Abstract
A heteroduplex tracking assay (HTA) was developed for genetic analyses of the hepatitis C virus (HCV) using single-stranded probes from the core (C)/E1 region. Nucleotide sequencing of reverse transcriptase (RT)-PCR products from 15 Italian dialysis patients confirmed the specificity and accuracy of the HTA genotyping method, which identified 5 of 15 (33.3%) 1b, 7 of 15 (46.7%) 3a, and 3 of 15 (20%) type 2 infections. The genotypes of an additional 12 HCV antibody-positive blood donors from different geographical locations were also in agreement with the genotypes determined by the Inno-LiPA HCV II kit (Innogenetics) and/or restriction fragment length polymorphism (RFLP). Isolates which had between 35 to 40% nucleotide divergence from control subtype 1a, 1b, 2a, 2b, or 3a standards could be typed. Surprisingly, HTA detected one 1b-2 coinfection which was missed by DNA sequencing. Three samples that were designated non-2a or 2b type 2 by HTA were found to be type 2a by both RFLP and direct nucleotide sequencing of the 5′ untranslated region. The genetic distance between patient type 2 and control 2a, 2b, and 2c isolates indicated that a new subtype was present in the population being studied. Serotyping (RIBA serotyping strip immunoblot assay kit) of 23 dialysis patients showed that the genotype could be determined in 6 of 8 (75%) C/E1 RT-PCR-negative and 15 of 23 (65.2%) RT-PCR-positive samples, indicating that the two tests complement each other.
Hepatitis C virus (HCV) is a single-stranded RNA virus (9) which is known to cause hepatitis and infects an estimated 100 million people worldwide (3). The HCV genome consists of 5′ and 3′ untranslated regions (UTR) that flank a single long open reading frame which encodes the structural (core [C], envelope 1 [E1], E2, and p7) and nonstructural (NS2, NS3, NS4a, NS4b, NS5a, and NS5b) proteins required for the manufacture of infectious virus (reviewed in references 21, 23, 38, and 43). Typical for RNA viruses, the mutation rate of HCV genomes is reported to be 1.44 × 10−3 per genome per site per year (28), and variants circulate in a quasispecies distribution within infected individuals (26, 41). Classification of viral genomes into genotypes, subtypes, and isolates based on the degree of nucleotide heterogeneity of different segments of the viral genome has been proposed (reviewed in reference 4; 33), although the relationship between genetic polymorphism and biological features of the different genotypes has not been clearly established for HCV (reviewed in reference 2). Since genotype may be a factor in designing and administering vaccines and therapeutic agents, reliable assays for hepatitis C are needed.
Since sequencing complete viral genomes is not practical, and many clinical laboratories do not have DNA-sequencing facilities, several methods have been described for determining HCV genotypes based on restriction enzyme-, PCR-, or hybridization-based analysis of the 5′ UTR (11, 36), core (30, 34), or NS5 (16, 33) region. Current methods for subtyping have limited accuracy either because they assess short, relatively conserved regions of the genome, such as the 5′ UTR, or because they can yield false results due to nonspecificity of PCR primers (25, 30). Although hybridization-based assays may be reasonably accurate (36), information about nonhybridizing samples can be obtained only by DNA sequencing. We describe the application of the hybridization-based heteroduplex tracking assay (HTA) (12–14) to HCV for genotyping and identifying molecular variants which might be missed by other genotyping methods. HTA involves hybridizing probes from known HCV subtypes to reverse transcriptase PCR (RT-PCR) products from the homologous sera or from unknown samples and electrophoresing the hybridization products on gels. The formation of a heteroduplex band on a gel indicates genotype and subtype. In the absence of genetic rearrangements, insertions, and/or deletions, the genetic relationship between isolates can be determined by the relative migration of the heteroduplexes on gels (12).
To develop and evaluate an HTA genotyping system for HCV, three methods of typing isolates (restriction fragment length polymorphism [RFLP], DNA sequencing, and RIBA serotyping strip immunoblot assay [SIA]) were performed on 23 well-preserved and carefully characterized serum samples from a cohort of Italian dialysis patients (5, 18, 39). Samples from an additional 12 blood donors from different countries (the United States, Egypt, and Holland) were also analyzed by HTA, and the results were compared to those obtained by the Inno-LiPA HCV II test (Innogenetics) and/or RFLP to establish the accuracy of HTA as a method for determining genotype.
MATERIALS AND METHODS
Patient population.
Twenty-six of 35 Italian patients who had been undergoing regular hemodialysis in the same unit since 1991 (5, 18, 39) tested positive for anti-HCV antibodies in August 1995 by the second- and third-generation Ortho HCV enzyme-linked immunosorbent assay (ELISA). Serum samples were collected, divided into aliquots, and stored at −80°C. Twenty-five patients were both Ortho ELISA 3.0 and RIBA 3.0 HCV SIA reactive, while one patient was indeterminate (C-22-P reactive). Twenty-three ELISA 3.0 and RIBA 3.0 SIA-reactive serum samples were available for this study. RT-PCR was performed on 15 of 23 samples with primers from both the 5′ UTR (modified from Han et al., [19] sense primer 93B, 5′ACCATGAATCACTCCCCT3′ and antisense primer JH51, 5′CCCAACACTACTGGGCTA3′) and C/E1 (described in Table 1). One, nine, and two RT-PCR (5′ UTR and C/E1)-positive blood donor serum samples from Egypt (a gift from J. Lau, University of Gainesville), the United States (Sacramento Blood Bank), and Holland (a gift from H. Reesink, C. van der Poel, and N. Lelie; CLB, Amsterdam, Holland), respectively, were analyzed by HTA.
TABLE 1.
HTA RT-PCR primers
| Primer | Sequence | Nucleotide coordinates (5′→3′)a | PCR round |
|---|---|---|---|
| C170Sb | CCTGGTTGCTCTTTCTCTATCT | 508–529 | I |
| E338A1 | GATGGCTTGTGGGATCCGGAG | 1032–1012 | I |
| E338A2a | GATGACCTCGGGGACGCGCAT | 1032–1012 | I |
| E338A2b | GACCAGTTCTGGAACACGAGC | 1032–1012 | I |
| E338A3a | CAAGGTCTGGGGTAAACGCAG | 1032–1012 | I |
| E320Ac | CCAGTTCATCATCATATCCCA | 978–958 | II |
| C179S1 | TGGCCCTGCTCTCTTGCTTGAC | 536–557 | II |
| C179S2 | TTGCTCTTCTGTCGTGCGTCAC | 536–557 | II |
| C179S3 | TTGCTCTGTTCTCTTGCTTAAT | 536–557 | II |
According to Choo et al. (10).
Universal sense primer for genotype determination.
Universal antisense primer for genotype determination.
RNA extraction, cDNA synthesis, and PCR.
RNA was extracted from 100 μl of sera or plasma on at least two different occasions with the Stratagene RNA isolation kit (no. 200345) according to the manufacturer’s instructions (8). A 25-μl cDNA reaction mixture contained 20% plasma RNA extract, 10 mM Tris-HCl (pH 8.3), 10 mM MgCl2, 10 mM dithiothreitol, 75 mM KCl, 1 mM (each) deoxynucleoside triphosphates, 25 U of RNasin (Promega), 5 U of avian myeloblastosis virus RT (no. 80855B; Gibco BRL), and 100 pmol of antisense primer (Table 1). The cDNA mixture was diluted with 50 μl of water, boiled for 5 min, quick-cooled on ice, and added to the PCR reagents, with final concentrations according to the Perkin PCR kit (no. N801-0055) instructions. cDNA primer E338A1, -2a, -2b, or -3a and a universal primer (C170S) were used in the first round of PCR (PCR I) to amplify subtypes 1 (a and b), 2a, 2b, and 3a HCV, respectively. One to ten microliters of the PCR I product was added to a second PCR (PCR II) mixture containing 100 pmol of universal antisense primer (320A) and type- or subtype-specific sense primers (C179S1, 2a, 2b, and 3a). All PCRs consisted of 40 cycles (94°C for 10 s, 55°C for 30 s, and 72°C for 30 s) performed in a Perkin-Elmer 9600 thermocycler, with the exception of type 1b PCR II, in which the annealing temperature was lowered to 45°C.
Single-stranded probes.
Initially, subtype-specific probes were obtained from RT-PCR products generated from RNA extracted from individuals infected with HCV of known genotype (1a, 1b, 2a, 2b, and 3a) as described above, with the exception of primer 320A, which was biotinylated. Subsequently, RT-PCR products used to synthesize the probes used in Fig. 1 were subcloned in the PCR 2.1 vector (Invitrogen) and used as templates for the biotinylated PCR products used to generate the single-stranded probes shown in Fig. 3 (lanes 1a, 1b, 2a, 2b, and 3a). Approximately 500 ng of biotinylated PCR II product was bound to 20 μl (200 μg) of streptavidin-coated magnetic beads (no. 112.05; Dynal M280) which were first washed in 100 μl of phosphate-buffered saline, pH 7.4–0.1% bovine serum albumin, followed by 100 μl of binding and 1× washing (B&W) buffer containing 5 mM Tris-HCl (pH 7.5), 0.5 mM EDTA, and 1.0 M NaCl and finally resuspended in 50 μl of 2× B&W buffer. After being gently mixed at room temperature for approximately 15 min, the beads were collected in a magnetic field, the supernatant was removed, and the beads were washed three times with 200 μl of 1× B&W buffer to eliminate unbound PCR products, primers, and deoxynucleoside triphosphates. Eight microliters of freshly prepared 0.1 M NaOH was added to the beads for 10 min at room temperature to denature the bound, double-stranded PCR products. Beads were again collected in a magnetic field, and the supernatant containing single-stranded DNA was removed and neutralized with 40 μl of water, 4 μl of 0.2 M HCl, and 1.0 μl of 1.0 M Tris-HCl, pH 8.0 (11a). Twenty nanograms of single-stranded DNA was labeled with 100 μCi of [γ-32P]ATP (3,000 Ci/mmol) with T4 kinase and passed over a G-50 column (no. 27.5335.01; Pharmacia) to remove excess [γ-32P]ATP. Biotinylated primers and single-stranded probes were stored at −20°C in aliquots for optimal results.
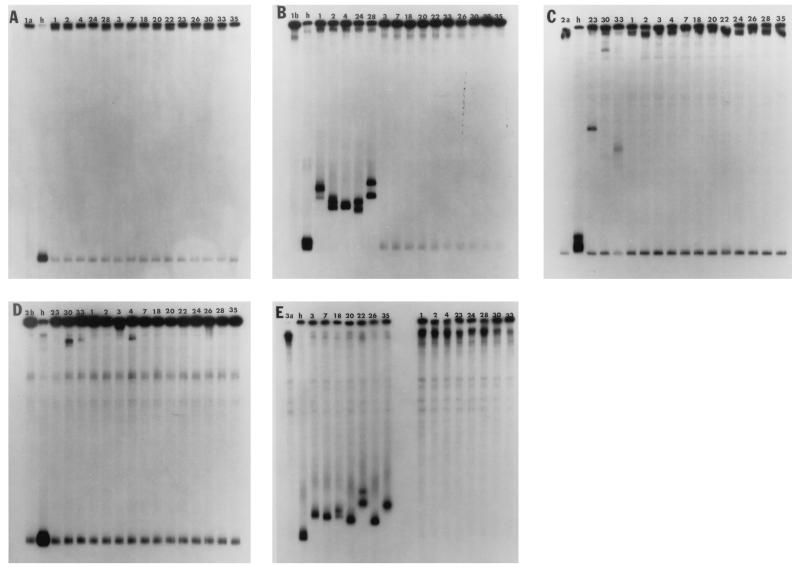
FIG. 1.
Single-stranded probes (lanes 1a, 1b, 2a, 2b, and 3a in panels A to E, respectively) were hybridized to PCR products from either the same control serum samples from which they were derived, forming homoduplexes (h), or from dialysis patient sera, forming heteroduplexes (numbered lanes), and were electrophoresed on MDE gels.
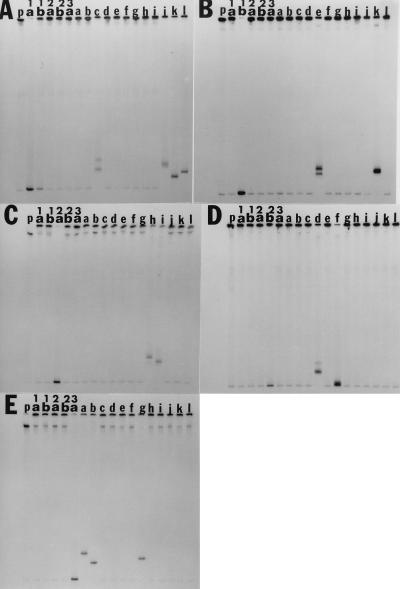
FIG. 3.
Single-stranded probes (1a, 1b, 2a, 2b, and 3a) were hybridized to either PCR products from which each single-stranded probe was derived (lanes 1a, 1b, 2a, 2b, 3a in panels A to E) or to HCV antibody-positive sera from the United States (lanes b to j), Egypt (lane a), and Holland (lanes k and l). The single-stranded probe alone is designated by the letter p.
HTA.
One to two microliters (0.5 ng) of single-stranded DNA probe (specific activity, 1 × 109 cpm/μg) was hybridized to a 100-fold molar excess of colinear, double-stranded PCR product from either the homologous standard (JK1a, -1b, -2a, -2b, or -3a) or patient sera in the following 10-μl reaction mixture: 4 μl of double-stranded PCR product (∼50 ng), 4 μl of 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 mM sodium citrate, 10 mM EDTA [pH 8.0]) and 2 μl of water. The reaction was denatured for 3 min at 95°C and annealed at 55°C for 2 h in a heat block. The entire sample was loaded onto a 1-mm-thick, 6% Hydrolink MDE gel (no. 4739-00; Baker) and electrophoresed for 16 h at 500 V. Gels were vacuum dried at 80°C on filter paper and exposed to X-ray film for 15 min. The relative migration of a heteroduplex was calculated as the distance (in millimeters) the heteroduplex migrated from the top of the gel divided by the distance (in millimeters) the homoduplex migrated from the top of the gel.
DNA sequencing.
PCR products and cloned DNA were purified by either Wizard PCR or Maxi prep (no. A7170 or A7270; Promega) (no. 12191; Qiagen) according to the manufacturer’s protocol and sequenced by standard methods. PCR products were cloned with the TA cloning vector (no. K2000-01; Invitrogen). A minimum of three clones were sequenced for each patient sample.
Phylogenetic trees.
The phylogenetic trees in Fig. 2 were constructed by pairwise, progressive alignment of the nucleotide sequences to one another with the computer software program Gene-Works Unweighted Pair Group Method with Arithmetic Mean (41).
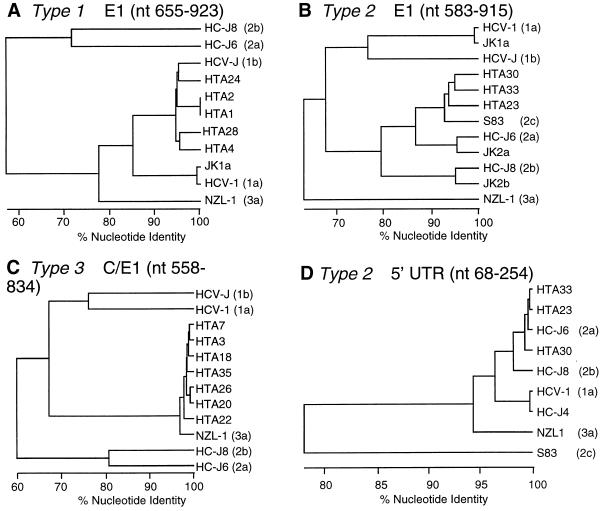
FIG. 2.
Phylogenetic analysis of partial C/E1, E1, or 5′ UTR nucleotide sequences from dialysis patients (HTA 1 to 4, 7, 18, 20 to 26, 28, 30, 33, and 35) and published subtypes 1a (HCV-1 [10]), 1b (HCV-J [22]), 2a (HC-J6 [29]), 2b (HC-J8 [29]), 2c (S83 [4]), and 3a (NZL-1 [31]). Nucleotide coordinates are according to reference 12.
RFLP analysis of 5′ UTR PCR products.
cDNA amplified with 5′ UTR primers (32) was digested as described by Davidson et al. (11) with ScrF1 to distinguish type 2 HCV subtypes. The MvaI/HinF1 fragment was used to determine genotype for the 12 blood donor samples. Digested DNA was electrophoresed on 6% polyacrylamide gels, stained with ethidium bromide, and visualized under UV light. There were no differences between the restriction fragment patterns of PCR products generated with primers described by Shimizu et al. (32) and those described by Davidson et al. (11), since the PCR primers are almost colinear.
Inno-LiPA HCV II test.
The Inno-LiPA HCV II test (Innogenetics), which is based on sequence heterogenetics in the 5′ UTR, was performed with samples from blood donors from the United States according to the manufacturer’s protocol.
RIBA HCV SIA.
Eight serotype-specific HCV peptide antigens, five of which are from the NS4 region of HCV subtypes 1a and 1b, 2a and 2b, and 3 and three of which are from the core region of types 1 and 2, were immobilized on a nitrocellulose solid support. An algorithm was defined such that serotype was determined by which type-specific NS4 peptide was reactive in the tested sera. In the absence of NS4 peptide reactivity, core peptide reactivity was considered for the interpretation of the results (15).
RESULTS
Specificity of HTA genotyping.
Single-stranded probes (nucleotides 536 to 978), derived from sera of known subtype, were hybridized to colinear RT-PCR products from 15 Italian dialysis patients and electrophoresed on MDE gels. As shown in Fig. 1A through E, each probe (lanes 1a, 1b, 2a, 2b, and 3a, respectively) hybridized to RT-PCR products from the homologous sera, forming a homoduplex (lane h). Heteroduplexes were observed for five samples hybridized to the JK1b probe (Fig. 1B, samples 1, 2, 4, 24, and 28), and seven samples hybridized to the JK3a probe (Fig. 1E, samples 3, 7, 18, 20, 22, 26, and 35), while there was no subtype 1a HCV (Fig. 1A). Probe-only lanes (1a, 1b, 2a, 2b, and 3a in Fig. 1 and p in Fig. 3) show the level of background, which can be observed in all lanes, when the probe is degrading. The specificity of HTA probes for their respective subtypes was demonstrated by the lack of cross-hybridization of the subtype-specific probes (JK1a, JK1b, JK2a, JK2b, and JK3a) to each other or to PCR products from more than one patient. Two exceptions were patient 4, who was most likely coinfected with 1b and type 2 HCV (Fig. 1B and D), and blood donor k (Fig. 3A and B), who had both 1a and 1b HCV.
The mean relative migration of subtype 1b isolates (0.79; range, 0.72 to 0.83) was less than and did not overlap with that of the 3a isolates (0.89; range, 0.86 to 0.93), indicating that the 1b isolates were less closely related to the 1b probe than the 3a isolates were to the 3a probe. Surprisingly, we found three samples which formed slowly migrating heteroduplexes, two of which hybridized to the JK2a and JK2b probes (samples 30 and 33 in Fig. 1C and D) and one which hybridized to the JK2a probe only (Fig. 1C, sample 23). Samples 23, 30, and 33 (HTA 23, 30, and 33) had relative migrations of 0.45, 0.1, and 0.55 to the JK2a probe, while HTA 30 and 33 had relative migrations of 0.12 and 0.1 with respect to the JK2b probe. The data indicated that HTA 23, 30, and 33 were substantially divergent from the JK2a and/or JK2b probes and suggested that they were most likely a non-2a, non-2b type 2 subtype.
DNA sequencing verifies HTA genotype and subtype assignments and confirms complexity within type 2.
The genotypes of all 15 PCR-positive Italian patients and subtype probes were verified by nucleotide sequencing (data not shown) of the partial C/E1 PCR products by direct sequencing and/or by sequencing cloned PCR products. Phylogenetic trees shown in Fig. 2A through C, which were derived from the nucleotide sequences, confirmed that isolates which were designated 1b (HTA 1, 2, 4, 24, and 28) or 3a (HTA 3, 7, 18, 20, 22, 26, and 35) segregated with the appropriate published 1b (HCV-J [22]) and 3a (NZ1-1 [31]) prototype sequences. The mean percentage of nucleotide divergence of 1b and 3a patient isolates from probes JK1a and JK3a of 10.5% (range, 8.7 to 11.6%) and 4.3% (range, 1.4 to 6.5%) was in agreement with the HTA prediction that the 3a isolates were related more closely to the JK3a probe than the 1b isolates were to the JK1b probe, based on the relative migration of the 1b and 3a heteroduplexes on gels. The C/E1 nucleotide sequence data indicated that (i) isolates with ≥1.4% nucleotide substitutions can be separated by HTA analysis, (ii) isolates within the subtypes 1b and 3a were <15% divergent, and (iii) HTA subtype probes do not cross-hybridize with isolates with greater than approximately 35 to 40% nucleotide divergence.
Since HTA 23, 30, and 33 were 22.8 to 27% divergent from JK2a, and HTA 30 and 33 were 33 to 34.5% divergent from JK2b, we compared the nucleotide sequences of HTA 23, 30, and 33 to a published 2c isolate (S83 [4]). HTA 23, 30, and 33 were 16.5, 14.7, and 18.6% divergent from S83 and segregated with S83 in Fig. 2B as expected. Interestingly, HTA 33, which was most divergent from S83, was more closely related to HTA 23 and 30 (11.7 and 15.6% nucleotide heterogeneity) than to either JK2a or JK2b, suggesting a greater complexity among type 2 isolates than among type 1 or type 3 isolates.
Although most patients had a single virus isolate or a tight cluster of virus isolates, which were not resolved on gels when hybridized to their respective probes, HTA 18 and 22 each had two bands which were not observed in the homoduplex or single-stranded probe control lane (Fig. 1E, lanes h and 3a). The data suggested that these patients each had at least two isolates with greater than 1.4% nucleotide heterogeneity. It is unlikely that the double bands seen in all lanes except one in Fig. 1B resulted from double-stranded probe molecules contaminating the single-stranded probe, since the result was reproducible with different preparations of probe and was not observed for the other probes generated under identical conditions.
Comparison of partial C/E1 HTA and RFLP.
We compared the genotype results obtained by partial C/E1 HTA with RFLP (5′ UTR) and direct nucleotide sequencing of 5′ UTR PCR products from all 15 patients. Subtype assignments were all in agreement except for HTA 23, 30, and 33, which were incorrectly identified as subtype 2a by RFLP with ScrFI (Table 2). A dendrogram of the 5′ UTR nucleotide sequences of HTA 23, 30, and 33 and published sequences (Fig. 2D) showed HTA 23, 30, and 33 as 2a, in agreement with RFLP results, but was inconsistent with the same analysis performed on the E1 sequence (Fig. 2B). The 5′ UTR analysis also inaccurately located 2c distant from subtypes 2a and 2b, a result which is most likely due to the small number of nucleotide substitutions in the 5′ UTR.
TABLE 2.
Comparison of serotyping, RFLP and HTA genotype analysis of sera from dialysis patients
| HTA sample no. | C/E1 PCR | Results according to RIBA HCV SIAe
|
Subtype bya:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| NS4 subtype
|
Core 1 | 2(I+J) | Geno- type | RFLP | HTA | ||||
| 1 | 2 | 3 | |||||||
| 1 | + | ++++ | − | ± | +++ | − | 1 | 1b | 1b |
| 2 | + | ± | − | − | − | − | NRb | 1b | 1b |
| 3 | + | − | − | − | ± | − | NR | 3a | 3a |
| 4 | + | − | − | − | − | − | NR | 1b | 1b |
| 5 | − | +++ | − | ± | ± | − | 1 | ND | ND |
| 6 | − | ++++ | ± | − | ++++ | ± | 1 | ND | ND |
| 7 | + | − | − | ± | − | − | NR | 3a | 3a |
| 18 | + | − | − | − | ++ | +++ | 2 | 3a | 3a |
| 19 | − | ++ | − | − | ± | ++ | 1 | ND | ND |
| 20 | + | ± | − | − | ++++ | ± | 1–3c | 3a | 3a |
| 22 | + | − | − | ++ | − | − | 3 | 3a | 3a |
| 23 | + | − | ± | − | − | + | 2 | 2a | 2d |
| 24 | + | ++++ | − | ++ | ++++ | ± | 1 | 1b | 1b |
| 26 | + | − | ± | ++++ | ++ | ++ | 3 | 3a | 3a |
| 27 | − | ± | − | − | ± | ± | NR | ND | ND |
| 28 | + | ++++ | − | ++ | ± | − | 1 | 1b | 1b |
| 29 | − | ++ | − | ± | − | − | 1 | ND | ND |
| 30 | + | − | ± | − | ± | ++++ | 2 | 2a | 2d |
| 31 | − | +++ | − | − | − | ++ | 1 | ND | ND |
| 32 | − | − | − | − | − | +++ | 2 | ND | ND |
| 33 | + | − | − | − | ± | ++ | 2 | 2a | 2d |
| 34 | − | − | ++ | ++ | − | − | UNT(2)c | ND | ND |
| 35 | + | − | − | ± | ++ | ± | 1–3c | 3a | 3a |
ND, not done.
NR, nonreactive.
UNT, untypeable.
Non-2a, non-2b type 2.
−, negative; ±, positive/negative; + to ++++, positive.
Serotyping analysis of patient sera.
The serotype of 15 of 23 (65.2%) patient serum samples could be determined by the RIBA HCV Serotyping SIA (Table 2). Four 5′ UTR-, C/E1-, RT-PCR-, and ELISA 3.0-positive samples, which were subtyped by HTA, were nonreactive. Seventy-five percent (6 of 8) of PCR-negative samples were assigned a serotype (Table 2), while 1 of 8 was nonreactive and 1 of 8 was untypeable. Two subtype 3a samples (HTA 20 and 35) were untypeable (designated either type 1 or 3). One sample (HTA 18) was a subtype 3a by HTA, RFLP, and DNA sequencing but was designated type 2 by serotyping. The discrepant sample may be attributed to a past infection or to a coinfection at a time when viremia was undetectable by PCR. Eight of 15 RT-PCR (53.3%) samples were concordant among all three assays (HTA, RFLP, and serotyping) with respect to genotype.
Comparison of genotype determined by HTA to those determined by Inno-LiPA HCV II and/or RFLP in blood donors from different geographical locations.
HCV genotypes determined by HTA were in agreement with those determined by RFLP and the Inno-LiPA HCV II test for all 12 samples (Table 3), irrespective of the country from which the serum or probe was obtained. For example, the 1b probe, which came from a Japanese serum sample, hybridized to 1b sera from the United States, Holland, and Italy (Fig. 3B, lanes e and k and Fig. 1B, lanes 1, 2, 4, 24, and 28), and the type 3a probe (of U.S. origin) hybridized to 3a samples from the United States, Egypt, and Italy (Fig. 3E, lanes a, b, and g and Fig. 1E, lanes 3, 7, 18, 20, 22, 26, and 35). The data shown in Fig. 3 indicated that the 1a and 1b probes failed to cross-hybridize to each other (panels A and B, lanes 1a and 1b), and therefore sample k, which hybridized to both probes, must have had a 1a-1b coinfection. HTA also showed that samples c (panel A), e (panel B), g and h (panel C), and d (panel D) had at least one viral variant. The cloned 2b probe in Fig. 3D was derived from blood donor serum sample f, and therefore a homoduplex can be seen at the bottom of the gel.
TABLE 3.
Comparison of Inno LiPA HCV II (IL-II), RFLP, and HTA genotype analyses of blood donor samples
| Sample | Country of origin | Genotype determined by:
|
||
|---|---|---|---|---|
| IL/II | RFLPa | HTA | ||
| a | Egypt | NTb | 3 | 3a |
| b | United States | 3a | 3 | 3a |
| c | United States | 1a | 1 | 1a |
| d | United States | 2b | 2 | 2b |
| e | United States | 1b | 1 | 1b |
| f | United States | 2b | 2 | 2b |
| g | United States | 3a | 3 | 3a |
| h | United States | 2a | 2 | 2a |
| i | United States | 2a | 2 | 2a |
| j | United States | 1a | 1 | 1a |
| k | Holland | NT | 1 | 1a, 1b |
| l | Holland | NT | 1 | 1a |
Subtype not determined.
NT, not tested.
DISCUSSION
Based on the work of Delwart et al., who previously established the validity of HTA for determining genotype and molecular evolution studies on human immunodeficiency virus type 1 (12–14), Gretch et al. applied HTA to the study of HCV quasispecies (42). Unfortunately, the primers used in quasispecies analysis were not appropriate for genotype determination due to a high degree of nucleotide variation in the amino terminus of E2 (20, 40). In order to develop an HTA genotyping method for HCV which would also be suitable for other purposes, RT-PCR primers were chosen from the carboxy terminus of core and the middle two-thirds of E1. A set of universal PCR primers were selected from highly conserved regions of C/E1, along with a set of primers which were derived from regions of substantial sequence heterogeneity to ensure subtype specificity.
DNA sequencing of the partial C/E1 PCR products from a cohort of Italian dialysis patients confirmed the accuracy of HTA genotype assignment in all 15 individuals but failed to identify type 2 sequences in a 1b-2 coinfected patient (HTA 4). Since direct DNA sequencing or sequencing a relatively small number of cloned PCR products, i.e., 3 to 30, identifies only predominant viral species, sensitive hybridization-based techniques are more capable of detecting low-frequency variants in a population. RFLP and Inno-LiPA HCV II verified C/E1 HTA genotyping of 12 type 1, 2, and 3 blood donors from different geographical locations, indicating that the source of the isolates or probes did not influence the accuracy of HTA. In addition, HTA detected a 1a-1b coinfection in sample k. The inability of certain assays to detect coinfections could be attributed to either the relative abundance and/or efficiency of amplifying different genomes in the sample. We found that single-stranded probes derived from sera containing a virus population of less than 1.4% nucleotide divergence were effective for genotyping; however, the use of single-stranded probes from cloned RT-PCR products guarantees that the pattern of heteroduplex molecules on MDE gels reflects the actual population of variants and avoids potential artifacts which may be more difficult to discern when double-stranded HTA probes (12–14, 42) or single-strand conformation polymorphism (17) is used.
The molecular definitions of genotype, subtype, and isolate were originally based on the nucleotide region of NS5 for types 1 to 3 HCV (33) (the most frequent genotypes found in North America, Europe, and parts of Asia) and was later found to be consistent for other segments of the HCV genome, including the E1 gene (4, 34, 37). For full-length E1, subtypes and genotypes were separated by approximately 24 to 31% and 33 to 46% nucleotide heterogeneity, respectively, while isolates within the same subtype were found to be ≤15% divergent (4). Recently, sequences of new isolates in Southeast Asia showed that these isolates were more broadly distributed with respect to percent nucleotide divergence than types 1 to 3 HCV and that the boundaries between type, subtype, and isolate might need to be reevaluated in these populations (1, 27, 35). The interpretation of HTA data for Southeast Asian isolates would be expected to be complicated by the same factors that have affected the genotype assignment of these isolates by direct nucleotide sequencing. Our partial C/E1 nucleotide sequencing data for 1b and 3a HCV showed that isolates within the same subtype were ≤15% divergent and had approximately 35 to 42% nucleotide heterogeneity between isolates of different genotypes, which is in good agreement with the results of Bukh et al. (4).
Consistent with previous studies showing that the relative migrations of the heteroduplexes are proportional to genetic distance (12), we found that the mean and range of the relative migration of 1b and 3a isolates were nonoverlapping and reflected the percentage of nucleotide divergence from their respective probes. Although the relative migration of individual isolates was not necessarily directly proportional to the percentage of nucleotide divergence from the probe, the slight differences in the observed relative migration of individual isolates may be due to the distribution of nucleotide substitutions in PCR fragments. In agreement with Delwart et al. (12), a minimum of 1.4% nucleotide divergence was sufficient to separate viral isolates in the HTA described in this study.
The substantially retarded migration of HTA 23, 30, and 33 in Fig. 1C and D suggested that they were non-2a, non-2b type 2 subtypes. These samples would have either been scored negative (6) or misassigned as either 2a or 2a-2b coinfections if standard PCR-based genotyping methods had been used, depending on the stringency of the assay. DNA sequence comparisons showed that HTA 23, 30, and 33 were 14.7 to 18.6% divergent from S83 (a 2c subtype known to be prevalent in Italy [6]). These results indicate that HTA 23, 30, and 33 (i) approached or exceeded the expected ∼15% divergence between isolates within the same subtype and (ii) were more closely related to each other than to other known type 2 isolates which have been sequenced in the C/E1 region. Interestingly, based on a calculation of the percentage of nucleotide divergence alone, HTA 23, 30, and 33 would be considered borderline type 2 compared to 2b isolates (33 to 34%), while they would clearly be classified as type 2 compared to type 2a or 2c (22.8 to 27.9% versus to 16.5 to 18.6%) HCV. Since percentage of nucleotide divergence is closely related to genetic distance as determined by phylogenetic analysis for types 1, 2, and 3 HCV (33, 34), we demonstrate that, as for HIV-1, HTA is useful for identifying evolutionary variants which can be sequenced and subjected to more sophisticated phylogenetic analyses. These findings also illustrate that viral evolution is a dynamic process and highlight the ambiguities inherent in the classification of HCV strictly on the basis of molecular sequence criteria.
A comparison of several genotyping methods demonstrated that the partial C/E1 HTA was the most comprehensive for the following reasons: (i) not only was subtype for patients and blood donors accurately assigned, but coinfections, which were missed by DNA sequencing and RFLP, were also identified; and (ii) in contrast to HTA, RFLP and direct sequencing of the 5′ UTR incorrectly identified all three type 2 samples (HTA 23, 30, and 33) as either type 2a or as 2a-2b due to inadequate nucleotide heterogeneity in the 5′ UTR. HTA has a further advantage in that this method visualizes intrasubtype variants, which tests such as RFLP and Inno-LiPa HCV II are not designed to detect. Such variants could be important in understanding and characterizing therapeutic drug escape mutants.
We also conclude that since 65.2% (15 of 23) of total and 75% (6 of 8) of PCR-negative dialysis patients were typeable by the RIBA HCV serotyping SIA, serotyping appears to have independent value in typing HCV and complements nucleic acid-based genotyping assays.
ACKNOWLEDGMENTS
We thank David Gretch, Jeff Wilson, Steve Polyak, and Kevin Crawford for technical assistance and helpful discussions. DNA sequencing was expertly performed by Chun Ting Lee. We also thank George Lamson for computer-generated dendrograms and Peter Anderson for preparation of the manuscript.
This work was supported by Chiron Vaccines.
REFERENCES
- 1.Adams N J, Chamberlain R W, Taylor L A, Davidson F, Lin C K, Elliott R M, Simmonds P. Complete coding sequence of hepatitis C virus genotype 6a. Biochem Biophys Biophys Res Commun. 1997;234:393–396. doi: 10.1006/bbrc.1997.6627. [DOI] [PubMed] [Google Scholar]
- 2.Alter H J. To C or not to C, these are the questions. Blood. 1995;85:1681–1695. [PubMed] [Google Scholar]
- 3.Alter M J. Epidemiology of hepatitis C in the West. Semin Liver Dis. 1995;15:5–14. doi: 10.1055/s-2007-1007259. [DOI] [PubMed] [Google Scholar]
- 4.Bukh J, Miller R H, Purcell R H. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin Liv Dis. 1995;15:41–63. doi: 10.1055/s-2007-1007262. [DOI] [PubMed] [Google Scholar]
- 5.Calabrese G, Vagelli G, Guaschino R, Gomella M. Transmission of anti-HCV within the household of haemodialysis patients. Lancet. 1991;337:1466. doi: 10.1016/0140-6736(91)92774-v. [DOI] [PubMed] [Google Scholar]
- 6.Cammarota G, Maggi F, Vatteroni M L, Da Prato L, Barsanti L, Bendinelli M, Pistello M. Partial nucleotide sequencing of six subtype 2c hepatitis C viruses detected in Italy. J Clin Microbiol. 1995;33:2781–2784. doi: 10.1128/jcm.33.10.2781-2784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan S-W, McOmish F, Holmes E C, Dow B, Peutherer J F, Follett E, Yap P L, Simmonds P. Analysis of a new hepatitis C virus type and its phylogenetic relationship to existing variants. J Gen Virol. 1992;73:1131–1141. doi: 10.1099/0022-1317-73-5-1131. [DOI] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1986;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Choo Q-L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 10.Choo Q-L, Richman K H, Han J H, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby A, Barr P J, Weiner A J, Bradley D W, Kuo G, Houghton M. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidson E, Simmonds P, Ferguson J C, Jarvis L M, Dow B C, Follett E A C, Seed C R, Krusius T, Lin C, Medgyesi G A, Kiyokawa H, Olim G, Duraisamy G, Cuypers T, Saeed A A, Teo D, Conradie J, Kew M C, Lin M, Nuchaprayoon C, Ndimbie O K, Yap P L. Survey of major genotypes and subtypes of hepatitis C virus using RFLP of sequences amplified from the 5′ non-coding region. J Gen Virol. 1995;76:1197–1204. doi: 10.1099/0022-1317-76-5-1197. [DOI] [PubMed] [Google Scholar]
- 11a.Delwart, E. Personal communication.
- 12.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rübsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay, Analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 13.Delwart E L, Sheppard H W, Walker B D, Goudsmit J, Mullins J I. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J Virol. 1994;68:6672–6683. doi: 10.1128/jvi.68.10.6672-6683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delwart E L, Busch M P, Kalish M L, Mosley J W, Mullins J I. Rapid molecular epidemiology of HIV-1 transmission. AIDS Res. 1995;11:1181–1193. doi: 10.1089/aid.1995.11.1081. [DOI] [PubMed] [Google Scholar]
- 15.Dixit V, Quan S, Martin P, Larson D, Brezina M, DiNello R, Sra K, Lau J Y N, Chien D, Kolberg J, Tagger A, Davis G, Polito A, Gitnick G. Evaluation of a novel serotyping system for hepatitis C virus: strong correlation with standard genotyping methodologies. J Clin Microbiol. 1995;33:2978–2983. doi: 10.1128/jcm.33.11.2978-2983.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enomoto N, Takada A, Nakao T, Date T. There are two major types of hepatitis C virus in Japan. Biochem Biophys Res Comm. 1990;170:1021–1025. doi: 10.1016/0006-291x(90)90494-8. [DOI] [PubMed] [Google Scholar]
- 17.Enomoto N, Kurosaki M, Tanaka Y, Marumo F, Sato C. Fluctuation of hepatitis C virus quasispecies in persistent infection and interferon treatment revealed by single-strand conformation polymorphism analysis. J Gen Virol. 1994;75:1361–1369. doi: 10.1099/0022-1317-75-6-1361. [DOI] [PubMed] [Google Scholar]
- 18.Guaschino R, Calabrese G, Bonelli F, Calvo L, Lazzaro A, Vagelli, G. G, Griva S. Should HCV RNA detection affect the isolation policy in dialysis units? Nephrol Dial Transplant. 1994;7:1070. [PubMed] [Google Scholar]
- 19.Han J H, Shyamala V, Richman K H, Brauer M J, Irvine B, Urdea M S, Tekamp-Olson P, Kuo G, Choo Q-L, Houghton M. Characterization of the terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5′ untranslated region and poly(A) tails at the 3′ end. Proc Natl Acad Sci USA. 1991;88:1711–1715. doi: 10.1073/pnas.88.5.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Ohkoshi S, Shimotohno K. Hypervariable regions in the putative glycoprotein of hepatitis C virus. Biochem Biophys Res Commun. 1991;175:220–228. doi: 10.1016/s0006-291x(05)81223-9. [DOI] [PubMed] [Google Scholar]
- 21.Houghton M. Hepatitis C viruses. In: Fields B N, Knipe D M, Howley P M, et al., editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1035–1057. [Google Scholar]
- 22.Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci USA. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolykhalov A A, Feinstone S M, Rice C M. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kostrikis L G, Bagdades E, Cao Y, Zhang L, Dimitriou D, Ho D D. Genetic analysis of human immunodeficiency virus type 1 strains from patients in Cyprus: identification of a new subtype designated subtype I. J Virol. 1995;69:6122–6130. doi: 10.1128/jvi.69.10.6122-6130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lau J Y N, Mizokami M, Kolberg J A, Davis G L, Prescott L E, Ohno T, Perrillo R P, Lindsay K L, Gish R G, Qian K P, Kohara M, Simmonds, P. P, Urdea M S. Application of six hepatitis C virus genotyping systems to sera from chronic hepatitis C patients in the United States. J Infect Dis. 1995;181:281–289. doi: 10.1093/infdis/171.2.281. [DOI] [PubMed] [Google Scholar]
- 26.Martell M, Esteban J I, Quer J, Genescà J, Weiner A, Esteban R, Guardia J, Gómez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellor J, Walsh E A, Prescott L E, Jarvis L M, Davidson F, Yap P L, Simmonds P the International HCV Collaborative Study Group. Survey of type 6 group variants of hepatitis C virus in Southeast Asia by using a core-based genotyping assay. J Clin Microbiol. 1996;34:417–423. doi: 10.1128/jcm.34.2.417-423.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okamoto H, Kojima M, Okada S, Yoshizawa H, Iizuka H, Tanaka T, Muchmore E E, Peterson D A, Ity Y, Mishiro S. Genetic drift of hepatitis C virus during an 8.2-year infection in a chimpanzee: variability and stability. Virology. 1992;190:894–899. doi: 10.1016/0042-6822(92)90933-g. [DOI] [PubMed] [Google Scholar]
- 29.Okamoto H, Kurai K, Okada S I, Yamamoto K, Lizuka H, Tanaka T, Fukada S, Tsuda F, Mishiro S. Full-length sequence of a hepatitis C virus genome having poor homology to reported isolates: comparative study of four distinct genotypes. Virology. 1992;188:331–341. doi: 10.1016/0042-6822(92)90762-e. [DOI] [PubMed] [Google Scholar]
- 30.Okamoto T, Sugiyama Y, Okada S. Typing hepatitis C virus by polymerase chain reaction with type-specific primers: application to clinical surveys and tracing infectious sources. J Gen Virol. 1992;73:673–679. doi: 10.1099/0022-1317-73-3-673. [DOI] [PubMed] [Google Scholar]
- 31.Sakamoto M, Akahane Y, Tsuda F, Tanaka T, Woodfield D G, Okamoto H. Entire nucleotide sequence and characterization of a hepatitis C virus of genotype V/3a. J Gen Virol. 1994;75:1761–1768. doi: 10.1099/0022-1317-75-7-1761. [DOI] [PubMed] [Google Scholar]
- 32.Shimizu Y K, Iwamoto A, Hijikata M, Purcell R H, Yoshikura H. Evidence for in vitro replication of hepatitis C virus genome in a human T cell line. Proc Natl Acad Sci USA. 1992;89:5477–5481. doi: 10.1073/pnas.89.12.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simmonds P, Holmes E C, Cha T-A. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol. 1993;74:2391–2399. doi: 10.1099/0022-1317-74-11-2391. [DOI] [PubMed] [Google Scholar]
- 34.Simmonds P, Smith D B, McOmish F, Yap P L, Kolberg J, Urdea M S, Holmes E C. Identification of genotypes of hepatitis C virus by sequence comparisons in the core, E1 and NS-5 regions. J Gen Virol. 1994;75:1053–1061. doi: 10.1099/0022-1317-75-5-1053. [DOI] [PubMed] [Google Scholar]
- 35.Simmonds P, Mellor J, Sakuldamrongpanich T, Nuchaprayoon C, Tanprasert S, Holmes E C, Smith D B. Evolutionary analysis of variants of hepatitis C virus found in Southeast Asia: comparison with classifications based upon sequence similarity. J Gen Virol. 1996;77:3013–3024. doi: 10.1099/0022-1317-77-12-3013. [DOI] [PubMed] [Google Scholar]
- 36.Stuyver L, Rossau R, Wyseur A, Duhamel M, Vanderborght B, Van Heuverswyn H, Maertens G. Typing of hepatitis C virus isolates and characterization of new subtypes using a line probe assay. J Gen Virol. 1993;74:1093–1102. doi: 10.1099/0022-1317-74-6-1093. [DOI] [PubMed] [Google Scholar]
- 37.Stuyver L, Van Arnhem W, Wyseur A, Hernandez F, Delaporte E, Maertens G. Classification of hepatitis C viruses based on phylogenetic analysis of the envelope 1 and nonstructural 5B regions and identification of five additional subtypes. Proc Natl Acad Sci USA. 1994;91:10134–10138. doi: 10.1073/pnas.91.21.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka T, Kato N, Cho M-J, Sugiyama K, Shimotohno K. Structure of the 3′ terminus of the hepatitis C virus genome. J Virol. 1996;70:3307–3312. doi: 10.1128/jvi.70.5.3307-3312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vagelli G, Calabrese G, Guaschino R, Gonella M. Effect of HCV positive patients isolation on HCV infection incidence in a dialysis unit. Nephrol Dial Transplant. 1992;7:1070. [PubMed] [Google Scholar]
- 40.Weiner A J, Brauer M J, Rosenblatt J, Richman K H, Tung J, Crawford K, Bonino F, Saracco G, Choo Q-L, Houghton M, Han J H. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology. 1991;180:842–848. doi: 10.1016/0042-6822(91)90104-j. [DOI] [PubMed] [Google Scholar]
- 41.Weiner A J, Thaler M M, Crawford K, Ching K, Kansopon J, Chien D Y, Hall J E, Hu F, Houghton M. A unique, predominant hepatitis C virus variant found in an infant born to a mother with multiple variants. J Virol. 1993;67:4365–4368. doi: 10.1128/jvi.67.7.4365-4368.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson J J, Polyak S J, Day T D, Gretch D R. Characterization of simple and complex hepatitis C virus quasispecies by heteroduplex gel shift analysis: correlation with nucleotide sequencing. J Gen Virol. 1995;76:1763–1771. doi: 10.1099/0022-1317-76-7-1763. [DOI] [PubMed] [Google Scholar]
- 43.Yamada N, Tanihara K, Takada A, Yorihuzi T, Tsutsumi M, Shimomura H, Tsuji T, Date T. Genetic organization and diversity of the 3′ noncoding region of the hepatitis C virus genome. Virology. 1996;223:255–261. doi: 10.1006/viro.1996.0476. [DOI] [PubMed] [Google Scholar]