Abstract
Metronidazole resistance among Prevotella spp. is rare. We report here the first case of bacteremia due to a high-level metronidazole-resistant Prevotella sp. responsible for treatment failure.
CASE REPORT
A 78-year-old man with a past history of non-insulin-dependent diabetes mellitus and diverticulitis was hospitalized because of fever and altered clinical status. Physical examination was normal and no abnormalities were revealed by a systematic computed tomography scan of the abdomen. Laboratory studies revealed a white blood cell count of 28,000 cells/mm3 (90% polymorphonuclear leukocytes), a C-reactive protein level of 139 mg/liter, and a blood glucose level of 2 g/liter. Three sets of blood culture samples were collected in BACTEC Plus Aerobic/F medium and BACTEC Plus Anaerobic/F medium (BACTEC 9000 system; Becton Dickinson, Le Pont de Claix, France). The patient was treated empirically with intravenous cefotaxime (1 g three times daily) and ofloxacin (200 mg twice daily). Blood cultures taken at the admission detected the presence of Escherichia coli (susceptible to both antibiotics used) and Streptococcus anginosus (susceptible to cefotaxime). Urine and lower respiratory tract samples remained negative after quantitative bacteriologic cultures. On day 7, the patient remained febrile and new blood cultures (three sets) were taken while intravenous metronidazole (500 mg three times daily) and teicoplanin (400 mg once daily) were added.
A strictly anaerobic, nonmotile, gram-negative rod was the only bacterium detected in the three anaerobic blood culture vials of the three sample sets taken on day 7. In vitro susceptibility testing was performed by using a standard disk diffusion method on brucella agar supplemented with 5% sheep blood, 1 mg of vitamin K1/liter, and 5 mg of hemin/liter (7). Metronidazole susceptibility was determined by using 16-μg metronidazole Neo-Sensitabs (Rosco Diagnostica, Taastrup, Denmark). According to the criteria of the Comité de l'Antibiogramme de la Société Française de Microbiologie for susceptibility testing of anaerobes (7), the isolate was considered susceptible to amoxicillin-clavulanate, piperacillin-tazobactam, cefoxitin, imipenem, chloramphenicol, clindamycin, and metronidazole after 48 h of incubation. It was also intermediate to cefotaxime and resistant to ofloxacin. β-lactamase production was demonstrated by using a nitrocefin test (Cefinase; bioMérieux, Marcy-l'Etoile, France). Using an Etest method (13), we found a MIC ratio of >500 for cefotaxime/cefotaxime-clavulanic acid and ceftazidime/ceftazidime-clavulanic acid, suggesting the presence of an extended-spectrum β-lactamase.
It has been shown that metronidazole resistance may be only detected after prolonged incubation time (9, 12). Plates were therefore incubated for additional 48 h. This permitted us to observe that small colonies began to appear inside the metronidazole inhibition zone after 72 h and became more numerous after 96 h. The same phenomenon was also observed within the ellipse area of a metronidazole Etest strip after 72 h. Colonies from within and outside the inhibition zone, respectively, designated AIP 261-03 and AIP 268-03, were subcultured without antibiotic, and MICs were determined by using an agar dilution method as recommended by the Clinical and Laboratory Standards Institute (CLSI; formerly the National Committee for Clinical Laboratory Standards) (21). Both populations were found to be intermediate to cefotaxime (32 mg/liter). For metronidazole, MICs of 2 and 64 mg/liter were found for AIP 268-03 and AIP 261-03, respectively. On day 14, the treatment was changed to piperacillin-tazobactam (4 g three times daily) and ciprofloxacin (400 mg twice daily). The patient became apyretic and was discharged home 3 weeks later.
The presence of 5-nitroimidazole (nim) resistance genes was investigated by PCR with universal primers (28). Positive control strains containing nim genes included Bacteroides fragilis 638R containing plasmid pIP417 (nimA), B. fragilis BF8 (nimB), B. fragilis 638R containing plasmid pIP419 (nimC), B. fragilis 638R containing plasmid pIP421 (nimD), and B. fragilis ARU 6881 (nimE) gave amplification products, whereas AIP 261-03, AIP 268-01, and B. fragilis ATCC 25285 (nim-negative and metronidazole-susceptible control strain) did not. For AIP 268-03, a significant increase of the metronidazole MIC (24 mg/liter) was observed after three passages in brucella broth supplemented with hemin and vitamin K1 and containing subinhibitory concentrations of metronidazole. When the same assay was performed with AIP 261-03, the MIC was also increased up to 256 mg/liter. For all metronidazole-resistant isolates, the metronidazole MICs remained unchanged after further four consecutive subcultures on drug-free medium.
AIP 261-03 and AIP 268-03 did not grow on unsupplemented brucella agar nor on brucella agar supplemented with vitamin K1. Both isolates grew on brucella agar supplemented with hemin, but a substantially decreased growth rate was observed for AIP 261-03 whose colonies of which were smaller than those of AIP 268-03 after 48 h of incubation. Both isolates gave nonpigmented colonies on brucella sheep blood agar.
Conventional tests (15, 17), as well as tests with the API 20A (bioMérieux), Rapid ID32A system (bioMérieux), and the RapID ANA II system (AES, Combourg, France) showed that AIP 261-03 and AIP 268-03 produced the same profile. They were susceptible to bile and resistant to kanamycin, vancomycin, and colistin. Production of catalase, indole, and lipase was not detected. Gelatin was not liquefied. Acid was produced from lactose and saccharose but not from cellobiose, salicin, xylose, and xylan. The organisms did not hydrolyze esculin. Positive reactions were obtained for β-galactosidase, α-glucosidase, β-glucosidase, α-galactosidase, α-fucosidase, and N-acetyl-β-glucosaminidase. For both isolates, the major metabolic end products were acetic acid, lactic acid, and succinic acid as assessed by quantitative gas chromatography (3). However, it is noteworthy that the resistant strain AIP 261-03 produced higher levels of lactic acid than the susceptible strain AIP 268-03, namely, 44.6 versus 11 mmol/liter, respectively.
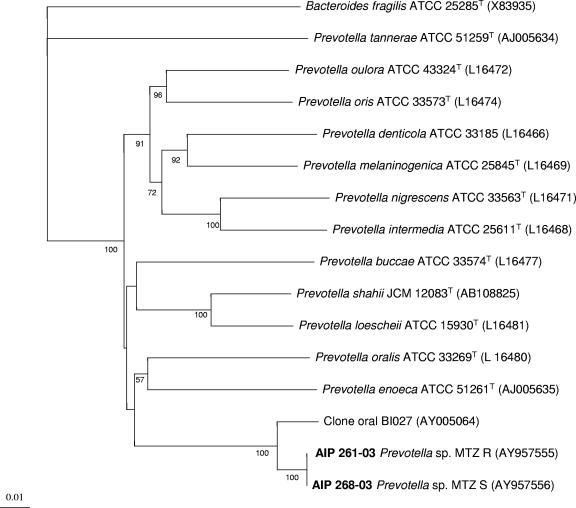
The cultural and biochemical characteristics were consistent with those of the genus Prevotella. 16S rRNA gene determination was performed as described previously (5). The 16S rRNA sequences obtained were compared to all eubacterial 16S rRNA sequences available in the GenBank database by using the multisequence Advanced BLAST comparison software from the National Center for Biotechnology Information (1). A phylogenetic tree was constructed from 1,380 unambiguous nucleotide positions by using the Fitch and Margoliash algorithm (11) and showed that both strains fell within the genus Prevotella (Fig. 1).
FIG. 1.
Dendrogram showing the phylogenetic positions of AIP 261-03 and AIP 268-03 within the genus Prevotella based on 16S rRNA sequences. The sequence of B. fragilis was used as an outgroup. The numbers above the branches are bootstrap percentages from 100 resampled data sets. The reference sequences were obtained from the GenBank and EMBL databases. Accession numbers are given in parentheses. The bar represents a 1% sequence difference.
A 16S rRNA sequence identity of 100% was found between the two strains. The highest sequence similarity value (99%) was then obtained with the 16S rRNA sequence of the oral clone BI027 (22). After which, sequence similarity values ranged from 89% (Prevotella oralis ATCC 33269T) to 80% (Prevotella tannerae ATCC 51259T). Thus, the strains AIP 261-03 and AIP 268-03 can be considered to represent two clones of a novel species within the genus Prevotella. Both strains exhibited the same pulsed-field gel electrophoresis pattern (CHEF DRIII; Bio-Rad) after DNA digestion performed by using XbaI as described by Chung et al. (6).
Prevotella spp. are anaerobic gram-negative bacilli that may be involved in various human infections, including infections of the head, neck, lower respiratory tract, central nervous system, and abdominal and female genital tract, and bacteremia (17). In humans, the sources of such infections are usually the oral cavity, as well as the intestinal and urogenital tracts, where these bacteria are prevalent in the local commensal flora. Despite a previous history of diverticulitis, an intestinal source was ruled out in our patient since the abdominal computed tomography scan and a further performed coloscopy were normal. Examination of the oropharynx revealed no abnormalities, and no other potential source of infection could be identified.
The treatment of such infections is commonly based on the use of metronidazole. For a long time, it has been considered that acquired resistance to this antibiotic was rare among anaerobes despite its extensive use. However, recent studies have shown that this resistance is not uncommon especially among Bacteroides spp. (12). Such resistance has also been observed in Prevotella spp. and gram-positive anaerobic bacteria (14, 19, 23, 26, 27). For most Prevotella spp. strains, a low-level resistance was observed, and only a single isolate of Prevotella melaninogenica with a high-level resistance (MIC of metronidazole: 32 mg/liter) has been reported by Lubbe et al. (19). Such a resistance level was also found in our isolate. For Bacteroides spp., it has been suggested that high-level metronidazole resistance may lead to eradication failure (25). This was also demonstrated in our patient who remained febrile while being treated with metronidazole for 7 days and who became apyretic 3 days after metronidazole was changed to piperacillin-tazobactam. It is believed that exposure to metronidazole can lead to the development of metronidazole-resistant mutants (12, 16). In contrast, in the present report, a metronidazole-resistant Prevotella sp. isolate was recovered before metronidazole was started in a patient that had not received previous 5-nitroimidazole therapy. This was also shown by other for metronidazole-resistant Bacteroides strains (24). We found that in our isolate metronidazole resistance was inducible and that, as shown by other with nim-positive Bacteroides spp. strains (12, 18), the mutant remained resistant after the removal of the antibiotic. Therefore, it may be possible that exposure to metronidazole in vivo has led to select a preexisting resistant subpopulation. However, this remains uncertain since no blood samples were taken while the patient was treated by metronidazole, and consequently no isolate was recovered after metronidazole treatment.
Among Bacteroides spp., 5-nitroimidazole resistance is most often related to the presence of nim genes, although nim gene-negative metronidazole-resistant strains have been reported (12, 18).
The proposed resistance mechanism conferred by the nim genes is that they encode a 5-nitroimidazole reductase, which possibly use ferredoxin as the electron donor (4). 5-Nitroimidazole is reduced to its amine derivative through a low redox potential reaction. For B. fragilis, it has been suggested that metronidazole resistance may also be due to alterations of different other enzymes involved in electron transfer reactions, particularly but not exclusively the pyruvate-oxidoreductase complex (2, 8, 10, 20). Decreased activity of pyruvate-oxidoreductase complex, which may be compensated for by the increase of the production of lactate dehydrogenase, causes alteration in the end products of glucose metabolism (2, 8, 20). This phenomenon induces elevated levels of lactate, as observed with our metronidazole-resistant strain. However, multiple pathways may be involved in the resistance to 5-nitroimidazole drugs, and further investigations are needed to determine the mechanism of metronidazole resistance in the new Prevotella species reported herein.
In the present study, metronidazole susceptibility was initially determined after 48 h. However, as seen here and in previous reports (9, 12), some slow-growing metronidazole-resistant clones could only be detected later. These would have been overlooked after 48 h of incubation using standard methods, including the agar dilution method. Therefore, given that this specific population may be responsible for treatment failure, we strongly recommend that plates be reexamined after 72 h to look for small colonies within the metronidazole inhibition zone. This case highlights the importance of careful antimicrobial susceptibility testing of anaerobes, especially in severe infections.
Nucleotide sequence accession numbers.
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA sequences of strains AIP 261-03 and AIP 268-03 are AY957555 and AY957556, respectively.
Acknowledgments
We thank Isabelle Scholtus and Marie Bedora-Faure for technical help.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Britz, M. L., and R. G. Wilkinson. 1979. Isolation and properties of metronidazole-resistant mutants of Bacteroides fragilis. Antimicrob. Agents Chemother. 16:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlier, J. P. 1985. Gas chromatography of fermentation products: its application in diagnosis of anaerobic bacteria. Bull. Inst. Pasteur 83:57-69. [Google Scholar]
- 4.Carlier, J. P., N. Sellier, M. N. Rager, and G. Reysset. 1997. Metabolism of 5-nitroimidazole in susceptible and resistant isogenic strains of Bacteroides fragilis. Antimicrob. Agents Chemother. 41:1495-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlier, J. P., G. K'ouas, A. Lozniewski, F. Sirveaux, P. Cailloux, and F. Mory. 2004. Osteosynthesis-associated bone infection caused by a nonproteolytic, nontoxigenic Clostridium botulinum-like strain. J. Clin. Microbiol. 42:484-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung, W. O., J. Gabany, G. R. Persson, and M. C. Roberts. 2002. Distribution of erm(F) and tet(Q) genes in 4 oral bacterial species and genotypic variation between resistant and susceptible isolates. J. Clin. Periodontol. 29:152-158. [DOI] [PubMed] [Google Scholar]
- 7.Comité de l'Antibiogramme de la Société Française de Microbiologie. 2004. Communiqué 2004. Société Française de Microbiologie, Paris, France.
- 8.Diniz, C. G., L. M. Farias, M. A. R. Carvalho, E. R. Rocha, and C. J. Smith. 2004. Differential gene expression in a Bacteroides fragilis metronidazole-resistant mutant. J. Antimicrob. Chemother. 54:100-108. [DOI] [PubMed] [Google Scholar]
- 9.Dublanchet, A. 1990. Bacteroides fragilis with reduced sensitivity to metronidazole. An unusual phenotypic expression. Med. Mal. Infect. 20(HS):113-116. [Google Scholar]
- 10.Edwards, D. I. 1993. Nitroimidazole drugs-action and resistance mechanisms. II. Mechanisms of resistance. J. Antimicrob. Chemother. 31:201-210. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package), version 3.5c. Department of Genetics, University of Washington, Seattle.
- 12.Gal, M., and J. S. Brazier. 2004. Metronidazole resistance in Bacteroides spp. carrying nim genes and the selection of slow-growing metronidazole-resistant mutants. J. Antimicrob. Chemother. 54:109-116. [DOI] [PubMed] [Google Scholar]
- 13.Handal, T., I. Olsen, C. B. Walker, and D. A. Caugant. 2004. β-lactamase production and antimicrobial susceptibility of subgingival bacteria from refractory periodontitis. Oral Microbiol. Immunol. 19:303-308. [DOI] [PubMed] [Google Scholar]
- 14.Hecht, D. W. 2004. Prevalence of antibiotic resistance in anaerobic bacteria: worrisome developments. Clin. Infect. Dis. 39:92-97. [DOI] [PubMed] [Google Scholar]
- 15.Holdeman, L. V., E. P. Cato, and W. E. C. Moore. 1977. Anaerobe laboratory manual, 4th ed. Virginia Polytechnic Institute and State University, Blacksburg, Va.
- 16.Ingham, H. R., S. Eaton, C. W. Venables, and P. C. Adams. 1978. Bacteroides fragilis resistant to metronidazole after long-term therapy. Lancet i:214. [DOI] [PubMed] [Google Scholar]
- 17.Jousimies-Somer, H., P. Summanen, D. M. Citron, E. J. Baron, H. M. Wexler, and S. M. Finegold. 2002. Wadsworth-KTL anaerobic bacteriology manual, 6th ed. Star Publishing Co., Belmont, Calif.
- 18.Löfmark, S., H. Fang, M. Hedberg, and C. Edlund. 2005. Inducible metronidazole resistance and nim genes in clinical Bacteroides fragilis group isolates. Antimicrob. Agents Chemother. 49:1253-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lubbe, M. M., P. L. Botha, and L. J. Chalkley. 1999. Comparative activity of eighteen antimicrobial agents against anaerobic bacteria isolated in South Africa. Eur. J. Clin. Microbiol. Infect. Dis. 18:46-54. [DOI] [PubMed] [Google Scholar]
- 20.Narikawa, S., T. Suzuki, M. Yamamoto, and M. Nakamura. 1991. Lactate dehydrogenase activity as a cause of metronidazole resistance in Bacteroides fragilis NCTC 11295. J. Antimicrob. Chemother. 28:47-53. [DOI] [PubMed] [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards. 2003. Methods for antimicrobial susceptibility testing of anaerobic bacteria, 6th ed. Approved standards M11-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 22.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phillips, I., C. Warren, E. Taylor, R. Timewell, and S. Eykyn. 1981. The antimicrobial susceptibility of anaerobic bacteria in a London teaching hospital. J. Antimicrob. Chemother. 8(Suppl. D):17-26. [DOI] [PubMed] [Google Scholar]
- 24.Rotimi, V. O., B. I. Duerden, V. Ede, and A. E. MacKinnon. 1979. Metronidazole-resistant Bacteroides from untreated patient. Lancet i:833. [DOI] [PubMed] [Google Scholar]
- 25.Rotimi, V. O., M. Khoursheed, J. S. Brazier, W. Y. Jamal, and F. B. Khodakhast. 1999. Bacteroides species highly resistant to metronidazole: an emerging clinical problem? Clin. Microbiol. Infect. 5:166-169. [DOI] [PubMed] [Google Scholar]
- 26.Sandoe, J. A. T., J. K. Struthers, and J. S. Brazier. 2001. Subdural empyema caused by Prevotella loescheii with reduced susceptibility to metronidazole. J. Antimicrob. Chemother. 47:366-367. [DOI] [PubMed] [Google Scholar]
- 27.Theron, M. M., M. N. Janse Van Rensburg, and L. J. Chalkley. 2004. Nitroimidazole resistance genes (nimB) in anaerobic gram-positive cocci (previously Peptostreptococcus spp.). J. Antimicrob. Chemother. 54:240-242. [DOI] [PubMed] [Google Scholar]
- 28.Trinh, S., and G. Reysset. 1996. Detection by PCR of the nim genes encoding 5-nitroimidazole resistance in Bacteroides spp. J. Clin. Microbiol. 34:2078-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]