Abstract
The DNA relatedness and restriction fragment length polymorphism patterns of whole-cell DNA and the general phenotypic properties of 14 isolates of the form species Candida parapsilosis representing three diverse genetic groups were compared. The data confirm the unrelatedness of the three groups at the species level.
In recent years, Candida parapsilosis, a form species of the Fungi Imperfecti defined by morphological and physiological properties, has been increasingly implicated as a cause of fungemia, endocarditis in neutropenic or severely debilitated patients, and candidal vaginitis (22). The species has been commonly associated with the skin and fingernails of humans (18), and it has been isolated from soil and fresh and marine waters (1, 3, 13).
Genetic heterogeneity has been reported within this species in several studies. Scherer and Stevens (16) divided their clinical isolates of C. parapsilosis into three groups on the basis of restriction fragment length polymorphisms (RFLPs). The data from this study suggested that C. parapsilosis is genotypically more heterogeneous than Candida albicans. Lehmann et al. (6), on the basis of randomly amplified polymorphic DNA profiles, also observed three distinct groups among isolates of C. parapsilosis. Group I and II isolates of the latter study shared the API 20C (Analytab, Plainview, N.Y.) code 6756171, whereas the two isolates in group III had the biotype code 6776171. Further studies of representative strains from these three groups indicated that each group included isolates with distinctive karyotypes (9). Lin et al. (8) examined 45 clinical isolates that were physiologically similar to C. parapsilosis as determined with the API 20C kit and reported the presence of three distinct isoenzyme-defined groups that were also distinguishable on the basis of sequence analysis of the internally transcribed spacer sequences flanking the 5.8S RNA gene.
In this investigation, we examined the nuclear DNA base composition, DNA-DNA reassociation, and DNA RFLPs of 13 isolates of C. parapsilosis from diverse sources (including representatives of groups I to III mentioned above) and the type strain.
MATERIALS AND METHODS
Yeast strains.
The strains examined in this study are listed in Table 1. They include the type strain of C. parapsilosis and representative strains of the three groups reported by Lin et al. (8). The standard morphological and physiological characterizations of the yeasts were performed according to the methods described by van der Walt and Yarrow (21).
TABLE 1.
Source and G+C content of isolates grouped within C. parapsilosis
| Code no.a | Source | Tmb (°C) ± SD (n = 2 or 3) | mol% G+C ± SD (n = 2 or 3) | Groupc |
|---|---|---|---|---|
| NRRL Y-10518 | Marine isolate | 86.3 ± 0.10 | 41.5 ± 0.24 | I |
| NRRL Y-9282 | Marine isolate | 86.2 ± 0.10 | 41.2 ± 0.24 | I |
| CBS 8181 | Rotten wood in Chile | 86.2 ± 0.07 | 41.2 ± 0.17 | I |
| BT.017 | Sprue, Tex. | 86.1 ± 0.07 | 40.9 ± 0.17 | I |
| CBS 2215 | Pickling vat, 10% brine | 86.1 ± 0.05 | 40.9 ± 0.12 | I |
| NRRL Y-10208 | Marine isolate | 85.9 ± 0.15 | 40.5 ± 0.37 | I |
| ATCC 22019T | Sprue, Puerto Rico | 85.9 ± 0.00 | 40.5 ± 0.00 | I |
| NRRL Y-10022 | Marine isolate | 85.8 ± 0.10 | 40.2 ± 0.24 | I |
| CBS 8050 | Epidermis of Porphyra tenera | 85.8 ± 0.00 | 40.2 ± 0.00 | I |
| NRRL Y-9536 | Antarctic Peninsula | 85.9 ± 0.05 | 40.5 ± 0.12 | II |
| MCO 452 | Human urine, Tex. (8) | 85.1 ± 0.10 | 38.5 ± 0.24 | II |
| MCO 448 | Human hand, Tacoma, Wash. (8) | 86.0 ± 0.15 | 40.7 ± 0.37 | III |
| MCO 429 | Clinical, Livermore, Calif. (8) | 86.0 ± 0.10 | 40.7 ± 0.24 | III |
| 96.026087 | Clinical, Connecticut | 85.5 ± 0.07 | 39.4 ± 0.17 | III |
ATCC, American Type Culture Collection; CBS, Centraalbureau voor Schimmelcultures; MCO, Medical College of Ohio; NRRL, Northern Regional Research Laboratory.
Tm, midpoint temperature.
Based on Lin et al. (8).
DNA base composition and reassociation studies.
DNA was isolated and purified by the method of Marmur (10) as modified by Meyer and Phaff (12). DNA base composition (mol% G+C) was determined by the thermal denaturation method as described by Marmur and Doty (11). The optical renaturation method of Seidler and Mandel (17) as modified by Kurtzman et al. (5) was employed for the DNA reassociation studies.
RFLPs.
DNA was extracted and purified by the methods of Su and Meyer (19) and Scherer and Stevens (16), and restriction enzyme analysis was performed with HinfI, HindIII, PvuII, and HaeIII restriction endonucleases (Boehringer Mannheim GmbH, Mannheim, Germany). The DNA fragments were separated in a 1% agarose gel in Tris-acetate-EDTA buffer (40 mM Tris-OH, 20 mM acetic acid, 2 mM Na2EDTA [pH 8.1]) (2).
RESULTS
Percent DNA relatedness divided the isolates into three distinct groups, each of which agreed with the molecular characteristics of the representative cultures of the groups established by Lin et al. (8). There was greater than 95% DNA relatedness among isolates within a group and less than 25% relatedness between isolates from different groups (Table 2). The G+C contents of the isolates were 40.2 to 41.5 mol% (group I), 38.5 to 40.5 mol% (group II), and 39.4 to 40.7 mol% (group III) (Table 1).
TABLE 2.
DNA relatedness among groups I, II, and III of C. parapsilosis
| Strain (group) | Strain compared | Group | % DNA relatedness |
|---|---|---|---|
| ATCC 22019T (I) | NRRL Y-10518 | I | 100.0 |
| NRRL Y-10022 | I | 100.0 | |
| NRRL Y-10208 | I | 100.0 | |
| CBS 8050 | I | 99.7 | |
| CBS 2215 | I | 98.3 | |
| NRRL Y-9282 | I | 97.3 | |
| BT.017 | I | 96.6 | |
| CBS 8181 | I | 95.5 | |
| NRRL Y-9536 | II | 24.6 | |
| MCO 429 | III | 21.6 | |
| MCO 452 | II | 9.6 | |
| MCO 448 | III | 9.5 | |
| NRRL Y-10022 (I) | 96.026087 | III | 10.8 |
| MCO 452 (II) | NRRL Y-9536 | II | 96.0 |
| MCO 448 | III | 21.3 | |
| MCO 429 | III | 18.7 | |
| MCO 448 (III) | MCO 429 | III | 100.0 |
| 96.026087 | III | 99.7 |
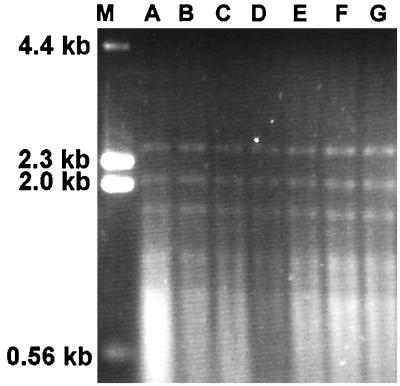
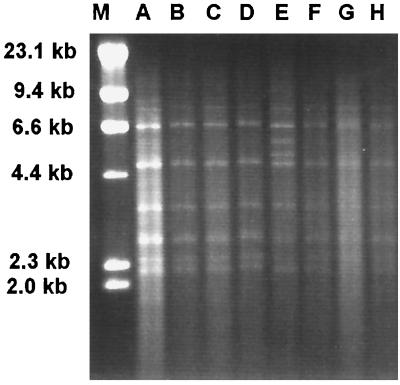
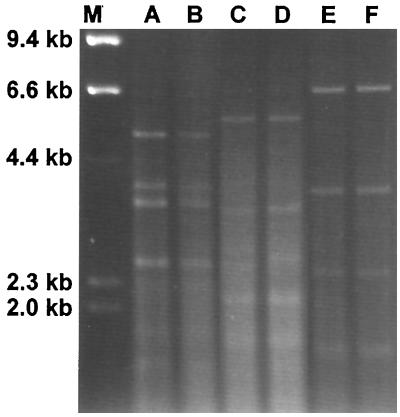
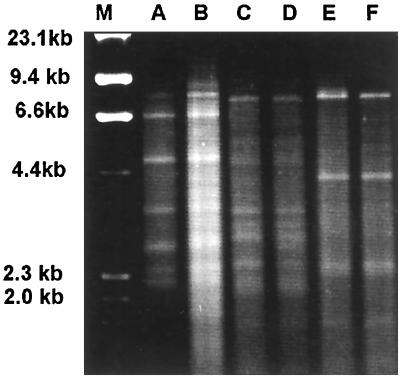
Group I isolates all had similar RFLP patterns after digestion with HinfI (Fig. 1). Polymorphisms were evident in this group after double digestion with HindIII and HaeIII (Fig. 2) and HindIII and PvuII (Fig. 3). The RFLP patterns of representative isolates of each group after digestion with HinfI (data not shown) and the double digestions demonstrated RFLP distinctions among the three groups (Fig. 4 and 5). No polymorphisms were observed with the restriction digest patterns of isolates of group II and group III.
FIG. 1.
C. parapsilosis group I RFLP patterns from HinfI digests. Lanes: M, λ DNA digested with HindIII; A, ATCC 22019; B, NRRL Y-10022; C, NRRL Y-10208; D, NRRL Y-9282; E, NRRL Y-10518; F, CBS 2215; G, CBS 8181.
FIG. 2.
C. parapsilosis group I RFLP patterns from HindIII-HaeIII digests. Lanes: M, λ DNA digested with HindIII; A, ATCC 22019; B, NRRL Y-10022; C, NRRL Y-10208; D, NRRL Y-10518; E, CBS 2215; F, NRRL Y-9282; G, CBS 8050; H, CBS 8181.
FIG. 3.
C. parapsilosis group I RFLP patterns from HindIII-PvuII digests. Lanes: M, λ DNA digested with HindIII; A, ATCC 22019; B, NRRL Y-10022; C, NRRL Y-10208; D, NRRL Y-10518; E, CBS 2215; F, NRRL Y-9282; G, CBS 8050; H, CBS 8181.
FIG. 4.
C. parapsilosis RFLP profiles from HindIII-HaeIII digests. Lanes: M, λ DNA digested with HindIII; A and B, group I ATCC 22019 and CBS 8050, respectively; C and D, group II MCO 452 and NRRL Y-9536, respectively; E and F, group III MCO 429 and MCO 448, respectively.
FIG. 5.
C. parapsilosis RFLP profiles from HindIII-PvuII digests. Lanes: M, λ DNA digested with HindIII; A and B, group I ATCC 22019 and CBS 8050, respectively; C and D, group II MCO 452 and NRRL Y-9536, respectively; E and F, group III MCO 429 and MCO 448, respectively.
DISCUSSION
Our DNA relatedness and RFLP analyses support the earlier divisions of isolates classified as C. parapsilosis into three separate groups. The low degree of DNA relatedness among the groups indicates that they represent distinct species. Although two of these groups (II and III) have been recognized only within the past decade, one of the group II cultures was originally isolated in 1966 from the Antarctic Peninsula (possibly from human or animal pollution). Our recent clinical isolates (1996) represented group I and group III. We do not have sufficient information for the phenotypic differentiation of these genetically distinct yeasts, nor can we currently discern if differences in their epidemiology exist.
The finding of genetically diverse groups among the form species of the imperfect yeasts is not unusual. Lehmann et al. (7) found two distinct genetic groups within Candida haemulonii. The phenotype of Candida famata is found among strains of Candida guilliermondii and several species of Debaryomyces (14). A variety of distinct genotypes are present within the general phenotype of Trichosporon cutaneum (15). Most recently, C. albicans was found to include a diverse genetic group which was designated a distinct species, Candida dubliniensis (20). The application of molecular procedures to yeast systematics has resulted in major reclassifications of many species and genera (4, 13). It is obvious that understanding the genetic diversity of the yeasts associated with human infections requires further study.
ACKNOWLEDGMENT
We offer special thanks to D. G. Ahearn, Georgia State University, Atlanta, Ga., for his guidance and recommendations in this research and the preparation of the manuscript.
REFERENCES
- 1.Ahearn, D. G. Yeasts pathogenic for humans. In C. P. Kurtzman and J. W. Fell (ed.), The yeasts, a taxonomic study, in press. Elsevier Science Publishers B.V., Amsterdam, The Netherlands.
- 2.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 3.Fell J W. Yeasts in oceanic regions. In: Jones E B C, editor. Recent advances in aquatic mycology. London, England: Elek Science; 1976. pp. 93–124. [Google Scholar]
- 4.Kurtzman C P. Molecular taxonomy of the yeasts. Yeast. 1994;10:1727–1740. doi: 10.1002/yea.320101306. [DOI] [PubMed] [Google Scholar]
- 5.Kurtzman C P, Smiley M J, Johnson C J, Wickerham L J, Fuson G B. Two new and closely related heterothallic species, Pichia amylophila and Pichia mississippiensis. Int J Syst Bacteriol. 1980;30:208–216. [Google Scholar]
- 6.Lehmann P F, Lin D, Lasker B A. Genotypic identification and characterization of species and strains within the genus Candida by using random amplified polymorphic DNA. J Clin Microbiol. 1992;30:3249–3254. doi: 10.1128/jcm.30.12.3249-3254.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehmann P F, Wu L-C, Pruitt W R, Meyer S A, Ahearn D G. Unrelatedness of groups of yeasts within the Candida haemulonii complex. J Clin Microbiol. 1993;31:1683–1687. doi: 10.1128/jcm.31.7.1683-1687.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin D, Wu L-C, Rinaldi M G, Lehmann P F. Three distinct genotypes within Candida parapsilosis from clinical sources. J Clin Microbiol. 1995;33:1815–1821. doi: 10.1128/jcm.33.7.1815-1821.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lott T J, Kuykendall R J, Welbel S F, Pramanik A, Lasker B A. Genomic heterogeneity in the yeast Candida parapsilosis. Curr Genet. 1993;23:463–467. doi: 10.1007/BF00312635. [DOI] [PubMed] [Google Scholar]
- 10.Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 11.Marmur J, Doty P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- 12.Meyer S A, Phaff H J. Deoxyribonucleic acid composition in yeasts. J Bacteriol. 1969;97:52–56. doi: 10.1128/jb.97.1.52-56.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer, S. A., R. W. Payne, and D. Yarrow.Candida Berkhout. In C. P. Kurtzman and J. W. Fell (ed.), The yeasts, a taxonomic study, in press. Elsevier Science Publishers B.V., Amsterdam, The Netherlands.
- 14.Nishikawa A, Tomomatsu H, Sugita T, Ikeda R, Shinoda T. Taxonomic position of clinical isolates of Candida famata. J Med Vet Mycol. 1996;34:411–419. doi: 10.1080/02681219680000731. [DOI] [PubMed] [Google Scholar]
- 15.Nishiura Y, Nakagawa-Yoshida K, Suga M, Shinoda T, Gueho E. Assignment and serotyping of Trichosporon species: the causative agents of summer-type hypersensitivity pneumonitis. J Med Vet Mycol. 1997;35:45–52. doi: 10.1080/02681219780000861. [DOI] [PubMed] [Google Scholar]
- 16.Scherer S, Stevens D A. Application of DNA typing methods to epidemiology and taxonomy of Candida species. J Clin Microbiol. 1987;25:675–679. doi: 10.1128/jcm.25.4.675-679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seidler R J, Mandel M. Quantitative aspects of deoxyribonucleic acid renaturation: base composition, state of chromosome replication, and polynucleotide homologies. J Bacteriol. 1971;106:608–614. doi: 10.1128/jb.106.2.608-614.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strausbaugh L J, Sewell D L, Ward T T, Pfaller M A, Heitzman T, Tjoelker R. High frequency of yeast carriage on hands of hospital personnel. J Clin Microbiol. 1994;32:2299–2300. doi: 10.1128/jcm.32.9.2299-2300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su C-S, Meyer S A. Characterization of mitochondrial DNA in various Candida species: isolation, restriction endonuclease analysis, size, and base composition. Int J Syst Bacteriol. 1991;41:6–14. doi: 10.1099/00207713-41-1-6. [DOI] [PubMed] [Google Scholar]
- 20.Sullivan D J, Westerneng T J, Haynes K A, Bennett D E, Coleman D C. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology. 1995;141:1507–1521. doi: 10.1099/13500872-141-7-1507. [DOI] [PubMed] [Google Scholar]
- 21.van der Walt J P, Yarrow D. Methods for the isolation, maintenance, classification and identification of yeasts. In: Kreger-van Rij N J W, editor. The yeasts: a taxonomic study. 3rd ed. Amsterdam, The Netherlands: Elsevier Science Publishers B. V.; 1984. pp. 45–104. [Google Scholar]
- 22.Vazquez J A, Beckley A, Donabedian S, Sobel J D, Zervos M J. Comparison of restriction enzyme analysis versus pulsed-field gradient gel electrophoresis as a typing system for Torulopsis glabrata and Candida species other than C. albicans. J Clin Microbiol. 1993;31:2021–2030. doi: 10.1128/jcm.31.8.2021-2030.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]