Abstract
Repetitive sequence-based PCR was compared to pulsed-field gel electrophoresis (PFGE) for the ability to discriminate Enterococcus faecalis isolates at the subspecies level. The BOXA2R primer, derived from repetitive sequences in Streptococcus pneumoniae, was applied to 41 isolates of E. faecalis collected from various sources. The REP1R-Dt and REP2-Dt primers, derived from the gram-negative repetitive extragenic palindromic element, were also applied to 18 selected isolates. Of the 41 isolates examined, 7 were β-lactamase producing and 8 were vancomycin resistant. By PFGE, 17 isolates had distinct patterns; the other 24 were classified into eight different clonal groups. By PCR using the BOXA2R primer, 16 isolates generated distinct patterns; the other 25 were classified into nine different clonal groups. There were only minor differences in the PCR results obtained by using the BOXA2R primer and the REP1R-Dt and REP2-Dt primers. Two isolates among vancomycin-resistant enterococci from the greater Houston, Tex., area were related by PFGE, distinct by PCR with the BOXA2R primer, and related by PCR with the REP1R-Dt and REP2-Dt primers. Clonal relationships among the remaining 39 isolates were similar by both PFGE and PCR. PCR reliably discriminated all epidemiologically unrelated isolates. Although PCR is less time consuming than PFGE, PCR results were more difficult to interpret than PFGE results, perhaps because fewer bands were generated by PCR than by PFGE and some PCR products were inconsistently seen.
Enterococci have become a leading cause of clinical infections, particularly of the nosocomial type. From 1986 to 1989, enterococci were isolated from 12% of the patients with nosocomial infections reported to the National Nosocomial Infection Surveillance system (15). In addition to the fact that they cause a significant proportion of nosocomial infections, resistance of enterococci to antimicrobial agents is another concern. Recovery of high-level aminoglycoside-resistant enterococci, vancomycin-resistant enterococci (VRE), and multiple-drug-resistant enterococci has been increasingly reported (1, 11, 12, 21). Understanding the epidemiology of enterococcal infections is an important part of dealing with these problematic organisms. In the past, laboratory typing of organisms of interest was based largely on phenotypic characteristics such as antibiogram, biotype, or phage type. These techniques have limitations in discriminatory power, sensitivity, reproducibility, and/or availability of reagents. More recently, molecular genetic techniques have been successfully applied to many species of bacteria. Among the available techniques, pulsed-field gel electrophoresis (PFGE) has gained wide acceptance as an excellent method for typing of bacteria at the subspecies level in terms of discriminatory power and reproducibility. It has been successfully applied to the epidemiologic evaluation of various bacterial species, including enterococci (4–6, 13, 20), and practical guidelines for interpretation of PFGE when evaluating possible short-term outbreaks of relatively small numbers of isolates in hospitals or communities have been published (19). However, PFGE is a time-consuming procedure and expensive equipment is needed.
Recently, PCR-based DNA fingerprinting of microorganisms has been developed by using a wide variety of techniques and primer designs. The basis of PCR-based DNA fingerprinting is that the primers can bind to specific regions of the DNA, and when this binding occurs in the proper orientation and within an optimum distance, species- or strain-specific amplification products may be generated. Primers such as REP1R-Dt and REP2-Dt, which are derived from the repetitive extragenic palindromic (REP) sequences found primarily in gram-negative bacteria (17, 24), have been used in the technique known as repetitive sequence-based PCR (rep-PCR) (23, 24, 27) for studying DNA fingerprints of many bacterial species. The BOX elements, identified in Streptococcus pneumoniae, are another example of interspersed repetitive DNA sequences consisting of three sequences—boxA, boxB, and boxC (10). The BOXA1R and BOXA2R primers are based on the boxA sequence and have been used in rep-PCR amplification of DNA from many bacterial species (7). Other REP-like sequence-based primers have also been used in the evaluation of particular outbreaks (9, 18, 25), and the results indicated that rep-PCR is a promising molecular epidemiologic technique.
In this study, we compared rep-PCR done with the BOXA2R primer and PFGE for ease of use and the ability to discriminate strains of Enterococcus faecalis, including VRE and β-lactamase-producing (Bla+) strains, which we obtained from various sources. The REP1R-Dt and REP2-Dt primers were also used with 18 selected isolates, and the results were compared with results obtained with the BOXA2R primer. We interpreted the PCR-generated patterns by using different criteria and evaluated how these interpretations correlated with epidemiological data, as well as the results of PFGE.
MATERIALS AND METHODS
Bacterial isolates.
Forty-one isolates of E. faecalis from the collection in our laboratory were examined (see Table 1). Most of these isolates were collected from 1979 to 1996 from different parts of the United States, Chile, Argentina, Belgium, and Thailand, and some have been previously studied by PFGE (4, 13, 14, 20, 22). Eight vancomycin-resistant E. faecalis isolates were collected from hospitals in the greater Houston, Tex., area; from Cleveland, Ohio; and from volunteers in Belgium (22). Seven Bla+ isolates were selected from our previous studies (14, 20). These bacteria were randomly selected with broad criteria based on clinical characteristics, antibiotic resistance profiles, geographic origin, and time period. To avoid bias when interpreting the results of PCR and PFGE, the designations of all bacteria were coded so that the authors were blind to the identity of each isolate until after interpretation of the PCR and PFGE results.
TABLE 1.
Results of rep-PCR and PFGE of E. faecalis
| Isolate no.a | Lab name (designation in previous study or original name) | Reference(s) | Origin, yr of collection | rep-PCR patternb
|
PFGE patternb,d | |
|---|---|---|---|---|---|---|
| REP1R-Dt and REP2-Dt primers | BOXA2R primer | |||||
| 1 | TX0614 (E228) | 14, 20 | Richmond, Va., 1990 | BR-1 | 1 | 5 |
| 2 | TX0640 (Fla1) | 14, 20 | Florida, 1990 | BR-1a | 1 | 5 |
| 3 | TX0637 (CH570) | 14, 20 | Pittsburgh, Pa., 1988 | BR-1a | 1a | 5 |
| 4 | TX0638 (DEL) | 14, 20 | Delaware, 1990 | BR-1a | 1a | 5 |
| 5 | TX0608 (E47) | 14, 20 | Richmond, Va., 1988 | BR-1 | 1b | 5 |
| 6 | TX0921 (HH22) | 14, 20 | Houston, Tex., 1981 | BR-1b | 1c | 5 |
| 7 | TX0630 (HG6280) | 14, 20 | Argentina, 1990 | BR-2 | 2 | 19 |
| 8 | TX2528 | 2, 3 | Houston, Tex., 1995 | VR-1 | 3 | V-1 |
| 9 | TX2512 | 3 | Houston, Tex., 1994 | 3 | V-1 | |
| 10 | TX2527 | 2, 3 | Houston, Tex., 1995 | VR-1 | 3 | V-1a |
| 11 | TX2537 | This study | Houston, Tex., 1995 | VR-2 | 3a | V-1a |
| 12 | TX2486 | 3 | Houston, Tex., 1994 | VR-2a | 4 | V-1b |
| 13 | TX2403 (A256) | 3 | Cleveland, Ohio, 1991 | VR-3 | 5 | V-2 |
| 14 | TX2505 | 22 | Belgium, 1992 | VR-4 | 6 | B-8 |
| 15 | TX2533 | This study | Lubbock, Tex., 1995 | 7 | A | |
| 16 | TX0046 (END11) | 20 | Boston, Mass., <1982c | 8 | E-1 | |
| 17 | TX0045 (END6) | 20 | Boston, Mass., <1980c | 8 | E-1 | |
| 18 | TX0048 (END27) | 20 | Boston, Mass., <1980c | 9 | E-3 | |
| 19 | TX0014 | This study | Houston, Tex., 1992 | 10 | K-1 | |
| 20 | TX0017 | This study | Houston, Tex., 1993 | 10 | K-1 | |
| 21 | TX0024 (MC02152) | 20 | Illinois, 1977 | R-5 | 11 | MC-1 |
| 22 | TX0052 (Linebarger) | This study | Springfield, Mass., 1993 | 12 | K-2 | |
| 23 | TX0913 (HH181) | 13 | Houston, Tex., 1980 | R-6 | 13 | H-1 |
| 24 | TX0910 (HH54) | 13 | Houston, Tex., 1980 | 13 | H-1 | |
| 25 | TX0919 (HH31) | 13 | Houston, Tex., 1980 | 14 | H-3 | |
| 26 | TX0854 (BE82) | 13 | Bangkok, Thailand, 1980 | 15 | B-1 | |
| 27 | TX0858 (BE86) | 13 | Bangkok, Thailand, 1980 | 15 | B-1 | |
| 28 | TH2450a | This study | Belgium, 1992 | 16 | F-1 | |
| 29 | TX2451a | This study | Belgium, 1992 | 16a | F-1a | |
| 30 | TX2449a | This study | Belgium, 1992 | R-7 | 17 | F-2 |
| 31 | TX2453h | This study | Belgium, 1992 | 18 | F-3 | |
| 32 | TX0224 (HG1081) | 20 | Argentina, 1991 | 19 | 17 | |
| 33 | TX0220 (HG1113) | 20 | Argentina, 1991 | 20 | 16 | |
| 34 | TX0226 (HG1090) | 20 | Argentina, 1991 | 21 | 12 | |
| 35 | TX0768 (CE13) | 13 | Chile, 1980 | 22 | C-1 | |
| 36 | TX0770 (CE30) | 13 | Chile, 1980 | 22 | C-1 | |
| 37 | TX0785 (CE K4) | 13 | Chile, 1980 | 22a | C-1b | |
| 38 | TX0784 (CE K1) | 13 | Chile, 1980 | 23 | C-3 | |
| 39 | ATCC 29212 | 4 | American Type Culture Collection, Rockville, Md. | 24 | VI | |
| 40 | OG1RF | 4 | <1978c | R-8 | 25 | VIII |
| 41 | JH2-2 | 4 | <1974c | R-9 | 26 | VII |
Isolates 1 to 7 are β-lactamase producing, and 8 to 15 are vancomycin resistant.
Related patterns are denoted by lowercase letters.
The organism was isolated at an unknown time prior to the year shown.
For isolates which were previously studied, the original designation of PFGE pattern names in the previous publication was used.
PFGE of genomic DNA.
Genomic DNA was prepared in agarose plugs and digested with SmaI (New England Biolabs, Inc., Beverly, Mass.) as previously described (13). Electrophoresis was performed by using clamped homogeneous electric fields (CHEF-DRII; Bio-Rad Laboratories, Richmond, Calif.) with ramped pulse times beginning with 5 s and ending with 30 s at 200 V for 28.5 to 30 h.
PCR conditions and primer selection.
Genomic DNA of all isolates was prepared by the hexadecyltrimethylammonium bromide DNA precipitation method (26). The genomic DNA was then resuspended in 50 μl of distilled water (dH2O), quantitated by a spectrophotometer (GeneQuant RNA/DNA Calculator; Pharmacia LKB Biochrom Ltd., Science Park, Cambridge, England), and kept at −20°C until PCR was performed. DNA from all 41 isolates was analyzed with the BOXA2Roligonucleotide sequence (5′-ACGTGGTTTGAAGAGATTTTCG-3′) (7),which was used as the single primer. A subset of 18 isolates was also analyzed with the REP1R-Dt and REP2-Dt primers in combination. The nucleotide sequences of the REP1R-Dt and the REP2-Dt primers are 5′-IIINCGNCGNCATCNGGC-3′ (24) and 5′-NCGNCTTATCNGGCCTAC-3′ (23), respectively.
PCR was performed by modification of published protocols (8, 24). A master mixture consisting of all reagents except genomic DNA and Taq DNA polymerase was prepared, aliquoted at 22 μl/tube in a UV sterile biohood, and stored at −20°C until PCR was performed. Each 22 μl of the master mixture contained 5 μl of 5× buffer (8), 2.5 μl of 10% dimethyl sulfoxide (Invitrogen, San Diego, Calif.), 1 μl (50 pmol) of the BOXA2R primer (Integrated DNA Technologies Inc., Coralcille, Iowa), 6.25 μl of a deoxynucleoside triphosphate mixture (each deoxynucleoside triphosphate at 0.625 mM in the final total volume) made from a stock of 10 mM dCTP, 10 mM dGTP, 10 mM dATP, and 10 mM dTTP (Perkin-Elmer Corp., Norwalk, Conn.), and 7.25 μl of dH2O. For PCR with the REP1R-Dt and REP2-Dt primers (Integrated DNA Technologies Inc.), we used 1 μl (50 pmol) of each primer and decreased the volume of dH2O to 6.25 μl. PCR was initiated by adding 1 μl (100 ng) of genomic DNA to each tube, and then all tubes were placed in a thermal cycler (Perkin-Elmer Corp.). The thermal cycler was then heated, and the temperature was kept stable at 80°C during addition of 2 μl (2 U) of Taq DNA polymerase (Fisher Scientific, Pittsburgh, Pa.) to the mixture, bringing the total final volume to 25 μl/tube. One drop of sterile mineral oil was placed on top of the reaction mixture. The PCR conditions included an initial denaturation step (95°C, 7 min) and then 35 cycles of denaturation (90°C, 30 s), annealing (40°C, 1 min), and extension (65°C, 8 min), followed by one final extension step at 65°C for 16 min. Five microliters of amplification products was electrophoresed at 45 V for 7 h in a 1.5% agarose gel (ultraPure; GIBCO BRL, Life Technologies, Inc., Gaithersburg, Md.) made with 1× TBE (10× TBE contains 0.89 M Tris, 0.89 M boric acid, and 0.025 M EDTA). The gels were stained with 0.5 μg of ethidium bromide per ml for 30 min, destained for 2 h, and then photographed with a UV light source.
Interpretation of PCR and PFGE.
Interpretation of PFGE patterns was done as suggested by Tenover et al. (19). The banding patterns of PCR products were independently classified by all of the authors of this report without knowledge of the identities or PFGE patterns. Isolates were considered identical and representative of a single strain if they showed identical PCR banding patterns. Isolates which differed from each other by one band (one missing or extra band) were classified as related isolates belonging to the same clonal group. If there were two or more band differences between isolates, we considered them different clonal groups. We also assessed how classifying patterns with a difference of up to two bands as related isolates would affect our PCR interpretations. The results were then compared to those obtained by PFGE.
RESULTS
PFGE.
By PFGE, 25 clonal groups were identified among the 41 E. faecalis isolates examined (Table 1). The pattern names of isolates which had been studied previously were the same as in original publications. There were 8 clonal patterns that contained multiple isolates (patterns 5, V-1, E-1, K-1, H-1, B-1, B-8, and C-1; total, 24 isolates) and 17 patterns of one isolate each that were all different. Six of seven Bla+ isolates were classified as belonging to the same clone; of these, four (no. 1, to 4) exhibited identical banding patterns and the other two clonally related isolates (no. 5 and 6) differed from others in the clonal group by one or two bands, as previously described (14). There were four clonal patterns among the eight VRE isolates examined. The five VRE isolates from the Houston area showed similar banding patterns (pattern V-1 and variants V-1a and V-1b) and were classified as related isolates. Other VRE isolates from Cleveland, Ohio; Lubbock, Tex.; and Belgium (one isolate each) were all classified as distinct strains.
rep-PCR.
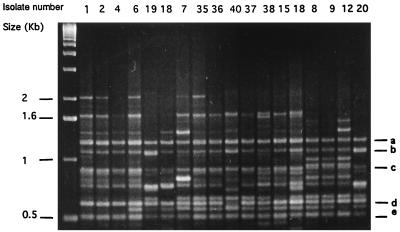
PCR products generated with the BOXA2R primer yielded 7 to 15 bands. The amplification products were mainly in the range of 0.5 to 3.0 kb. Five bands ranging from 0.5 to 1.4 kb, (labeled a to e in Fig. 1) appeared in all isolates. By our criterion, by which isolates showing patterns with differences of two or more bands were considered different, 26 clonal groups were identified among the 41 isolates. Eight clones contained multiple isolates (total, 23 isolates), and the other 18 isolates were all distinct strains. Six Bla+ isolates were classified as clonally related by rep-PCR with the BOXA2R primer (pattern 1 and variant 1a, lanes 3 and 4; variant 1b, lane 5; variant 1c, lane 6) (Fig. 1), while one Bla+ isolate from Argentina showed different banding patterns. For the eight VRE isolates, five different clonal groups were observed. There were two clonal groups among the five VRE isolates from the Houston area, including one with four related isolates (pattern 3 and variant 3a) and one different strain (pattern 4). Isolates from different geographical areas showed different banding patterns, except six Bla+ isolates from different states in the United States which were known to be related in a previous study.
FIG. 1.
Agarose gel showing PCR patterns of 18 E. faecalis isolates generated with the BOXA2R primer. The numbers above the lanes correspond to the isolate numbers in Table 1. The designations a to e represent DNA fragments that appeared in all isolates.
Differences in band intensity at matching positions among some isolates (no. 1, 2, 4, and in Fig. 1) made interpretation somewhat difficult, although all were classified as clonally related by PCR after repeated experiments. After PCR pattern interpretation, the identities of these isolates were disclosed; all of these isolates had been classified as clonally related by PFGE.
Comparison of techniques.
Most isolates classified as identical, related, or different by PCR with the BOXA2R primer were also classified in the same manner by PFGE (Table 2). However, two VRE isolates (no. 11 and 12) from different hospitals in the Houston area were different from each other only by PCR. These two isolates showed differences of three to five bands by PFGE; therefore, they were closely related or possibly related by this method (19).
TABLE 2.
Comparison of clonal relationships among 41 isolates of E. faecalis by rep-PCR with the BOXA2R primer and by PFGE
| Distinct patterns by both PFGE and PCRa | Identical patterns by both PFGE and by PCR | Related patterns by both PFGE and by PCR | Related patterns by PFGE, identical patterns by PCR | Related patterns by PFGE, distinct patterns by PCR |
|---|---|---|---|---|
| 7, 13, 14, 15, 18, 21, 22, 25, 30, 31, 34, 38, 39, 40, 41 | 1 and 2 | 6 and 1, 1-band difference by both PFGE and by PCR 5 and 1, 1-band difference by both PFGE and by PCR 35 (36) and 37, 1-band difference by both PFGE and by PCR 9 and 11, 1-band difference by PCR, 2-band difference by PFGE | 16 and 17, 1-band difference by PFGE 10 and 8 (9), 1-band difference by PFGE 28 and 29, 4- to 6-band difference by PFGE | 11 and 12, 3-band difference by PCR, 3- to 5-band difference by PFGE |
| 3 and 4 | ||||
| 8 and 9 | ||||
| 19 and 20 | ||||
| 23 and 24 | ||||
| 26 and 27 | ||||
| 35 and 36 |
Each isolate showed unique banding patterns which were different from each other and different from those of all other isolates.
We also evaluated the effect of classifying the patterns with a difference of up to two bands by PCR as related. In this case, isolates 37 (Chile, 1980), 19 (Boston, Mass., before 1980), and 16 (Lubbock, Tex., 1995) (Fig. 1) were placed in the same clonal group by rep-PCR with the BOXA2R primer but classified as different strains by PFGE. Similarly, isolates 7, 34, 35, 36, and 40, were placed in the same clonal groups by rep-PCR with the BOXA2R primer, while only isolates 35 and 36 were placed in the same clonal group by PFGE.
We also used the REP1R-Dt and REP2-Dt primers in the same PCR protocol to study the DNA fingerprints of 18 selected isolates and to compare the results to those obtained with the BOXA2R primer and with PFGE (Table 1). Among these selected isolates, seven were Bla+, six were VRE, and five were other, unrelated isolates. Compared to the BOXA2R primer, approximately the same number of bands was obtained. Some Bla+ isolates had extra bands among the larger fragments, but if amplification products larger than 3 kb were excluded, all Bla+ isolates except isolate 7 were classified as clonally related, as they were by the BOXA2R primer and by PFGE (Table 1). Among six VRE examined, classification by the BOXA2R and the REP1R-Dt and REP2-Dt primers was different for three isolates which were all classified as related by PFGE. With the REP1R-Dt and REP2-Dt primers, isolates 10 and 11 were not classified as being in the same clone as they showed a difference of three bands with these primers while they were classified as clonally related by the BOXA2R primer and PFGE. Isolates 11 and 12 were clonally related by PCR with the REP1R-Dt and REP2-Dt primers and PFGE but not by PCR with the BOXA2R primer. Isolates 8 and 10 were classified as belonging to the same clone by both sets of primers, as well as PFGE. Other isolates classified by PCR with the BOXA2R primer as different strains were also classified as different by PCR with the REP1R-Dt and REP2-Dt primers.
DISCUSSION
DNA fingerprinting of bacteria by rep-PCR has been extensively investigated in the last few years, typically by comparing banding patterns obtained from genomic DNA of the organisms by visual inspection without complicated mathematical calculation (27). However, there are no consensus methods or criteria for interpretation of the banding patterns obtained from the reactions. The criterion that was used in one study was that isolates were considered to belong to the same group if their PCR patterns differed by no more than two bands (27). In our study, we found that if a two-band difference had been allowed for the isolates to be considered related, more isolates unrelated by either epidemiology or by PFGE would have been misclassified into the same clonal groups. When stricter criteria were used, i.e., when a difference of only one band was allowed for isolates to be classified as related, the interpretation of rep-PCR results was more similar to that of those obtained by PFGE. This might be related to the lower number of bands for comparison obtained by PCR than by PFGE and the fact that some of these bands were common to all isolates. We do not known whether these bands are actually the same, but because they appeared in all isolates, they were not useful for distinguishing the organisms based on banding patterns. Further steps, such as restriction endonuclease digestion or DNA sequencing, might have revealed some degree of similarity among these fragments. Although we might obtain more information, the procedure would be more complicated and the applicability of PCR might be more limited.
Because there appear to be fewer REP-like sequences in gram-positive bacteria (24), it was suggested that primers derived from repetitive sequences in gram-positive bacteria such as S. pneumoniae might increase the resolving power of rep-PCR (23). In this study, clonal relationships among E. faecalis isolates revealed by both sets of primers were similar. This finding is similar to the recent observation of Koeuth et al. (7) that DNA fingerprints generated by the REP1R-Dt and REP2-Dt primers and the BOXA1R primer yielded similar dendrograms.
In PCRs with the REP1R-Dt and REP2-Dt primers and the BOXA2R primer, some amplification products larger than 3.0 kb were not uniformly seen among related isolates in the group of Bla+ isolates. This phenomenon was also noted in the study of Woods et al. (27). These bands could result from nonuniform amplification of larger DNA, and they were excluded from the interpretation of PCR patterns in our study.
Reproducibility of the results obtained with PCR was sometimes problematic, as inconsistency of banding patterns was observed in different reactions when the same genomic DNA was used. Variation in the amount of any component of the reaction might be one factor resulting in artifactual variation in banding patterns. A master mixture prepared from the same batch of reagents was used to ensure the consistency of the PCR mixtures, and this helped to attain reproducibility of banding patterns. However, the ability of the master mixture to support amplification reactions decreased in only 1 week (16); therefore, we performed PCR as soon as possible after preparation of the master mixture. In addition, in the early phase of our experiments, we found variability in banding patterns when all components of the reactions were mixed before the thermal cycler was started. To minimize this problem, the thermal cycler was warmed before starting the amplification cycles, as this procedure could reduce false priming which might occur in the initial round of the reactions (16).
In conclusion, the results of rep-PCR were generally concordant with those of PFGE in identifying clonal relationships among E. faecalis isolates. The procedure is less time consuming and less costly than PFGE. Given that PCR is increasingly available in health care facilities, rep-PCR using the BOXA2R primer could be a useful technique for studying DNA fingerprints of E. faecalis, particularly when devices for PFGE are not available. However, there may be more difficulty in interpretation of banding patterns because of differences in band intensity. In such circumstances, PFGE would be an appropriate technique for verifying the result of PCR. Therefore, interpretation of PCR results should be done with caution and with consideration of epidemiological data. Further studies with various bacterial species may provide more understanding of this technique, including pitfalls of the procedure and criteria for interpretation of results.
ACKNOWLEDGMENTS
We thank Teresa M. Coque for verifying the designation of E. faecalis isolates and identification of VRE, Thearith Koeuth and James R. Lupski for providing PCR protocols and reagents for our first round of rep-PCR, and Christian Brauning for assistance in taking photographs of agarose gels by computer and preparing some primers.
K.M. is supported by a grant from the Ministry of University Affairs, Thailand.
REFERENCES
- 1.Antalek M D, Mylotte J M, Lesse A J, Sellick J A. Clinical and molecular epidemiology of Enterococcus faecalis bacteremia, with special reference to strains with high-level resistance to gentamicin. Clin Infect Dis. 1995;20:103–109. doi: 10.1093/clinids/20.1.103. [DOI] [PubMed] [Google Scholar]
- 2.Coque T M, Murray B E. Identification of Enterococcus faecalis strains by DNA hybridization and pulsed-field gel electrophoresis. J Clin Microbiol. 1995;33:3368–3369. doi: 10.1128/jcm.33.12.3368-3369.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coque T M, Tomayko J F, Ricke S C, Okhyusen P C, Murray B E. Vancomycin-resistant enterococci from nosocomial, community, and animal sources in the United States. Antimicrob Agents Chemother. 1996;40:2605–2609. doi: 10.1128/aac.40.11.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordillo M E, Singh K V, Murray B E. Comparison of ribotyping and pulsed-field gel electrophoresis for subspecies differentiation of strains of Enterococcus faecalis. J Clin Microbiol. 1993;31:1570–1574. doi: 10.1128/jcm.31.6.1570-1574.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gouby A, Carles-Nurit M-J, Bouziges N, Bourge G, Mesnard R, Bouvet P J M. Use of pulsed-field gel electrophoresis for investigation of hospital outbreaks of Acinetobacter baumannii. J Clin Microbiol. 1992;30:1588–1591. doi: 10.1128/jcm.30.6.1588-1591.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harsono K D, Kaspar C W, Luchansky J B. Comparison and genomic sizing of Escherichia coli O157:H7 isolates by pulsed-field gel electrophoresis. Appl Environ Microbiol. 1993;59:3141–3144. doi: 10.1128/aem.59.9.3141-3144.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koeuth T, Versalovic J, Lupski J R. Differential subsequence conservation of interspersed repetitive Streptococcus pneumoniae BOX elements in diverse bacteria. Genome Res. 1995;5:408–418. doi: 10.1101/gr.5.4.408. [DOI] [PubMed] [Google Scholar]
- 8.Kogan S C, Doherty M, Gitschier J. An improved method for prenatal diagnosis of genetic disease by analysis of amplified DNA sequences: application to hemophilia A. N Engl J Med. 1987;317:985–990. doi: 10.1056/NEJM198710153171603. [DOI] [PubMed] [Google Scholar]
- 9.Liu P Y F, Lau Y J, Hu B S, Shyr J M, Shi Z Y, Tsai W S, Lin Y H, Tseng C Y. Analysis of clonal relationships among isolates of Shigella sonnei by different molecular typing methods. J Clin Microbiol. 1995;33:1779–1783. doi: 10.1128/jcm.33.7.1779-1783.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin B, Humbert O, Camara M, Guenzi E, Walker J, Mitchell T, Andrew P, Prudhomme M, Alloing G, Hakenbeck R, Morrison D A, Boulnois J G, Claverys J P. A highly conserved repeated DNA element located in the chromosome of Streptococcus pneumoniae. Nucleic Acids Res. 1992;20:3479–3483. doi: 10.1093/nar/20.13.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mederski-Samoraj B D, Murray B E. High-level resistance to gentamicin in clinical isolates of enterococci. J Infect Dis. 1983;147:751–757. doi: 10.1093/infdis/147.4.751. [DOI] [PubMed] [Google Scholar]
- 12.Montecalvo M A, Horowitz H, Gedris C, Carbonaro C, Tenover F C, Issah A, Cook P, Wormser G P. Outbreak of vancomycin-, ampicillin-, and aminoglycoside-resistant Enterococcus faecium bacteremia in an adult oncology unit. Antimicrob Agents Chemother. 1994;38:1363–1367. doi: 10.1128/aac.38.6.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray B E, Singh K V, Heath J D, Sharma B R, Weinstock G M. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray B E, Singh K V, Markowitz S M, Lopardo H A, Patterson J E, Zervos M J, Rubeglio E, Eliopoulos G M, Rice L B, Goldstein F W, Jenkins S G, Caputo G M, Nasnass R, Moore L S, Wong E S, Weinstock G M. Evidence for clonal spread of a single strain of β-lactamase-producing Enterococcus faecalis to six hospitals in five states. J Infect Dis. 1991;163:780–785. doi: 10.1093/infdis/163.4.780. [DOI] [PubMed] [Google Scholar]
- 15.Schaberg, D. R., D. H. Culver, and R. P. Gaynes. 1991. Major trends in the microbial etiology of nosocomial infection. Am. J. Med. 91(Suppl. 3B):72S–75S. [DOI] [PubMed]
- 16.Sirko D A, Ehrlich G D. Laboratory facilities, protocol, and operations. In: Ehrlich G D, Greenberg S J, editors. PCR-based diagnostics in infectious disease. Cambridge, England: Blackwell Scientific Publications; 1994. pp. 19–43. [Google Scholar]
- 17.Stern M J, Ames G F L, Smith M H, Robinson E C, Higgins C F. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell. 1984;37:1015–1026. doi: 10.1016/0092-8674(84)90436-7. [DOI] [PubMed] [Google Scholar]
- 18.Struelens M J, Bax R, Depalno A, Quint W G, Belkum A V. Concordant clonal delineation of methicillin-resistant Staphylococcus aureus by macrorestriction analysis and polymerase chain reaction genome fingerprinting. J Clin Microbiol. 1993;31:1964–1970. doi: 10.1128/jcm.31.8.1964-1970.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomayko J F, Murray B E. Analysis of Enterococcus faecalis isolates from intercontinental sources by multilocus enzyme electrophoresis and pulsed-field gel electrophoresis. J Clin Microbiol. 1995;33:2903–2907. doi: 10.1128/jcm.33.11.2903-2907.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uttley A H, Collins C H, Naidoo J, George R C. Vancomycin-resistant enterococci. Lancet. 1988;i:57–58. doi: 10.1016/s0140-6736(88)91037-9. [DOI] [PubMed] [Google Scholar]
- 22.Van der Auwera P, Pensart N, Korten V, Murray B E, Leclercq R. Influence of oral glycopeptides on the fecal flora of human volunteers: selection of highly glycopeptide-resistant enterococci. J Infect Dis. 1996;173:1129–1136. doi: 10.1093/infdis/173.5.1129. [DOI] [PubMed] [Google Scholar]
- 23.Versalovic J, Kapur V, Mason E O, Jr, Snah U, Koeuth T, Lupski J R, Musser J M. Penicillin-resistant Streptococcus pneumoniae strains recovered in Houston: identification and molecular characterization of multiple clones. J Infect Dis. 1993;167:850–856. doi: 10.1093/infdis/167.4.850. [DOI] [PubMed] [Google Scholar]
- 24.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verweij P E, Belkum A V, Melchers W J G, Voss A, Hoogkamp-Korstanje J A A, Meis J F G M. Interrepeat fingerprinting of third-generation cephalosporin-resistant Enterobacter cloacae isolated during an outbreak in a neonatal intensive care unit. Infect Control Hosp Epidemiol. 1995;16:25–29. doi: 10.1086/646998. [DOI] [PubMed] [Google Scholar]
- 26.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, David D M, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Brooklyn, N.Y: Green Publishing Associates; 1994. pp. 2.4.1–2.4.2. [Google Scholar]
- 27.Woods C R, Versalovic J, Koeuth T, Lupski J R. Analysis of relationships among isolates of Citrobacter diversus by using DNA fingerprints generated by repetitive sequence-based primers in the polymerase chain reaction. J Clin Microbiol. 1992;30:2921–2929. doi: 10.1128/jcm.30.11.2921-2929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]