.
To assess the impact of using the IC, we used the data in Table 1 to calculate test sensitivity and specificity. Two calculations were performed. First, the IC results were ignored and all PCR results were interpreted as positive or negative based upon the initial test result. Second, the IC results were used to identify inhibitory specimens and PCR results were interpreted as positive, negative, or inhibitory based on the repeat test result. Taking the IC results into account increased sensitivity for all tests (Table 2). The increase in sensitivity ranged from 0.9 (for the N. gonorrhoeae test performed on swab specimens) to 5.9% (for the N. gonorrhoeae test performed on urine specimens). In contrast, test specificity was not affected by taking the IC results into account (Table 2). This is expected because inhibited, uninfected specimens are interpreted as PCR negative when the IC results are ignored. These same specimens are also interpreted as PCR negative if they give a valid negative result when retested.
TABLE 2.
Enhancement of test sensitivity using IC
| COBAS AMPLICOR test and specimen | Sensitivity (%)
|
Specificity (%)
|
||
|---|---|---|---|---|
| Without IC | With IC | Without IC | With IC | |
| C. trachomatis | ||||
| Swab | 94.8 (110/116) | 97.4 (111/114) | 99.0 (832/840) | 99.0 (771/779) |
| Urine | 92.0 (104/113) | 94.0 (110/117) | 99.1 (844/852) | 99.0 (828/836) |
| N. gonorrhoeae | ||||
| Swab | 93.9 (108/115) | 94.8 (109/115) | 98.0 (814/831) | 97.9 (775/792) |
| Urine | 83.8 (98/117) | 89.7 (104/116) | 98.9 (828/837) | 98.8 (818/828) |
| M. tuberculosis | ||||
| Smear positive | 98.7 (151/153) | 98.7 (152/154) | 90.0 (27/30) | 89.7 (26/29) |
| Smear negative | 73.6 (92/125) | 75.6 (93/123) | 99.0 (4027/4068) | 99.0 (3906/3947) |
| HCV | 72.6 (752/1036) | 78.2 (802/1025) | 98.2 (912/929) | 98.2 (903/920) |
Using the IC increased test sensitivity for two reasons. First, additional infected specimens were detected upon retesting specimens that were initially inhibitory. A set of 66 (1 C. trachomatis swab, 6 C. trachomatis urine specimens, 1 N. gonorrhoeae swab, 6 N. gonorrhoeae urine specimens, 1 M. tuberculosis smear-positive specimen, 1 M. tuberculosis smear-negative specimen, and 50 HCV specimens) inhibitory specimens (Table 1) were misclassified as PCR negative when the IC results were ignored but were classified as true positive based on a positive retest result. Of the 66 specimens, 58 (1 C. trachomatis urine specimen, 1 N. gonorrhoeae swab, 5 N. gonorrhoeae urine specimens, 1 M. tuberculosis smear-negative specimen, and 50 HCV specimens) were positive by culture or EIA and were, therefore, classified as false negative when the IC results were ignored. The other eight specimens (one C. trachomatis swab, five C. trachomatis urine specimens, one N. gonorrhoeae urine specimen, and one M. tuberculosis smear-positive specimen) were negative by culture and were, therefore, classified as true negative when the IC results were ignored. These eight specimens, which were confirmed by PCR for an alternate target, would have gone undetected had the IC not been used.
Second, using the IC increased sensitivity because repeatedly inhibitory specimens are uninterpretable and were, therefore, excluded from the sensitivity calculations. When the IC results were ignored, these specimens were classified as false negative. Included in this category were 3 C. trachomatis swab specimens, 1 C. trachomatis urine specimen, 2 N. gonorrhoeae urine specimens, 2 M. tuberculosis smear-negative specimens, and 11 HCV specimens (Table 1).
DISCUSSION
The results of this study demonstrate that incorporating an IC into PCR-based tests increases sensitivity by enabling the user to identify and retest samples inhibitory to PCR. Furthermore, a positive IC result indicates that amplification has occurred and thus provides assurance that negative test results are truly negative. The COBAS AMPLICOR tests evaluated in this study exhibited low rates of inhibition. Because inhibition was infrequent, use of the IC resulted in only a relatively modest improvement in test sensitivity.
The IC can also be used to monitor competition between multiple targets in multiplex PCR tests. A negative IC result in specimens that are positive for one target indicates that competition and/or inhibition reduced amplification efficiency below the threshold required to generate a positive result from a low-level target. In such specimens, negative results for the other targets are considered invalid. The data presented here demonstrate that competition will not cause a false-negative result unless the concentration of one target is 104-fold greater than the concentration of the second target.
The IC is used at a concentration of 20 copies per test sample to monitor amplification at the limit of test sensitivity. PCR inhibitors decrease amplification efficiency, thereby reducing the amount of PCR product generated from each target molecule. A high target load can compensate for reduced amplification efficiency, yielding enough product to generate a positive signal. If used at a higher concentration, the IC might not detect weak inhibition that could cause false-negative results at extremely low target loads.
For routine clinical applications, the laboratory can maximize test sensitivity by using an IC to monitor amplification in every specimen. Nevertheless, it may be possible to use an IC selectively to increase efficiency without sacrificing performance. For example, the fully automated COBAS AMPLICOR system can be programmed to perform additional tests based on the outcome of an initial test. Thus, a COBAS AMPLICOR user can easily detect the IC only in specimens that test negative for the primary target(s). Using this algorithm will not compromise test performance because the IC result has no effect on the interpretation of positive specimens.
An IC can also be used selectively when past experience has demonstrated very low rates of inhibition. Use of an IC can be reserved for specific specimen or patient conditions where inhibition is more likely to occur; examples include specimens contaminated with blood or other interfering substances and urogenital specimens from pregnant women (11). An IC should also be selectively employed when the probability of infection is high and the consequence of a false-negative result is severe. An example is testing acid-fast bacillus smear-positive patients for M. tuberculosis, where a false-negative test result would lead the clinician to conclude that the patient has a nontuberculosis mycobacterial infection.
Even when the IC was used, test sensitivity was not 100%. Thus, factors other than inhibition must have caused false-negative results. Sample-to-sample variation is one source of false-negative results. Localization of infection may also be a factor when testing female urine samples for C. trachomatis and N. gonorrhoeae because PCR and culture are performed on specimens collected from different anatomical sites. Low target concentration represents a second potential source of false-negative results, especially when testing for M. tuberculosis where the volume of sample used for PCR testing is 5% of that used for culture. Finally, the reference test result could actually be false positive. This is almost certainly a factor in HCV testing where a positive EIA result for anti-HCV antibody indicates prior exposure but does not serve as a marker for current infection.
Inhibition has been shown to affect the sensitivity of LCR-based amplification tests (2, 4–6, 11, 22, 25), TMA tests (17, 21), and a nucleic acid sequence-based amplification test (18). It is, however, difficult to determine the frequency of inhibition because the commercially available LCR- and TMA-based tests lack an IC. Consequently, inhibition can be evaluated only in reference test-positive specimens that are expected to give positive amplification test results. Inhibition cannot be assessed in reference test-negative specimens since they are expected to give negative test results. The frequency of inhibitory, positive specimens can be estimated from the frequency reference test-positive, amplification test-negative specimens. This provides an upper limit on the inhibition rate in reference test-positive specimens; the actual inhibition rate may be lower because false-negative results may be caused by factors other than inhibition. The frequency of LCR false-negative results in urogenital specimens were 2% (7 of 345) (6), 7% (3 of 44) (22), 12% (10 of 87) (5), 13% (7 of 54) (25), 22% (2 of 9) (2), and 60% (15 of 25) (11). In some instances, specimens were retested at full strength and after being diluted. Approximately one-half of the retested false-negative specimens became positive (some required dilution, while others did not); thus, the false-negative results in these specimens can be attributed to inhibition. These observations suggest that the actual LCR inhibition rate ranged from 1 to 30%. The frequency of TMA false-negative results was 10% (6 of 60) (21); 5 of the 6 false-negative specimens were positive when retested, which suggests the inhibition rate in positive specimens was 8%. For both LCR and TMA, the inhibition rate for all specimens is probably higher; weak inhibition may go undetected in positive specimens that contain relatively high concentrations of target.
In addition to monitoring amplification in qualitative PCR tests, an IC can be used as an internal quantitation standard (QS) for quantitative PCR tests (19, 27). In quantitative PCR tests, the QS is added to each clinical specimen at a known copy number and is carried through all test processes. The QS controls for specimen-to-specimen differences in recovery during processing, amplification efficiency, and detection efficiency to permit the accurate quantitation of target nucleic acid in each specimen. The amount of target nucleic acid in a specimen is calculated by multiplying the target-specific signal by the ratio of QS input copy number to QS signal.
In summary, we have developed an IC that is introduced into each test sample and is coamplified with target nucleic acid from the clinical specimen. We have used the IC to demonstrate that the frequency of inhibition in COBAS AMPLICOR PCR tests ranged from 1 to 10%. Although the frequency of inhibition is low, an IC has been incorporated into COBAS AMPLICOR and AMPLICOR tests to assure the integrity of negative results. Furthermore, by using the IC, we identified additional infections that would otherwise have gone undetected.
ACKNOWLEDGMENTS
We thank E. Dragon, T. White, and R. Pinnola for helpful comments during preparation of this manuscript. J. Q. Yang and A. Wang provided clinical data for the COBAS AMPLICOR C. trachomatis and N. gonorrhoeae tests and IC cutoff data. Ann Butcher, K. Gutekunst, C. Young, V. Tevere, R. Sun, G. Colucci, M. Pedrocchi, H.-J. Burkardt, J. Vincelette, M. Bogard, J. Schirm, A. Bouza, R. Alonso, R. Antinozzi, and T. Fenner provided clinical data for the COBAS AMPLICOR M. tuberculosis and HCV tests.
REFERENCES
- 1.Bass C A, Jungkind D L, Silverman N S, Bondi J M. Clinical evaluation of a new polymerase chain reaction assay for detection of Chlamydia trachomatis in endocervical specimens. J Clin Microbiol. 1993;31:2648–2653. doi: 10.1128/jcm.31.10.2648-2653.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassiri M, Hu H Y, Domeika M A, Burczak J, Svensson L O, Lee H H, Mardh P A. Detection of Chlamydia trachomatis in urine specimens from women by ligase chain reaction. J Clin Microbiol. 1995;33:898–900. doi: 10.1128/jcm.33.4.898-900.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauwens J E, Clark A M, Loeffelholz M J, Herman S A, Stamm W E. Diagnosis of Chlamydia trachomatis urethritis in men by polymerase chain reaction assay of first-catch urine. J Clin Microbiol. 1993;31:3013–3016. doi: 10.1128/jcm.31.11.3013-3016.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chernesky M A, Chong S, Jang D, Luinstra K, Sellors J, Mahony J B. Ability of commercial ligase chain reaction and PCR assays to diagnose Chlamydia trachomatis infections in men by testing first-void urine. J Clin Microbiol. 1997;35:982–984. doi: 10.1128/jcm.35.4.982-984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chernesky M A, Lee H, Schachter J, Burczak J D, Stamm W E, McCormack W M, Quinn T C. Diagnosis of Chlamydia trachomatis urethral infection in symptomatic and asymptomatic men by testing first-void urine in a ligase chain reaction assay. J Infect Dis. 1994;170:1308–1311. doi: 10.1093/infdis/170.5.1308. [DOI] [PubMed] [Google Scholar]
- 6.Ching S, Lee H, Hook III E W, Jacobs M R, Zenilman J. Ligase chain reaction for detection of Neisseria gonorrhoeae in urogenital swabs. J Clin Microbiol. 1995;33:3111–3114. doi: 10.1128/jcm.33.12.3111-3114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dale B, Dragon E A. Polymerase chain reaction in infectious disease diagnosis. Lab Med. 1994;25:637–641. [Google Scholar]
- 8.DiDomenico N, Link H, Knobel R, Caratsch T, Weschler W, Loewy Z G, Rosenstraus M. COBAS AMPLICOR™: a fully automated RNA and DNA amplification and detection system for routine diagnostic PCR. Clin Chem. 1996;42:1915–1923. [PubMed] [Google Scholar]
- 9.Dutilh B, Bebear C, Rodriguez P, Vekris A, Bonnet J, Garret M. Specific amplification of a DNA sequence common to all Chlamydia trachomatis serovars using the polymerase chain reaction. Res Microbiol. 1989;140:7–16. doi: 10.1016/0923-2508(89)90053-3. [DOI] [PubMed] [Google Scholar]
- 10.Greenfield L, White T J. Sample preparation methods. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: American Society for Microbiology; 1993. pp. 122–137. [Google Scholar]
- 11.Jensen I P, Thorsen P, Moller B R. Sensitivity of ligase chain reaction assay of urine from pregnant women for Chlamydia trachomatis. Lancet. 1997;349:329–330. doi: 10.1016/s0140-6736(05)62829-2. [DOI] [PubMed] [Google Scholar]
- 12.Jungkind D, DiRenzo S, Beavis K G, Silverman N S. Evaluation of automated COBAS AMPLICOR PCR system for detection of several infectious agents and its impact on laboratory management. J Clin Microbiol. 1996;34:2778–2783. doi: 10.1128/jcm.34.11.2778-2783.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kellogg D E, Sninsky J J, Kwok S. Quantitation of HIV1 proviral DNA relative to cellular DNA by the polymerase chain reaction. Anal Biochem. 1990;189:202–208. doi: 10.1016/0003-2697(90)90108-l. [DOI] [PubMed] [Google Scholar]
- 14.Kellogg J A, Seiple J W, Klinedinst J L, Stroll E S, Cavanaugh S H. Improved PCR detection of Chlamydia trachomatis by using an altered method of specimen transport and high-quality endocervical specimens. J Clin Microbiol. 1995;33:2765–2767. doi: 10.1128/jcm.33.10.2765-2767.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loeffelholz M J, Lewinski C A, Silver S R, Purohit A P, Herman S A, Buonagurio D A, Dragon E A. Detection of Chlamydia trachomatis in endocervical specimens by polymerase chain reaction. J Clin Microbiol. 1992;30:2847–2851. doi: 10.1128/jcm.30.11.2847-2851.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu S-D Y, Kao S-Y, Silver S, Purohit A, Longiaru M, White T J. Abstracts of the 91st General Meeting of the American Society for Microbiology 1991. Washington, D.C: American Society for Microbiology; 1991. Detection of Neisseria gonorrhoeae and Chlamydia trachomatis in a combined system by PCR, abstr. C-115; p. 361. [Google Scholar]
- 17.Manterola J M, Gamboa F, Lonca J, Matas L, Ruiz Manzano J, Rodrigo C, Ausina V. Inhibitory effect of sodium dodecyl sulfate in detection of Mycobacterium tuberculosis by amplification of rRNA. J Clin Microbiol. 1995;33:3338–3340. doi: 10.1128/jcm.33.12.3338-3340.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morre S A, Sillekens P, Jacobs M V, van Aarle P, de Blok S, van Gemen B, Walboomers J M M, Meijer C J L M, van den Brule A J C. RNA amplification by nucleic acid sequence-based amplification with an internal standard enables reliable detection of Chlamydia trachomatis in cervical scrapings and urine samples. J Clin Microbiol. 1996;34:3108–3114. doi: 10.1128/jcm.34.12.3108-3114.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulder J, McKinney N, Christopherson M, Sninsky J, Greenfield L, Kwok S. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J Clin Microbiol. 1994;32:292–300. doi: 10.1128/jcm.32.2.292-300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noonan K E, Beck C, Holzmayer T A, Chin J E, Wunder J S, Andrulis I L, Gazdar A F, Willman C L, Griffith B, von Hoff D D, Roninson I B. Quantitative analysis of MDR1 (multidrug resistance) gene expression in human tumors by polymerase chain reaction. Proc Natl Acad Sci USA. 1990;87:7160–7164. doi: 10.1073/pnas.87.18.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasternack R, Vuorinen P, Miettinen A. Evaluation of the Gen-Probe Chlamydia trachomatis transcription-mediated amplification assay with urine specimens from women. J Clin Microbiol. 1997;35:676–678. doi: 10.1128/jcm.35.3.676-678.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasternack R, Vuorinen P, Pitkäjärvi T, Koskela M, Miettinen A. Comparison of manual Amplicor PCR, Cobas Amplicor PCR, and LCx assays for detection of Chlamydia trachomatis infection in women by using urine specimens. J Clin Microbiol. 1997;35:402–405. doi: 10.1128/jcm.35.2.402-405.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 24.Stary A, Tomazic-Allen S, Choueiri B, Burczak J, Steyrer K, Lee H. Comparison of DNA amplification methods for the detection of Chlamydia trachomatis in first-void urine from asymptomatic military recruits. Sex Transm Dis. 1996;23:97–102. doi: 10.1097/00007435-199603000-00002. [DOI] [PubMed] [Google Scholar]
- 25.van Doornum G J J, Buimer M, Prins M, Henquet C J M, Coutinho R A, Plier P K, Tomazic-Allen S, Hu H, Lee H. Detection of Chlamydia trachomatis infection in urine samples from men and women by ligase chain reaction. J Clin Microbiol. 1995;33:2042–2047. doi: 10.1128/jcm.33.8.2042-2047.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verkooyen R P, Luijendijk A, Huisman W M, Goessens W H F, Kluytmans J A J W, van Rijsoort-Vos J H, Verbrugh H A. Detection of inhibitors in cervical specimens by using the AMPLICOR Chlamydia trachomatis assay. J Clin Microbiol. 1996;34:3072–3074. doi: 10.1128/jcm.34.12.3072-3074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang A W, Doyle M V, Mark D F. Quantitation of mRNA by the polymerase chain reaction. Proc Natl Acad Sci USA. 1989;86:9717–9721. doi: 10.1073/pnas.86.24.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Chang S Y, MacMullen G, Huang D, Kwok S, Spadoro J. Program and abstracts of the San Diego Conference: the genetic revolution. Washington, D.C: American Society for Clinical Chemistry; 1994. Use of internal control quantitative standards to quantify target nucleic acid sequences by PCR, poster abstract; p. 12. [Google Scholar]
- 29.White T J, Madej R, Persing D H. The polymerase chain reaction: clinical applications. Adv Clin Chem. 1992;29:161–196. doi: 10.1016/s0065-2423(08)60224-3. [DOI] [PubMed] [Google Scholar]
- 30.Wiesenfeld H C, Uhrin M, Dixon B W, Sweet R L. Diagnosis of male Chlamydia trachomatis urethritis by polymerase chain reaction. Sex Transm Dis. 1994;21:268–271. doi: 10.1097/00007435-199409000-00004. [DOI] [PubMed] [Google Scholar]