Abstract
We evaluated a novel three-dimensional microarray (PamChip microarray) system to detect the presence of levofloxacin-related resistance mutations and the mecA gene. The results were compared to those obtained for 27 Staphylococcus aureus isolates by conventional DNA sequencing or PCR methods. Hybridization and fluorescence detection were performed using an FD10 system designed for PamChip microarray under conditions optimized for each target/probe on the array. In dilution series analysis using multiplex PCR samples, the sensitivity of the microarray was about 10 times greater than that of conventional PCR methods. A high level of data reproducibility was also confirmed in those analyses. Various point mutations in quinolone resistance-determining regions detected by our system corresponded perfectly to the results obtained by conventional DNA sequencing. The results of the mecA gene detection using our system also corresponded to the PCR method; that is, signal/band was detected in all isolates of methicillin-resistant S. aureus, and no signal/band was detected in any isolate of methicillin-susceptible S. aureus. In conclusion, our novel three-dimensional microarray system provided rapid, specific, easy, and reproducible results for the simultaneous detection of levofloxacin resistance and the mecA gene in S. aureus.
Effective antibiotic therapy for infections caused by Staphylococcus aureus can be difficult because of resistance to various antibiotics, including β-lactams, aminoglycosides, and fluoroquinolones (1, 11, 12, 15). β-Lactam and fluoroquinolone resistance is generally detected by established conventional susceptibility testing. The conventional susceptibility testing method is not complex or time consuming in terms of hands-on time, although it takes 2 or 3 days for the result. Methicillin resistance is derived from acquisition of the mecA gene encoding penicillin-binding protein 2′ by methicillin-resistant S. aureus (MRSA) (18, 19, 25, 29). The presence of the mecA gene is easily detected by the conventional PCR method (18, 19, 25). To remove PCR sizing artifacts, however, a more specific method, such as either hybridization or restriction digestion following PCR, is required. Fluoroquinolone resistance is conferred mainly by codon mutations in the quinolone resistance-determining regions (QRDRs) of the gyrA and gyrB (gyrAB) genes encoding subunits of DNA gyrase and of the grlA and grlB (grlAB) genes encoding subunits of DNA topoisomerase IV (6, 7, 11, 13). Point mutations in the QRDRs of the gyrA, gyrB, grlA, and grlB genes have been linked to the degree of fluoroquinolone resistance in MRSA (11, 22, 23). These point mutations are usually determined by conventional DNA sequencing as a gold standard method. In clinical laboratories, however, the ability to genetically determine fluoroquinolone resistance is limited by the complex and time-consuming nature of conventional DNA sequencing.
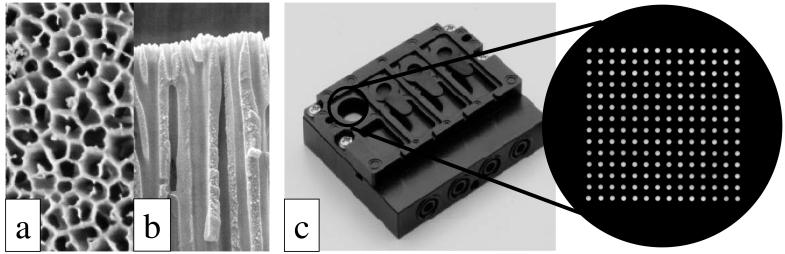
Recently, some new genotyping methods, such as the microarray technique, degenerate high-performance liquid chromatography, real-time PCR, TaqMan assay, and Invader assay, have been developed (2, 5, 8, 14, 32). The DNA microarray technique could have the ability to simultaneously analyze multiple-point mutations or deleted and/or inserted regions. In particular, the newly developed PamChip microarray (PamGene International, Hertogenbosch, The Netherlands) is a unique three-dimensional (3-D) flowthrough platform for kinetic hybridization reactions (30). Unlike general microarrays that employ a two-dimensional (2-D) substrate such as a glass slide, the PamChip microarray has long branching capillaries that bind probe DNA molecules onto a solid 3-D structure substrate (Fig. 1). The reactive surface of this substrate is several hundredfold larger than that of a 2-D substrate. A hybridization and image capture station, FD10 (Olympus Corporation, Tokyo, Japan), ensures an optimal solution-driven reaction on the PamChip microarray by repeated pumping of the sample (9, 17). In combination, the PamChip microarray and the FD10 system carry out semiautomatic, rapid, and specific hybridization reactions (17). Furthermore, the system is designed for the simultaneous analysis of four arrays.
FIG. 1.
Configuration of PamChip microarray. Representative magnified views of the upper phase (a) and a cross section of the flowthrough 3-D-structure substrate (b) and a magnified view of the array and an outward view of the PamChip microarray (c) are shown.
Kinetic hybridization is required for the simultaneous detection of multiple-point mutations and the presence of DNA sequences within a single microarray, because each target/probe has a different melting temperature (Tm). One set of conditions for an array containing multiple targets/probes may not be optimal for each target/probe on the array. One main advantage of the PamChip microarray system compared to other microarray formats, including 2-D arrays, is the flexibility for the optimization of hybridization conditions for each target/probe on the array using changes in specific reagents or in temperature.
In this study, to confirm the utility of the PamChip microarray system in the detection of levofloxacin resistance and the mecA gene, we compared the results obtained by the microarray method with those obtained by conventional DNA sequencing and PCR methods. On the microarray, the multiple-point mutations in the QRDRs were detected with sequencing by hybridization (SBH), and the presence of the mecA gene was detected with reverse dot blot hybridization (RDBH), in S. aureus isolates. The applicability of the system for future diagnosis is also discussed.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
Twenty-four levofloxacin-resistant MRSA isolates (MICs of >4 μg/ml) and three methicillin-susceptible S. aureus (MSSA) isolates were used in this study (see Table 3). The MICs for all MRSA isolates were determined by an agar dilution method described by CLSI (formerly NCCLS), which was used in our previous report (11). The clinical isolates were obtained from different patients in Hamamatsu city hospitals between 2001 and 2004. Bacteria were stored at −70°C in heart infusion broth (Nissui Pharmaceutical, Tokyo, Japan) containing 20% glycerol. Subsequently, bacteria were inoculated on heart infusion agar plates (Nissui Pharmaceutical) and incubated at 37°C overnight.
TABLE 3.
Results of the genotyping analysis for S. aureus by PamChip microarray
| Strain | Mutation ina:
|
mecA resultb | |||||
|---|---|---|---|---|---|---|---|
|
grlA codon(s) (product[s]):
|
grlB codon 451 (P) (product) |
gyrA codon(s) (product[s]):
|
|||||
| 47 (Y) | 79 (D) 80 (S) | 84 (E) | 84 (S) 85 (S) | 88 (E) | |||
| MRSA | |||||||
| HU2000- | |||||||
| 62S | TAT(Si) | GAC(Wt)TTC(F) | AAA(K) | CCA(Wt) | TTA(L)CCT(P) | GAA(Wt) | P |
| 63S | TAT(Si) | GAC(Wt)TAC(Y) | AAA(K) | CCA(Wt) | TTA(L)TCT(Wt) | GAA(Wt) | P |
| 64 | TAT(Si) | GAC(Wt)TAC(Y) | GAA(Wt) | CCA(Wt) | TTA(L)TCT(Wt) | GAA(Wt) | P |
| 86S | TAT(Si) | GAC(Wt)TTC(F) | GAA(Wt) | CCA(Wt) | TCA(Wt)TCT(Wt) | AAC(K) | P |
| 91S | TAT(Si) | GAC(Wt)TTC(F) | GAA(Wt) | CCA(Wt) | TCA(Wt)TCT(Wt) | AAC(K) | P |
| 92 | TAT(Si) | GAC(Wt)TTC(F) | GAA(Wt) | CCA(Wt) | TCA(Wt)TCT(Wt) | AAC(K) | P |
| 120 | TAT(Si) | GAC(Wt)TAC(Y) | GAA(Wt) | CCA(Wt) | TTA(L)TCT(Wt) | GAA(Wt) | P |
| 129 | TAT(Si) | GAC(Wt)TTC(F) | GAA(Wt) | TCA(S) | TTA(L)TCT(Wt) | GAA(Wt) | P |
| 133 | TAT(Si) | GAC(Wt)TAC(Y) | GAA(Wt) | CCA(Wt) | TTA(L)TCT(Wt) | GAA(Wt) | P |
| 134 | TAT(Si) | GAC(Wt)TAC(Y) | GAA(Wt) | CCA(Wt) | TTA(L)TCT(Wt) | GAA(Wt) | P |
| 135 | TAT(Si) | GAC(Wt)TTC(F) | GAA(Wt) | CCA(Wt) | TCA(Wt)TCT(Wt) | AAC(K) | P |
| 136 | TAT(Si) | GAC(Wt)TAC(Y) | GAA(Wt) | CCA(Wt) | TTA(L)TCT(Wt) | GAA(Wt) | P |
| 156 | TAT(Si) | GAC(Wt)TTC(F) | GAA(Wt) | TCA(S) | TTA(L)TCT(Wt) | GAA(Wt) | P |
| 177 | TAT(Si) | GAC(Wt)TTC(F) | GAA(Wt) | CCA(Wt) | TCA(Wt)TCT(Wt) | AAC(K) | P |
| 178 | TAT(Si) | GAC(Wt)TTC(F) | GAA(Wt) | TCA(S) | TTA(L)TCT(Wt) | GAA(Wt) | P |
| 199 | TAT(Si) | GAC(Wt)TTC(F) | GAA(Wt) | CCA(Wt) | TCA(Wt)TCT(Wt) | AAC(K) | P |
| 214 | TAT(Si) | GAC(Wt)TTC(F) | GAA(Wt) | CCA(Wt) | TCA(Wt)TCT(Wt) | AAC(K) | P |
| 228 | TAT(Si) | GAC(Wt)TTC(F) | GAA(Wt) | TCA(S) | TTA(L)TCT(Wt) | GAA(Wt) | P |
| 240 | TAT(Si) | GAC(Wt)TAC(Y) | AAA(K) | CCA(Wt) | TTA(L)TCT(Wt) | GAA(Wt) | P |
| SHH- | |||||||
| 41 | TAT(Si) | GAC(Wt)TTC(F) | GAA(Wt) | TCA(S) | TTA(L)TCT(Wt) | GAA(Wt) | P |
| 43 | TAT(Si) | GAC(Wt)TAC(Y) | GAA(Wt) | CCA(Wt) | TTA(L)TCT(Wt) | GAA(Wt) | P |
| 44 | TAT(Si) | GAC(Wt)TTC(F) | GAA(Wt) | TCA(S) | TTA(L)TCT(Wt) | GAA(Wt) | P |
| 45 | TAT(Si) | GAC(Wt)TTC(F) | GAA(Wt) | TCA(S) | TTA(L)TCT(Wt) | GAA(Wt) | P |
| 46 | TAT(Si) | GAC(Wt)TAC(Y) | GAA(Wt) | CCA(Wt) | TTA(L)TCT(Wt) | GAA(Wt) | P |
| MSSA | |||||||
| RN4220 | TAT(Si) | GAC(Wt)TCC(Wt) | GAA(Wt) | CCA(Wt) | TCA(Wt)TCT(Wt) | GAA(Wt) | N |
| Taku | TAT(Si) | GAC(Wt)TCC(Wt) | GAA(Wt) | CCA(Wt) | TCA(Wt)TCT(Wt) | GAA(Wt) | N |
| NCTC8325 | TAT(Si) | GAC(Wt)TCC(Wt) | GAA(Wt) | CCA(Wt) | TCA(Wt)TCT(Wt) | GAA(Wt) | N |
D, Asp; E, Glu; F, Phe; K, Lys; L, Leu; P, Pro; S, Ser; Y, Tyr.
P, positive; N, negative.
DNA techniques.
Genomic DNA of S. aureus was extracted according to a method previously reported (10, 21). Extracted DNA was applied to a 0.2-ml PCR tube to perform PCR. Primers used for the amplification of the QRDRs of the gyrA, gyrB, grlA, and grlB genes were prepared as previously reported (11). PCR was performed with Ex Taq DNA polymerase (Takara Biomedicals, Ohtsu, Japan). To determine point mutations in the QRDRs of the gyrAB and grlAB genes by conventional DNA sequencing, amplified products of all isolates were directly sequenced with a BigDye Terminator cycle-sequencing FS ready reaction kit and an ABI PRISM 310 genetic analyzer (Applied Biosystems, Foster City, CA).
Conventional PCR for mecA gene detection was performed under conditions the same as those for the multiplex PCR mentioned below, excluding primers for gyrAB and grlAB gene amplifications. Amplified fragments were checked by electrophoresis using a LabChip 7500 assay kit and an Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA).
Multiplex PCR.
To detect point mutations in the QRDRs of the gyrAB and grlAB genes and the presence of the mecA gene on the microarray, targets were amplified simultaneously by multiplex PCR. Sets of multiplex PCR primers are shown in Table 1. For amplifications of gyrAB and grlAB genes, multiplex primer sets were designed from DNA sequences in public databases by using the primer analysis software OLIGO version 6 (Molecular Biology Insights, Cascade, CO). For all PCR primers, Tm values were adjusted within the range of 54.7 to 60.5°C, and the GC content ranged from 30% to 55%. Multiplex PCR was performed in a solution containing 0.5 μM of each 5′-fluorescein isothiocyanate-labeled primer (Sigma-Aldrich Japan, Tokyo, Japan), 200 μM deoxynucleoside triphosphate, and 2.5 U of Ex Taq DNA polymerase for 40 cycles of 94°C for 1 min, 50°C for 2 min, and 72°C for 1 min. To establish the validity of the multiplex PCR method, amplified fragments were checked using a LabChip 7500 kit and an Agilent 2100 bioanalyzer. After the multiplex PCR condition was established, PCR products were hybridized directly on the microarray.
TABLE 1.
Multiplex PCR primers
| Target gene | Primer | Sequence (5′→3′) | Length (base) | Tm (°C) | GC % | Size (bp) |
|---|---|---|---|---|---|---|
| grlAB | grl-1 | AACGAAATCTAAATTGGGTA | 20 | 55.3 | 30 | 1,457 |
| grl-2 | GGAAATCTTGATGGCAATAC | 20 | 58.4 | 40 | ||
| gyrAB | gyr-1 | TGAAGTAACACGTCGTAAAT | 20 | 54.7 | 35 | 1,255 |
| gyr-2 | AGGTAAGACTGACGGCTCTC | 20 | 60.0 | 55 | ||
| mecAa | GMECAR-1 | ACTGCTATCCACCCTCAAAC | 20 | 60.5 | 50 | 163 |
| GMECAR-2 | CTGGTGAAGTTGTAATCTGG | 20 | 57.1 | 45 |
Primers for amplification of the mecA gene were previously described by Mehrotra et al. (18).
Probe design.
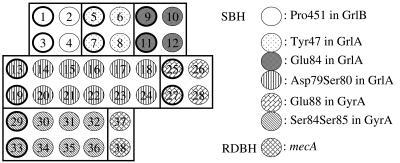
Eighteen common genotypes in the QRDRs for our microarray analysis were selected based on our previous report and those of others (4, 11, 14, 26, 27, 28). Sense and antisense oligonucleotide probes from 20 to 26 bases were used to determine specific point mutations in the QRDRs of the gyrAB and grlAB genes and to detect mecA-specific DNA sequences (Table 2). Sets of perfect-match probes for the wild type (Wt) and for specific mutants were prepared to determine the genotype with respect to each point mutation. The specific point mutations reported previously were at codons 84, 85, and 88 in gyrA; at codons 47, 79, 80, and 84 in grlA; and at codon 451 in grlB. Thirty-six probes were used for SBH analysis, the Tm values for these probes were adjusted within the range of 52 to 56°C, and the GC content was 30% to 50%. Twelve sense and antisense probes were designed to detect a single-point mutation at a single codon, with 24 probes for double-point mutations at a single or tandem codon. These point mutations were presented in the centers of the probe sequences. The point mutations, which involved an 8-base interval between codons 85 and 88 in gyrA and a 10-base interval between codons 80 and 84 in grlA, were detected using specific probes. To detect the mecA sequence, probes for RDBH were derived from 26-base sequences in the mecA gene. These 38 probes were spotted onto the PamChip microarray according to the layout shown in Fig. 2. The microarray was designed to determine eight point mutation sites, including eighteen genotypes in the QRDRs, and to detect the DNA sequence of the mecA gene simultaneously.
TABLE 2.
The 38 probes for PamChip microarray
| Probe no. | Sequence (5′→3′)a | Length | Tm (°C) | GC % | Strandb | Mutationc | Gene |
|---|---|---|---|---|---|---|---|
| 1 | GCGATATTACCATTACGTGG | 20 | 54 | 45 | S | P451(Wt) | grlB |
| 2 | GCGATATTATCATTACGTGG | 20 | 52 | 40 | S | P451S | grlB |
| 3 | CCACGTAATGGTAATATCGC | 20 | 54 | 45 | AS | P451(Wt) | grlB |
| 4 | CCACGTAATGATAATATCGC | 20 | 52 | 40 | AS | P451S | grlB |
| 5 | GTCGTATTTTATACGCAATGTATTC | 25 | 53 | 32 | S | Y47(Wt) | grlA |
| 6 | GTCGTATTTTATATGCAATGTATTC | 25 | 52 | 28 | S | Y47Y(Si) | grlA |
| 7 | GAATACATTGCGTATAAAATACGAC | 25 | 53 | 32 | AS | Y47(Wt) | grlA |
| 8 | GAATACATTGCATATAAAATACGAC | 25 | 52 | 28 | AS | Y47Y(Si) | grlA |
| 9 | TCAGTGTACGAAGCAATGGT | 20 | 54 | 45 | S | E84(Wt) | grlA |
| 10 | TCAGTGTACAAAGCAATGGT | 20 | 52 | 40 | S | E84K | grlA |
| 11 | ACCATTGCTTCGTACACTGA | 20 | 54 | 45 | AS | E84(Wt) | grlA |
| 12 | ACCATTGCTTTGTACACTGA | 20 | 52 | 40 | AS | E84K | grlA |
| 13 | ACATGGAGACTCCTCAGTGT | 20 | 56 | 50 | S | D79(Wt)S80(Wt) | grlA |
| 14 | ACATGGAGACTACTCAGTGT | 20 | 54 | 45 | S | D79(Wt)S80Y | grlA |
| 15 | ACATGGAGACTTCTCAGTGT | 20 | 54 | 45 | S | D79(Wt)S80F | grlA |
| 16 | ACATGGAGTCTCCTCAGTGT | 20 | 56 | 50 | S | D79VS80(Wt) | grlA |
| 17 | ACATGGAGTCTACTCAGTGT | 20 | 54 | 45 | S | D79VS80Y | grlA |
| 18 | ACATGGAGTCTTCTCAGTGT | 20 | 54 | 45 | S | D79VS80F | grlA |
| 19 | ACACTGAGGAGTCTCCATGT | 20 | 56 | 50 | AS | D79(Wt)S80(Wt) | grlA |
| 20 | ACACTGAGTAGTCTCCATGT | 20 | 54 | 45 | AS | D79(Wt)S80Y | grlA |
| 21 | ACACTGAGAAGTCTCCATGT | 20 | 54 | 45 | AS | D79(Wt)S80F | grlA |
| 22 | ACACTGAGGAGACTCCATGT | 20 | 56 | 50 | AS | D79VS80(Wt) | grlA |
| 23 | ACACTGAGTAGACTCCATGT | 20 | 54 | 45 | AS | D79VS80Y | grlA |
| 24 | ACACTGAGAAGACTCCATGT | 20 | 54 | 45 | AS | D79VS80F | grlA |
| 25 | CTATTTATGAAGCAATGGTACG | 22 | 52 | 36 | S | E88K(Wt) | gyrA |
| 26 | CTATTTATAACGCAATGGTACG | 22 | 52 | 36 | S | E88K | gyrA |
| 27 | CGTACCATTGCTTCATAAATAG | 22 | 52 | 36 | AS | E88K(Wt) | gyrA |
| 28 | CGTACCATTGCGTTATAAATAG | 22 | 52 | 36 | AS | E88K | gyrA |
| 29 | CTCATGGTGACTCATCTATTTAT | 23 | 53 | 34 | S | S84(Wt)S85(Wt) | gyrA |
| 30 | CTCATGGTGACTCACCTATTTAT | 23 | 55 | 39 | S | S84(Wt)S85P | gyrA |
| 31 | CTCATGGTGACTTATCTATTTAT | 23 | 51 | 30 | S | S84LS85(Wt) | gyrA |
| 32 | CTCATGGTGACTTACCTATTTAT | 23 | 53 | 34 | S | S84LS85P | gyrA |
| 33 | ATAAATAGATGAGTCACCATGAG | 23 | 53 | 34 | AS | S84(Wt)S85(Wt) | gyrA |
| 34 | ATAAATAGGTGAGTCACCATGAG | 23 | 55 | 39 | AS | S84(Wt)S85P | gyrA |
| 35 | ATAAATAGATAAGTCACCATGAG | 23 | 51 | 30 | AS | S84LS85(Wt) | gyrA |
| 36 | ATAAATAGGTAAGTCACCATGAG | 23 | 53 | 34 | AS | S84LS85P | gyrA |
| 37 | GTATGGCATGAGTAACGAAGAATATA | 26 | 55 | 34 | S | mecA | |
| 38 | TATATTCTTCGTTACTCATGCCATAC | 26 | 55 | 34 | AS | mecA |
DNA sequences of the codons with point mutations are underlined.
S, sense strand; AS, antisense strand.
D, Asp; E, Glu; F, Phe; K, Lys; L, Leu; P, Pro; S, Ser; V, Val; Y, Tyr; Wt, wild type.
FIG. 2.
The probe spot layout of the microarray layout for S. aureus genotyping. Spot numbers correspond to the probe numbers in Table 2. The probes, which identify point mutations, are surrounded with squares. Spots enclosed by thick lines were used to detect the Wt (numbers 1, 3, 5, 7, 9, 11, 13, 19, 25, 27, 29, and 33), while those enclosed by thin lines were used to detect the mutants.
Microarray analysis.
To confirm the sensitivity and reproducibility of the assay using the microarray system, multiplex PCR products amplified from two MRSA isolates (HU2000-62S and -64) were used as hybridization samples in a dilution series. The dilution concentrations were 1/10, 1/50, and 1/100. Those were presumed by using a LabChip 7500 kit and an Agilent 2100 bioanalyzer.
Hybridization and fluorescence detection were performed automatically by the FD10 system. Before hybridization, each array (test site) of the microarray was washed with 0.1% Tween 20 for a single pumping cycle. Multiple amplified PCR products were denatured for 3 min at 94°C and cooled on ice. The denatured PCR product (35 μl) was mixed with 15 μl of 20× standard saline phosphate EDTA (SSPE) at the test site of the microarray, which was preheated to 50°C, and hybridization was initiated immediately. In flowthrough hybridization, the liquid flow rate was 25 μl/s for 30 cycles (totaling approximately 5 min) at 50°C. To optimize the SBH analysis, kinetic analysis of the microarrays was performed with postwashing steps using 50 μl of SSPE buffers at five different concentrations: 6× (960 mM NaCl), 3× (480 mM NaCl), 1× (160 mM NaCl), 0.5× (80 mM NaCl), and 0.05× (8 mM NaCl). All postwashing steps were performed for five pumping cycles. Images of the array were then captured automatically and analyzed using the image analysis software of the FD10 system. To confirm the reproducibility of this microarray system, duplicate experiments were carried out for the samples, each one of which was amplified and hybridized on different days.
RESULTS
Multiplex PCR.
Multiplex PCR was designed to amplify three regions, including the mecA gene and the QRDRs of the gyrAB and grlAB genes (Table 1). Each multiplex PCR product was amplified as a single band of the predicted size for the gyrAB, grlAB, and mecA genes (1,255, 1,457, and 163 bp, respectively). The amounts of DNA fragments amplified by 40 cycles of multiplex PCR were sufficient for microarray analysis.
Microarray analysis.
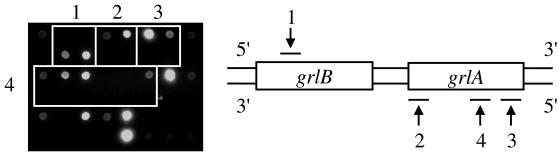
PCR products from the mecA gene were hybridized efficiently with both sense and antisense strand probes in all isolates. On the other hand, those from the gyrAB and grlAB genes were hybridized with either sense or antisense strand probes. In all isolates, hybridization signals for Pro451(Wt) and Pro451Ser in GrlB were observed only in an antisense strand probe, while signals for the following in GrlA were observed only in a sense strand probe: Tyr47(Wt) and Tyr47(Si) (silent mutation); Asp79(Wt)Ser80(Wt), Asp79(Wt)Ser80Tyr, Asp79(Wt)Ser80Phe, Asp79ValSer80(Wt), Asp79ValSer80Tyr, and Asp79ValSer80Phe; and Glu84(Wt) and Glu84Lys in GrlA (Fig. 3).
FIG. 3.
Hybridization image in MRSA HU2000-228. Areas surrounded by squares indicate spots of probe sets for the detection of each specific point mutation on the microarray. Square 1, Pro451 in GrlB was observed in antisense strand probes; square 2, Tyr47 in GrlA in sense strand probes; square 3, Glu84 in GrlA in sense strand probes; square 4, Asp79Ser80 in GrlA in sense strand probes. The illustration to the right indicates the region of each probe in the grlAB gene and the strand for the detection of point mutations in the grlAB gene.
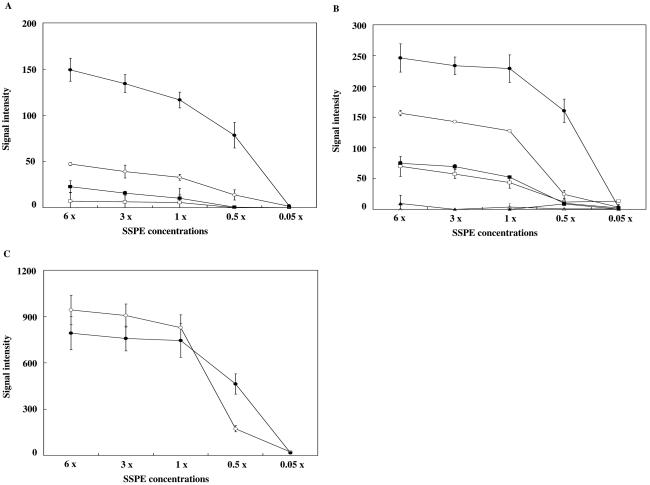
To optimize the assay conditions for each target/probe on the array, the fluorescence signal detection step, i.e., the postwashing step, was performed at five different sodium chloride concentrations. Images of the fluorescence signals were captured kinetically, and quantitative values for these signals were calculated by the FD10 system in MRSA HU2000-62S, -64, -86S, and -129 and in S. aureus RN4220. Kinetic data for MRSA HU2000-62S are shown in Fig. 4. Although nonspecific signals were observed in all antisense probes for Glu84 and Asp79Ser80 in GrlA in the range of 1× to 6× SSPE, specific signals were detected at 0.5× SSPE. All sense or antisense probes for Ser84Ser85 in GyrA, Tyr47 in GrlA, and Pro451 in GrlB were detected at 6× SSPE. No signal was detected at 0.05× SSPE. These kinetic analyses showed the recognition of significant hybridization signals for all probes except those for Glu88(Wt) and Glu88Lys in GyrA with two postwashing conditions: 0.5× and 6× SSPE. In both 0.5× and 6× SSPE, PCR products of the grlAB genes were cross-hybridized with the probe for Glu88(Wt) in GyrA (data not shown). The Tyr47(Si) genotype was preserved in all isolates of MRSA, according to the DNA sequencing analysis. The intensities of the cross-hybridized signals were corrected by considering the signal intensities as ratios relative to the signal intensity of Tyr47(Si). The signal intensity cross-hybridized with the probe for Glu88(Wt) was found to be 0.5 ± 0.1 times that for Tyr47(Si) in MRSA HU2000-62S, -64, -86S, and -129. Cross-hybridization signals were then corrected using the formula Ts = OGlu − Cf · OTyr [Ts, true signal intensity for Glu88(Wt); OGlu, observed signal intensity for Glu88(Wt); Cf, correction factor of 0.5; OTyr, observed signal intensity for Tyr47(Si)]. Alignment of the DNA sequences of the probe for Glu88(Wt) in GyrA and those of the PCR product for the grlAB gene showed that continuous homology with 11 bp was contained in the centers of the probes. Cross-hybridization occurred in all S. aureus strains. The genetic relationship between GyrA and GrlA is very close; that is, the amino acid residues from 71 to 92 in the QRDRs of GyrA are homologous to those from 67 to 96 in GrlA (approximately 80% amino acid homology). Ser84 and Glu88 in GyrA corresponded to Ser80 and Glu84, respectively, in GrlA. The similarities of these DNA sequences could account for the cross-hybridization observed in the SBH analysis. On the other hand, PCR products for the grlAB gene showed no cross-hybridization in any S. aureus strains by use of a probe for Glu88Lys containing 8 bp of continuous homology (data not shown). The signal at Glu88 was corrected using image analysis software to determine the genotype exactly.
FIG. 4.
Kinetic fluorescence signal intensities in MRSA HU2000-62S. (A) Ser84Ser85: Ser84(Wt)Ser85(Wt), TCATCT (□); Ser84(Wt)Ser85Pro, TCACCT (▪); Ser84LeuSer85(Wt), TTATCT (○); Ser84LeuSer85Pro, TTACCT (•). (B) Asp79Ser80: Asp79(Wt)Ser80(Wt), GATTCC (□); Asp79(Wt)Ser80Tyr, GACTAC (○); Asp79(Wt)Ser80Phe, GACTTC (•); Asp79ValSer80(Wt), GTCTCC (▴); Asp79ValSer80Tyr, GTCTAC (▵); Asp79ValSer80Phe, GTCTCC (▪). (C) Glu84: Glu84(Wt), GAA (○); Glu84Lys, AAA (•). Fluorescence signal intensities calculated from 0.05×-to-6× SSPE postwashing steps are shown. Duplicate experiments were carried out for each sample.
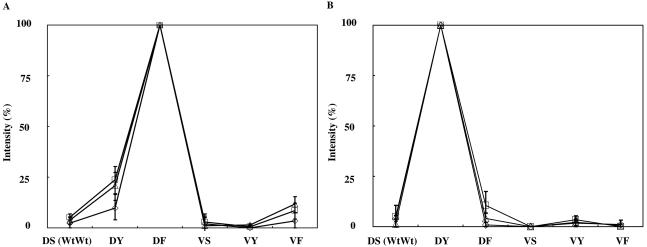
To examine the assay sensitivity and reproducibility around the limit concentration for genotyping, hybridization experiments using three diluted multiplex PCR products (1/10, 1/50, and 1/100) derived from two isolates were performed. Diluted samples were hybridized to the PamChip microarray in triplicate. In total, 18 arrays (test sites) of six microarrays were used. Hybridization signals were clear and detectable on the microarrays in all diluted samples (Fig. 5), whereas in the samples diluted to 1/50 and 1/100, PCR-amplified fragment bands could not be detected with a LabChip 7500 kit. The concentrations of amplified fragments of the gyrAB and grlAB genes in 1/100-diluted samples of MRSA HU2000-62S and -64 were estimated as 11.9 to 31.5 pM and 14.0 to 51.0 pM, respectively. The signal intensity ratios between perfect-match probe and mismatch probes always showed similar profiles (Fig. 5).
FIG. 5.
Sensitivity and reproducibility of the microarray assay around the limit concentration for mutation detection. Representative results obtained with three diluted multiplex PCR products from two isolates are shown. (A) Asp79Ser80 in MRSA HU2000-62S. (B) Asp79Ser80 in MRSA HU2000-64. Abbreviations for GrlA products: Wt, wild type; DS, Asp79(Wt)Ser80(Wt); DY, Asp79(Wt)Ser80Tyr; DF, Asp79(Wt)Ser80Phe; VS, Asp79ValSer80(Wt); VY, Asp79ValSer80Tyr; VF, Asp79ValSer80Phe. Dilution series of PCR products are 1/10 (□), 1/50(▵), and 1/100 (⋄). The percentages of the fluorescence signal intensities of genotypes related to Asp79Ser80 at 0.5× SSPE were calculated by the following formula: observed signal on probe/highest signal in the same target probes × 100.
Evaluation of clinical isolates.
Microarray analysis was performed under both 0.5× SSPE and 6× SSPE postwashing conditions in 24 MRSA isolates and 3 S. aureus isolates in duplicate. In the RDBH analysis for the mecA gene, a signal intensity at 0.5× SSPE above a predetermined threshold (>3,000 with a 0.2-s shutter speed) was considered positive for the presence of the DNA sequence. All MRSA isolates showed positive signals for the mecA gene, and all MSSA isolate signals were negative. These results perfectly corresponded to those obtained by the conventional PCR method (Table 3). In the SBH analysis of the QRDRs, the probe that produced the strongest signal intensity among the probe sets for the Wt and the mutants was judged a perfect-match sequence. Point mutations in the QRDRs determined by the PamChip microarray system corresponded to those determined by the DNA sequencer and were associated with amino acid substitutions such as Ser84Leu, Ser85Pro, and Glu88Lys in GyrA; Ser80Phe, Ser80Tyr, and Glu84Lys in GrlA; and Pro451Ser in GrlB (Table 3). Double-point mutations such as those corresponding to Ser84Leu and Ser85Pro in GyrA were detected using only a single probe. After calculation of the cross-hybridization signal of Glu88(Wt) in GyrA, the genotype corresponding to Glu88Lys was identified in seven isolates of MRSA HU2000-86S, -91S, -92, -135, -177, -199, and -214. In all isolates of S. aureus examined, the microarray system reliably identified the point mutation and the presence of the mecA gene (Table 3).
DISCUSSION
In this study we evaluated a novel three-dimensional microarray (PamChip microarray) system for the rapid and easy detection of levofloxacin resistance and the mecA gene in S. aureus. The microarray was designed to determine eight point mutation sites in the QRDRs of the gyrAB and grlAB genes and to detect the DNA sequence of the mecA gene simultaneously. All clinical isolate results of SBH and RDBH analysis with the microarray system corresponded to those of conventional DNA sequencing or PCR methods. Notably, double-point mutations, located near one another in the probes, could be analyzed by the microarray system. It could be difficult to detect them simultaneously using the TaqMan or Invader assay methods (2, 32).
Diluted PCR products in three different concentrations (1/10, 1/50, and 1/100) were analyzed with the PamChip microarray. Similar fluorescence intensity ratios were shown between the perfect-match probe and mismatch probes at all concentrations, whereas signal intensities were not always consistent with the theoretical density ratio. These results indicated that reproducibility around the limit concentration for signal detection is almost satisfactory and suitable for our microarray analysis. Moreover, the limit concentration for the signal detection of this system was about 10 times greater than that of conventional PCR methods (less than several pM).
The number of clinical isolates was too small to discuss analytical sensitivity and specificity for the microarray assay exactly. In future, it will be necessary to test a large number of isolates. Wang et al. (31) showed that 179 clinical isolates of levofloxacin-resistant MRSA in Japan each had one of the following genotype combinations: (i) Ser80Phe or Ser80Tyr in GyrA and Ser84Leu in GrlA, (ii) Ser80Phe or Ser80Tyr in GrlA and Glu88Lys in GyrA, and (iii) Glu84Lys in GrlA and Ser84Leu in GyrA. All of these genotypes of MRSA were included in this study. Our microarray system could have a possibility for a high level of analytical sensitivity in the detection of the levofloxacin resistance of MRSA isolates in Japan.
PCR primers for the amplification of the mecA gene, previously described by Mehrotra et al. (18), were used for our study as components of multiplex PCR or conventional PCR. In this previous report, they showed that the specificity of mecA gene detection using the PCR primers was 94% (18/19) for MRSA isolates. In mecA gene detection, the results with our microarray analysis corresponded to those obtained by conventional PCR (24/24). The specificity seemed to be almost equivalent to that of our microarray analysis.
Real-time PCR using the evaluation of melting curve analyses for point mutations is easy, fast, less costly, and comprehensive (5, 14); however, it could have artifacts due to the melting curve profile. This method suits mutation analysis of a single amplicon or of a few amplicons. Microarray analysis using specific hybridization can simultaneously analyze multiple mutation sites in combination with a multiplex PCR technique. As other several-point mutations in the QRDRs of the gyrAB and grlAB genes have been reported previously (8, 22, 23), the addition of probes to detect these mutations will make our system more comprehensive. Moreover, our microarray assay system has advantages in terms of the optimization of hybridization conditions for each target/probe within a single array.
In kinetic analysis, the stringency of the postwashing conditions was increased by decreasing the sodium chloride concentrations. To detect the point mutations associated with Glu84 and Asp79Ser80 in GrlA, the concentration of SSPE was reduced to 0.5× because of high Tm values. Specific signals for Pro451 in GrlB were detected at 6× SSPE, while those for Glu84 in GrlA were detected at 0.5× SSPE, although the calculated Tm range of the probes for Pro451 in GrlB was similar to that for Glu84 in GrlA (52 to 54°C). An accurate calculation of Tm values may be difficult for a probe with a mismatch site to the target DNA sequence. Accordingly, kinetic analysis is required under differing hybridization conditions, such as temperatures, or the use of multiple probes with different Tm values for a given point mutation may be required. Our results suggested that the PamChip microarray system was advantageous for the determination of specific hybridization conditions using kinetic analysis.
Hybridization steps were performed semiautomatically by the FD10 system. The total analysis time is within approximately 3.5 h: DNA extraction for 1 h by hand, PCR for 2 h, and automatic microarray analysis for 25 min (hybridization for 8 min, two washing steps for 12 min, and the data analysis step for 5 min). The PamChip microarray greatly reduces the time necessary for the hybridization analysis. Clinically, there is already a 24-h time addition if the process for microarray begins with the isolation of an organism from culture. It also requires PCR amplification of the target region as in other genetic methods based on PCR. To reduce the total assay time for genotyping and contamination errors, the PCR step ideally should be omitted in future. In this study, to specifically amplify the target DNA sequences, multiplex PCR was performed using a minimum of three primer sets designed with the gyrAB, grlAB, and mecA genes. Since the amounts of amplified products by multiplex PCR were less than those produced by single PCR in some isolates, 40 cycles were required for the stable multiplex amplification of DNA, and the products were used for microarray analysis.
Southern et al. (24) noted that short targets are better able to hybridize with oligonucleotide probes in a microarray than large targets are and stated that, ideally, targets and probes should have the same lengths. In our study, the combination of a 163-bp target for the mecA gene and 26 bases of specific probe resulted in hybridization that was efficient in comparison with that seen with a 1,255-bp target for the gyrAB gene or a 1,457-bp target for the grlAB gene and their short specific probes (20 to 25 bases). Hybridization signals for the gyrAB and grlAB targets were detected on either a sense or an antisense strand probe. Moreover, although the same target was used to detect the grlA and the grlB genes, hybridization signals for probes in the grlA gene were observed in the sense strand, and those for probes in the grlB gene were in the antisense strand (Fig. 3). This phenomenon suggests that the hybridization efficiencies for sense and antisense strand probes differ. This is in agreement with previous reports that the difference in signal intensities may depend on the secondary structures of probes and targets (3, 16, 20, 24). Therefore, the secondary structures of probes and targets are one of the important factors in microarray analysis.
We have not yet performed an assay using raw specimens. If plural genotypes of S. aureus are included in raw specimens, it may be difficult to detect them. Further, such evaluations with raw specimens are needed.
In conclusion, the unique features of the three-dimensional microarray (PamChip microarray) system provided a rapid, specific, easy, and reproducible detection of levofloxacin resistance and mecA resistance in S. aureus simultaneously. Flexibility for the optimization of hybridization conditions for each target nucleic acid and probe could be suitable for multiplex detection on the array. Our results suggest that this microarray technique has the potential for application in clinical microbiology laboratories in the future.
Acknowledgments
This study was supported by a Grant-in-Aid for Scientific Research (17790353) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
REFERENCES
- 1.Centers for Disease Control and Prevention. 1999. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota 1997-1999. Morb. Mortal. Wkly. Rep. 48:707-710. [PubMed] [Google Scholar]
- 2.Cooksey, R. C., B. P. Holloway, M. C. Oldenburg, S. Listenbee, and C. W. Miller. 2000. Evaluation of the Invader assay, a linear signal amplification method, for identification of mutations associated with resistance to rifampin and isoniazid in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 44:1296-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong, F., H. T. Allawi, T. Anderson, B. P. Neri, and V. I. Lyamichev. 2001. Secondary structure prediction and structure-specific sequence analysis of single-stranded DNA. Nucleic Acids Res. 29:3248-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang, H., and G. Hedin. 2003. Rapid screening and identification of methicillin-resistant Staphylococcus aureus from clinical samples by selective-broth and real-time PCR assay. J. Clin. Microbiol. 41:2894-2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrero, L., B. Cameron, B. Manse, D. Lagneaux, J. Crouzet, A. Famechon, and F. Blanche. 1994. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol. Microbiol. 13:641-653. [DOI] [PubMed] [Google Scholar]
- 7.Ferrero, L., B. Cameron, and J. Crouzet. 1995. Analysis of gyrA and grlA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 39:1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hannachi-M'Zali, F., J. E. Ambler, C. F. Taylor, and P. M. Hawkey. 2002. Examination of single and multiple mutations involved in resistance to quinolones in Staphylococcus aureus by a combination of PCR and denaturing high-performance liquid chromatography (DHPLC). J. Antimicrob. Chemother. 50:649-655. [DOI] [PubMed] [Google Scholar]
- 9.Hokaiwado, N., M. Asamoto, K. Tsujimura, T. Hirota, T. Ichihara, T. Satoh, and T. Shirai. 2004. Rapid analysis of gene expression changes caused by liver carcinogens and chemopreventive agents using a newly developed three-dimensional microarray system. Cancer Sci. 95:123-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horii, T., K. Yokoyama, S. Barua, T. Odagiri, N. Futamura, T. Hasegawa, and M. Ohta. 2000. The staphylokinase gene is located in the structural gene encoding N-acetylmuramyl-l-alanine amidase in methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 185:221-224. [DOI] [PubMed] [Google Scholar]
- 11.Horii, T., Y. Suzuki, A. Monji, M. Morita, H. Muramatsu, Y. Kondo, M. Doi, A. Takeshita, T. Kanno, and M. Maekawa. 2003. Detection of mutations in quinolone resistance-determining regions in levofloxacin- and methicillin-resistant Staphylococcus aureus: effects of the mutations on fluoroquinolone MICs. Diagn. Microbiol. Infect. Dis. 46:139-145. [DOI] [PubMed] [Google Scholar]
- 12.Ida, T., R. Okamoto, C. Shimauchi, T. Okubo, A. Kuga, and M. Inoue. 2001. Identification of aminoglycoside-modifying enzymes by susceptibility testing: epidemiology of methicillin-resistant Staphylococcus aureus in Japan. J. Clin. Microbiol. 39:3115-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito, H., H. Yoshida, M. Bogaki-Shonai, T. Niga, H. Hattori, and S. Nakamura. 1994. Quinolone resistance mutations in the DNA gyrase gyrA and gyrB genes of Staphylococcus aureus. Antimicrob. Agents Chemother. 38:2014-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lapierre, P., A. Huletsky, V. Fortin, F. J. Picard, P. H. Roy, M. Ouellette, and M. G. Bergeron. 2003. Real-time PCR assay for detection of fluoroquinolone resistance associated with grlA mutations in Staphylococcus aureus. J. Clin. Microbiol. 41:3246-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowy, F. D. 2003. Antimicrobial resistance: the example of Staphylococcus aureus. J. Clin. Investig. 111:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luebke, K. J., R. P. Balog, and H. R. Garner. 2003. Prioritized selection of oligodeoxyribonucleotide probes for efficient hybridization to RNA transcripts. Nucleic Acids Res. 31:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maekawa, M., T. Nagaoka, T. Taniguchi, H. Higashi, H. Sugimura, K. Sugano, H. Yonekawa, T. Satoh, T. Horii, N. Shirai, A. Takeshita, and T. Kanno. 2004. Three-dimensional microarray compared with PCR-single-strand conformation polymorphism analysis and DNA sequencing for mutation analysis of K-ras codons 12 and 13. Clin. Chem. 50:1322-1327. [DOI] [PubMed] [Google Scholar]
- 18.Mehrotra, M., G. Wang, and W. M. Johnson. 2000. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J. Clin. Microbiol. 38:1032-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murakami, K., W. Minamide, K. Wada, E. Nakamura, H. Teraoka, and S. Watanabe. 1991. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J. Clin. Microbiol. 29:2240-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peplies, J., F. O. Glockner, and R. Amann. 2003. Optimization strategies for DNA microarray-based detection of bacteria with 16S rRNA-targeting oligonucleotide probes. Appl. Environ. Microbiol. 69:1397-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Schmitz, F. J., B. Hofmann, B. Hansen, S. Scheuring, M. Luckefahr, M. Klootwijk, J. Verhoef, A. Fluit, H. P. Heinz, K. Kohrer, and M. E. Jones. 1998. Relationship between ciprofloxacin, ofloxacin, levofloxacin, sparfloxacin and moxifloxacin (BAY 12-8039) MICs and mutations in grlA, grlB, gyrA and gyrB in 116 unrelated clinical isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 41:481-484. [DOI] [PubMed] [Google Scholar]
- 23.Schmitz, F. J., M. E. Jones, B. Hofmann, B. Hansen, S. Scheuring, M. Luckefahr, A. Fluit, J. Verhoef, U. Hadding, H. P. Heinz, and K. Kohrer. 1998. Characterization of grlA, grlB, gyrA, and gyrB mutations in 116 unrelated isolates of Staphylococcus aureus and effects of mutations on ciprofloxacin MIC. Antimicrob. Agents Chemother. 42:1249-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Southern, E., K. Mir, and M. Shchepinov. 1999. Molecular interactions on microarrays. Nat. Genet. 21:5-9. [DOI] [PubMed] [Google Scholar]
- 25.Strommenger, B., C. Kettlitz, G. Werner, and W. Witte. 2003. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J. Clin. Microbiol. 41:4089-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi, H., T. Kikuchi, S. Shoji, S. Fujimura, A. B. Lutfor, Y. Tokue, T. Nukiwa, and A. Watanabe. 1998. Characterization of gyrA, gyrB, grlA and grlB mutations in fluoroquinolone-resistant clinical isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 41:49-57. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka, M., Y. X. Zhang, H. Ishida, T. Akasaka, K. Sato, and I. Hayakawa. 1995. Mechanisms of 4-quinolone resistance in quinolone-resistant and methicillin-resistant Staphylococcus aureus isolates from Japan and China. J. Med. Microbiol. 42:214-219. [DOI] [PubMed] [Google Scholar]
- 28.Tokuem, Y., K. Sugano, D. Saito, T. Noda, H. Ohkura, Y. Shimosato, and T. Sekiya. 1994. Detection of novel mutations in the gyrA gene of Staphylococcus aureus by nonradioisotopic single-strand conformation polymorphism analysis and direct DNA sequencing. Antimicrob. Agents Chemother. 38:428-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Utsui, Y., and T. Yokota. 1985. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 28:397-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Beumingen, R., H. van Damme, P. Boender, N. Bastiaensen, A. Chan, and T. Kievits. 2001. Fast and specific hybridization using flow-through microarrays on porous metal oxide. Clin. Chem. 47:1931-1933. [Google Scholar]
- 31.Wang, T., M. Tanaka, and K. Sato. 1998. Detection of grlA and gyrA mutations in 344 Staphylococcus aureus strains. Antimicrob. Agents Chemother. 42:236-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson, D. L., S. R. Abner, T. C. Newman, L. S. Mansfield, and J. E. Linz. 2000. Identification of ciprofloxacin-resistant Campylobacter jejuni by use of a fluorogenic PCR assay. J. Clin. Microbiol. 38:3971-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]