Abstract
Pseudallescheria boydii (anamorph Scedosporium apiospermum) is the species responsible for human scedosporiosis, a fungal infection with a high mortality rate and which is difficult to treat. Recently, it has been demonstrated that high genetic variation exists within this species. We have performed a morphological and molecular study involving numerous strains of clinical or environmental origins and from different countries. The analysis of partial sequences of the β-tubulin (two loci) and calmodulin genes and the internal transcribed spacer region of the rRNA gene has demonstrated that P. boydii is a species complex. The combined analysis of the sequences of the four loci of 60 strains has showed the presence of 44 haplotypes in the ingroup. Three species morphologically related to P. boydii sensu stricto, i.e., Pseudallescheria angusta, Pseudallescheria ellipsoidea, and Pseudallescheria fusoidea, which had previously been considered synonyms, could be differentiated genetically from P. boydii in our study. It is relevant that two of the three strains now included in P. ellipsoidea have caused invasive infections. The species Pseudallescheria minutispora and Scedosporium aurantiacum are clearly phylogenetically separated from the other species studied and are here proposed as new. Morphological features support this proposal. All the strains included in S. aurantiacum species have a clinical origin, while those included in P. minutispora are environmental. Further studies are needed to demonstrate whether all the species included in the P. boydii complex have different clinical spectra and antifungal susceptibility.
Pseudallescheria boydii (anamorph Scedosporium apiospermum) is a ubiquitous ascomycetous fungus that causes a wide array of human infections that can affect practically all the organs of the body (8). These infections have been known for a long time, but in recent years, a marked increase in severe invasive infections has been noticed, mainly in immunocompromised hosts. The treatment of these infections has not yet been resolved, and the mortality rate is very high (3, 17). One of the most typical features of this species, which is very rare in other pathogenic fungi, is its ability to develop sexual structures on routine culture media. The presence of spherical ascomata (cleistothecia) and fusiform or ellipsoidal ascospores allows easy identification of this species and its differentiation from the other species of Scedosporium, Scedosporium prolificans, whose sexual state still remains unknown.
On the basis of nuclear DNA-DNA reassociation, some studies have proved that important genetic variation exists in P. boydii. Gueho and de Hoog (10) found three infraspecific ecological and clinical groups. Rainer et al. (16) reported the existence of five different small-subunit rRNA gene sequence lengths. Random amplified polymorphic DNA studies also demonstrated that numerous and very different genotypes can be found (7). Other authors have reported considerable differences with respect to growth and sporulation (4, 5, 9). In addition, a high variability in antifungal susceptibility of the different isolates and in their clinical response has been observed (1, 2). All these data seem to suggest that P. boydii is probably a species complex. In recent years, application of the phylogenetic species concept in different biological species of pathogenic fungi has revealed phylogenetic lineages that reflected species divergence (12, 13) and the existence of cryptic species. These putative cryptic species in P. boydii can show different pathological behavior and different antifungal susceptibility, so their delimitation and characterization are key in order to choose the appropriate treatment of the severe infections caused by these fungi.
This paper reports the results of a combined phenotypic and phylogenetic study of numerous clinical and environmental strains, including several fresh isolates, of the P. boydii species complex and the description of two new species.
MATERIALS AND METHODS
Fungal isolates.
Sixty isolates of Pseudallescheria boydii and relatives from environmental or clinical sources were included in the study (Table 1). Clinical isolates were provided by different reference culture collections (Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands [CBS], Facultat de Medicina i Ciències de la Salut, Reus, Spain, [FMR], The BCCM/IHEM Biomedical Fungi and Yeasts Collection, Brussels, Belgium [IHEM], Mycotheque de l'Universite Catholique de Louvain, Belgium [MUCL], Collection of the Institute for Tropical Medicine, Antwerp, Belgium [RV], and Robert Koch Institute, Berlin, Germany [RKI]) or different physicians. Environmental isolates were generally fresh isolates recovered by the authors from soil samples from different geographical regions, and others were also provided by different culture collections. In addition, reference strains of each of the species Pseudallescheria angusta, Pseudallescheria ellipsoidea, and Pseudallescheria fusoidea were included in the study. Isolates were stored at 4 to 7°C until morphological or molecular studies were performed.
TABLE 1.
Isolates included in the study and their origins
| Isolate | Source | GenBank accession no.
|
|||
|---|---|---|---|---|---|
| BT2 | TUB | CAL | ITS | ||
| FMR 4072 | River sediment, Tordera River, Spain | AJ889592 | AJ890122 | AJ890216 | AJ888384 |
| FMR 4167 | Otitis, Valladolid, Spain | AJ888952 | AJ889580 | AJ890152 | AJ888385 |
| FMR 6694 | Cerebral abscess, Barcelona, Spain | AJ888953 | AJ889581 | AJ890153 | AJ888386 |
| FMR 6697 | Sputum, Madrid, Spain | AJ888954 | AJ889582 | AJ890154 | AJ888387 |
| FMR 6918 | Garden soil, Barcelona, Spain | AJ888955 | AJ889583 | AJ890155 | AJ888433 |
| FMR 6919 | Garden soil, Barcelona, Spain | AJ888956 | AJ889985 | AJ890156 | AJ888388 |
| FMR 6920 | Garden soil, Barcelona, Spain | AJ888957 | AJ889986 | AJ890157 | AJ888434 |
| FMR 6921 | Garden soil, Barcelona, Spain | AJ888958 | AJ889987 | AJ890158 | AJ888389 |
| FMR 6922 | Garden soil, Barcelona, Spain | AJ888959 | AJ889988 | AJ890159 | AJ888390 |
| FMR 7884 | Transplant, Madrid, Spain | AJ889594 | AJ890126 | AJ890209 | AJ888391 |
| FMR 7885 | Pleural liquid, Madrid, Spain | AJ888960 | AJ889989 | AJ890160 | AJ888392 |
| FMR 8521 | Forest soil, Montsia, Spain | AJ889853 | AJ890007 | AJ890178 | AJ888430 |
| FMR 8522 | Forest soil, Deltebre, Spain | AJ889854 | AJ890008 | AJ890179 | AJ888431 |
| FMR 8530 | Cultivated soil, Deltebre, Spain | AJ889857 | AJ890011 | AJ890182 | AJ888406 |
| FMR 8532 | Cultivated soil, Montsia, Spain | AJ889858 | AJ890012 | AJ890183 | AJ888407 |
| FMR 8534 | Cultivated soil, Montsia, Spain | AJ889859 | AJ890013 | AJ890185 | AJ888408 |
| FMR 8535 | Soil, Buenos Aires, Argentina | AJ889860 | AJ890014 | AJ890186 | AJ888409 |
| FMR 8537 | Soil, Buenos Aires, Argentina | AJ889861 | AJ890110 | AJ890187 | AJ888410 |
| FMR 8538 | Soil, Buenos Aires, Argentina | AJ889862 | AJ890111 | AJ890188 | AJ888443 |
| FMR 8539 | Soil, Buenos Aires, Argentina | AJ889863 | AJ890112 | AJ890189 | AJ888411 |
| FMR 8540 | Soil, Buenos Aires, Argentina | AJ889864 | AJ890113 | AJ890190 | AJ888412 |
| FMR 8541 | Soil, Buenos Aires, Argentina | AJ889605 | AJ890128 | AJ890215 | AJ888413 |
| FMR 8619 | Keratitis, Brazil | AJ889584 | AJ890115 | AJ890192 | AJ888416 |
| FMR 8620 | Keratitis, Brazil | AJ889585 | AJ890116 | AJ890193 | AJ888417 |
| FMR 8621 | Cystic fibrosis, Barcelona, Spain | AJ889586 | AJ890117 | AJ890194 | AJ888418 |
| FMR 8622 | Foot skin, Barcelona, Spain | AJ889587 | AJ890118 | AJ890195 | AJ888419 |
| FMR 8623 | Leukemic patient, Barcelona, Spain | AJ889596 | AJ890125 | AJ890210 | AJ888427 |
| FMR 8625 | Leukemic patient, Zaragoza, Spain | AJ889588 | AJ890119 | AJ890196 | AJ888420 |
| FMR 8630 | Ulcer of ankle, Santiago de Compostela, Spain | AJ889597 | AJ890133 | AJ890219 | AJ888440 |
| RV 43605 | Human, Zaire | AJ888951 | AJ889579 | AJ890341 | AJ888383 |
| IHEM 14263 | Cystic fibrosis, patient 1, Angers, France | AJ888961 | AJ889990 | AJ890161 | AJ888436 |
| IHEM 14268 | Cystic fibrosis, patient 4, Giens, France | AJ888962 | AJ889991 | AJ890162 | AJ888393 |
| IHEM 14354 | Cystic fibrosis, patient 7, Giens, France | AJ888963 | AJ889992 | AJ890163 | AJ888437 |
| IHEM 14358 | Cystic fibrosis, patient 9, Tours, France | AJ888964 | AJ889993 | AJ890164 | AJ888438 |
| IHEM 14451 | Cystic fibrosis, patient 3, Giens, France | AJ888965 | AJ889994 | AJ890165 | AJ888394 |
| IHEM 14462 | Cystic fibrosis, patient 8, Tours, France | AJ888966 | AJ889995 | AJ890166 | AJ888395 |
| IHEM 14464 | Cystic fibrosis, patient 8, Tours, France | AJ888967 | AJ889996 | AJ890167 | AJ888396 |
| IHEM 14467 | Cystic fibrosis, patient 9, Tours, France | AJ888968 | AJ889997 | AJ890168 | AJ888397 |
| IHEM 14638 | Cystic fibrosis, patient 1, Angers, France | AJ888969 | AJ889998 | AJ890169 | AJ888398 |
| IHEM 14754 | Cystic fibrosis, patient 8, Tours, France | AJ888970 | AJ889999 | AJ890170 | AJ888399 |
| IHEM 14758 | Cystic fibrosis, patient 1, Angers, France | AJ889846 | AJ890000 | AJ890171 | AJ888400 |
| IHEM 15144 | Cystic fibrosis, patient 8, Tours, France | AJ889847 | AJ890001 | AJ890172 | AJ888401 |
| IHEM 15149 | Cystic fibrosis, patient 5, Giens, France | AJ889848 | AJ890002 | AJ890173 | AJ888402 |
| IHEM 15458 | Cystic fibrosis, patient 6, Giens, France | AJ889599 | AJ890135 | AJ890221 | AJ888441 |
| IHEM 15579 | Cystic fibrosis, patient 2, Angers, France | AJ889600 | AJ890136 | AJ890222 | AJ888439 |
| IHEM 15642 | Cystic fibrosis, patient 4, Giens, France | AJ889849 | AJ890003 | AJ890174 | AJ888403 |
| MUCL 6106 | Forest soil, Haasrode, Belgium | AJ889850 | AJ890004 | AJ890175 | AJ888404 |
| MUCL 8302 | Soil, Germany | AJ889855 | AJ890009 | AJ890180 | AJ888442 |
| MUCL 8522 | Humic soil, Baarn, the Netherlands | AJ889589 | AJ890120 | AJ890208 | AJ888421 |
| MUCL 14009 | Forest soil, Yangambi, Zaire | AJ889851 | AJ890005 | AJ890176 | AJ888422 |
| MUCL 14092 | Forest soil, Yangambi, Zaire | AJ889602 | AJ890130 | AJ890213 | AJ888429 |
| MUCL 18784 | Treated wood (Coelocarpon preussi), Ivory Coast | AJ889856 | AJ890010 | AJ890181 | AJ888405 |
| MUCL 20263 | Greenhouse soil, Herverlee, Belgium | AJ889852 | AJ890006 | AJ890177 | AJ888423 |
| MUCL 29258 | Fuel oil, Antwerpen, Belgium | AJ889593 | AJ890123 | AJ890217 | AJ888424 |
| RKI 2782/95 | Trauma and sepsis, Hamburg, Germany | AJ889598 | AJ890134 | AJ890220 | AJ888432 |
| RKI 2956/93 | Bronchoalveolar lavage fluid, Berlin, Germany | AJ889865 | AJ890114 | AJ890191 | AJ888415 |
| CBS 101.22 | Mycetoma, Texas | AJ889590 | AJ890121 | AJ890207 | AJ888435 |
| CBS 106.53 | Goat dung, Aligarh, India | AJ889601 | AJ890131 | AJ890212 | AJ888428 |
| CBS 254.72 | Half-digested sewage tank, Ohio | AJ889604 | AJ890129 | AJ890214 | AJ888414 |
| CBS 311.72 | Brown sandy soil, Tsintsabis, Namibia | AJ889603 | AJ890132 | AJ890218 | AJ888425 |
| CBS 418.73 | Soil, Tadzjikistan | AJ889595 | AJ890124 | AJ890211 | AJ888426 |
| FMR 7294 | Blood, Australia | AJ889591 | AJ890127 | AJ890223 | AJ888444 |
Isolation from soil.
Soil samples were collected mainly from the superficial layer of soil (mainly garden soils) by using sterilized polyethylene bags. These bags were closed by rubber bands and labeled. Suspensions of this material were cultured on the selective medium Dichloran Rose-Bengal chloramphenicol agar (Oxoid, United Kingdom) with benomyl added at a final concentration of 10 μg/ml and incubated at room temperature. When typical colonies of P. boydii were observed, we tried to isolate them in pure culture.
DNA extraction, amplification, and sequencing.
DNA was extracted and purified directly from fungal colonies according to the Fast DNA kit protocol (Bio101, Vista, Calif.), with a minor modification that consisted of the homogenization step repeated three times with a FastPrep FP120 instrument (Thermo Savant, Holbrook, N.Y.). The DNA was quantified with GeneQuant pro (Amersham Pharmacia Biotech, Cambridge, England). The internal transcribed spacer (ITS) region of the nuclear rRNA gene was amplified with the primer pair ITS5 and ITS4 (21), a fragment of the nuclear gene calmodulin (CAL) was amplified with the degenerated primer pair CL1 and CL2A (14), and two regions within the β-tubulin gene, BT2 and TUB, were amplified using the degenerated primer pair BT2-F (5′-GG(CT)AACCA(AG)AT(ATC)GGTGC(CT) GC(CT)-3′) and BT2-R (5′-ACCCTC(AG)GTGTAGTGACCCTTGGC-3′) and TUB-F/TUB-R (6), respectively.
The PCR mixture (25 μl) included 20 to 60 ng of fungal DNA template, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, and 2.5 mM MgCl2 (10× Perkin-Elmer buffer II plus MgCl2 solution; Roche Molecular Systems, Branchburg, N.J.), 100 μM each deoxynucleoside triphosphate (Promega, Madison, Wis.), 1 μM of each primer, and 1.5 U of AmpliTaq DNA polymerase (Roche). The amplification program included an initial denaturation step at 94°C for 5 min followed by 35 cycles of denaturation at 95°C for 30 s, annealing for 1 min at 50°C (ITS), 55°C (CAL and TUB), or 60°C (BT2), and extension for 1 min at 72°C. A final extension step at 72°C for 7 min was included at the end of the amplification. After PCR, the products were purified with a GFXTM PCR DNA purification kit (Pharmacia Biotech, Cerdanyola, Spain) and stored at −20°C until they were used in sequencing.
The protocol for sequencing was the Taq DyeDeoxy Terminator Cycle Sequencing kit (Applied Biosystems, Gouda, The Netherlands). Reactions were run with a 310 DNA sequencer (Applied Biosystems). The consensus sequences were obtained using the Autoassembler program (Applied Biosystems).
Phylogenetic analysis.
The sequences were aligned using the Clustal X (version 1.8) computer program (19) followed by manual adjustments with a text editor. Most-parsimonious tree (MPT) analyses were performed by using PAUP* version 4.0b10 (18). One hundred heuristic searches were conducted with random sequence addition and tree bisection reconnection branch-swapping algorithms, collapsing zero-length branches and saving all minimal-length trees (MulTrees) on different data sets. Scedosporium prolificans (FMR 7294) and Pseudallescheria africana (CBS 311.72) were chosen as the outgroup. Regions of sequences with ambiguous alignments were excluded from all analyses (ITS, positions 58 and 59; BT2, positions 90 to 126), and gaps were treated as missing data. Support for internal branches was assessed using a heuristic parsimony search of 500 bootstrapped data sets. The combined data set was tested for incongruence with the partition homogeneity test (PHT), as implemented in PAUP*. To avoid detecting incongruence that is expected within lineages, partition homogeneity tests were restricted to data sets containing only 20 individuals that represented the main lineages (CBS 254.72, FMR 4072, FMR 4167, FMR 6694, FMR 6697, FMR 6920, FMR 6921, FMR 7884, FMR 8532, FMR 8540, FMR 8541, FMR 8623, FMR 8625, FMR 8630, IHEM 14268, IHEM 14467, IHEM 15458, MUCL 14009, RKI 2956/93, RKI 2782/95, and RV 43605).
To test alternative phylogenetic relationships, the Kishino-Hasegawa maximum-likelihood ratio test (11) was performed, as implemented in PAUP*.
Morphological study.
The fungi were subcultured on potato dextrose agar (PDA; Difco Laboratories, Detroit, Mich.) for macroscopic examination and growth rates at 25, 37, 40, 42, 45, and 50°C in darkness. For the study of microscopic characteristics, they were cultivated on oatmeal agar (OA) (30 g oat flakes, 1 g MgSO4 · 7H2O, 1.5 g KH2PO4, 15 g agar, 1 liter tap water). The microscopic features were determined by making wet mounts with lactic acid, which were then examined under a light microscope (Leitz Dialux 20).
Nucleotide sequence accession numbers.
All the sequences obtained were deposited in the GenBank database. Accession numbers are shown in Table 1.
RESULTS
Phylogeny.
Sixty isolates of P. boydii and relatives were chosen to examine species limits and evolutionary relationships among them. With the primers used, we were able to amplify and sequence 522 bp, 419 bp, 549 bp, and 570 bp of the ITS, BT2, TUB, and CAL loci, respectively. Of the 2,060 nucleotides sequenced, 386 characters (18.73%) were parsimony informative in the different P. boydii isolates. The lowest number was 42 in the ITS region, and the highest was 160 in CAL region. Sequences of the four region genes were analyzed phylogenetically as separate and combined data sets.
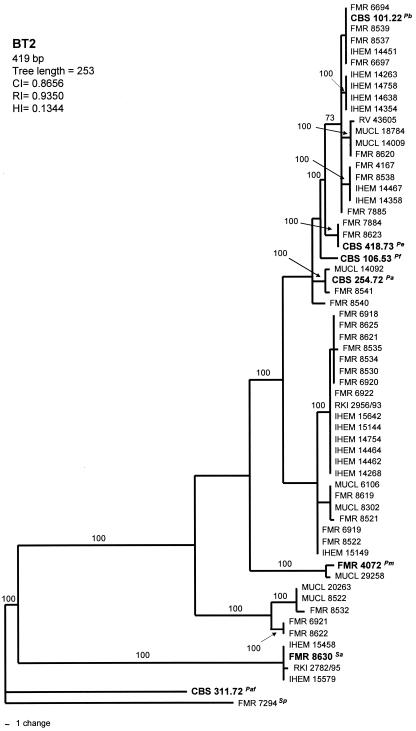
Phylogenetic analysis of the 419-bp BT2 data set yielded 12 MPT, which resulted in a total of 25 haplotypes (Fig. 1). There were 246 constant, 93 parsimony-informative, and 77 variable parsimony-uninformative characters in this fragment. The type strains of P. fusoidea, P. ellipsoidea and P. angusta were interspersed with the isolates of P. boydii. Four main highly supported clades (100%) were shown: the basal one comprising 4 European clinical isolates; two other small clades made up of 5 and 2 almost exclusively environmental isolates; and the biggest one, which comprised the 49 remaining isolates. However, inside the latter, another six terminal branches were present, each of them supported by a 100% bootstrap.
FIG. 1.
One of the 12 most-parsimonious trees obtained from heuristic searches based on BT2 sequence. Bootstrap support values above 70% are indicated at the nodes. Type strains are indicated with boldface type. P. africana and S. prolificans were used as outgroups. Pb, P. boydii; Pe, P. ellipsoidea; Pf, P. fusoidea; Pa, P. angusta; Pm, P. minutispora; Sa, S. aurantiacum; Paf, P. africana; Sp, S. prolificans; CI, consistency index; RI, retention index; HI, homoplasy index.
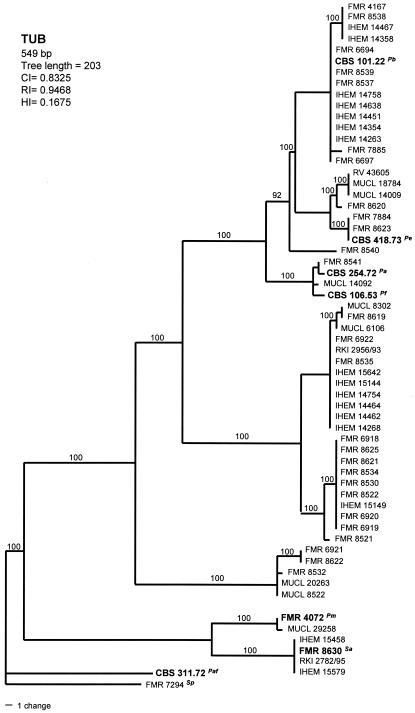
Parsimony analysis of the TUB data set yielded 420 MPT with 203 steps in length, in which 18 nodes received 100% bootstrap support. There were 420 constant, 90 parsimony-informative, and 39 variable parsimony-uninformative characters in this set. These trees resulted in a total of 22 different haplotypes (Fig. 2). Although the tree topology was slightly different from that of the previously mentioned locus, the three small, most-basal clades formed were also maintained here. Two of these clades, formed by four clinical and two environmental isolates, respectively, were the most phylogenetically distant.
FIG. 2.
One of the 420 most-parsimonious trees obtained from heuristic searches based on TUB sequence. Bootstrap support values above 90% are indicated at the nodes. Type strains are indicated with boldface type. P. africana and S. prolificans were used as outgroups. Pb, P. boydii; Pe, P. ellipsoidea; Pf, P. fusoidea; Pa, P. angusta; Pm, P. minutispora; Sa, S. aurantiacum; Paf, P. africana; Sp, S. prolificans; CI, consistency index; RI, retention index; HI, homoplasy index.
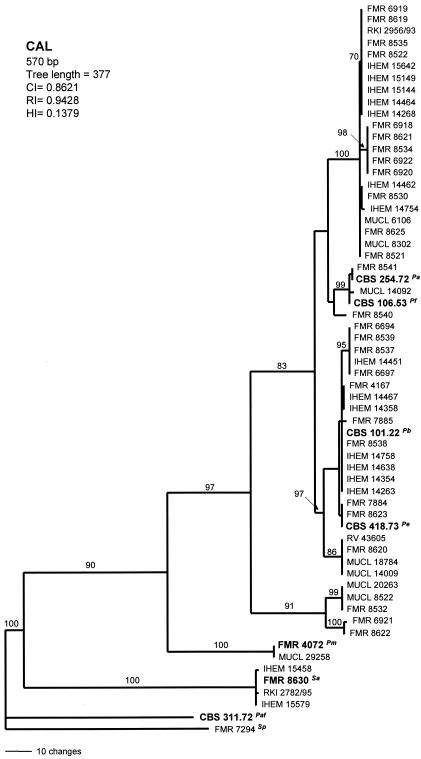
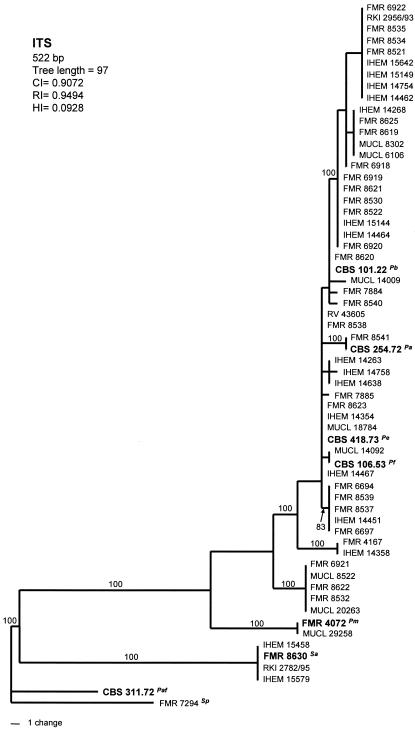
Analysis of CAL and ITS sequences yielded a single MPT of 377 steps in length and 5,000 MPT of 97 steps in length, respectively. The numbers of haplotypes observed were 21 in the CAL tree (Fig. 3) and 19 in the tree based on ITS sequences (Fig. 4). Both trees showed similar topologies to that of the BT2 tree. The two above-mentioned basal clades were also placed here away from the other isolates. Overall, the ITS rRNA gene data set is considerably less informative for phylogenetic reconstruction than the other three markers.
FIG. 3.
The single most-parsimonious tree obtained from heuristic searches based on CAL sequence. Bootstrap support values above 70% are indicated at the nodes. Type strains are indicated with boldface type. P. africana and S. prolificans were used as outgroups. Pb, P. boydii; Pe, P. ellipsoidea; Pf, P. fusoidea; Pa, P. angusta; Pm, P. minutispora; Sa, S. aurantiacum; Paf, P. africana; Sp, S. prolificans; CI, consistency index; RI, retention index; HI, homoplasy index.
FIG. 4.
One of the 5,000 most-parsimonious trees obtained from heuristic searches based on ITS sequence. Bootstrap support values above 80% are indicated at the nodes. Type strains are indicated with boldface type. P. africana and S. prolificans were used as outgroups. Pb, P. boydii; Pe, P. ellipsoidea; Pf, P. fusoidea; Pa, P. angusta; Pm, P. minutispora; Sa, S. aurantiacum; Paf, P. africana; Sp, S. prolificans; CI, consistency index; RI, retention index; HI, homoplasy index.
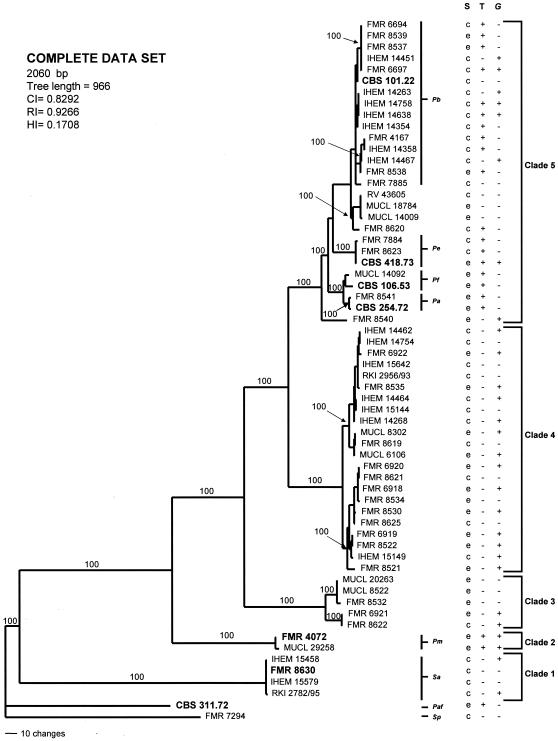
The result of the partition homogeneity test showed that the sequence data sets for the four loci were congruent (P = 0.07) and could therefore be combined. A total of 2,496 MPT were produced from a heuristic search using the combined data set of 2,060 characters from the four loci (Fig. 5). From these characters, 1,440 were constant, 386 were parsimony informative, and 234 were variable parsimony noninformative. Clustering was similar to that observed in the particular trees of the different genes analyzed. A total of 44 haplotypes were shown. Most nodes in the combined analysis showed increased clade support as measured by bootstrapping (20 nodes with 100% bootstrap support). As within the ITS, TUB, and CAL gene trees, two clades were identified as the basal-most lineages (clades 1 and 2), each of them with a bootstrap support of 100%. Phylogenetic analysis of the remaining monophyletic ingroup taxa (bootstrap, 100%) identified a basal clade (clade 3) and two bigger clades (clades 4 and 5), all them with 100% bootstrap support. The type strains of P. boydii, P. ellipsoidea, P. fusoidea, and P. angusta were placed in clade 5.
FIG. 5.
One of the 2,496 most-parsimonious trees obtained from heuristic searches based on analysis produced from the combined data set. Bootstrap support values of 100% are indicated at the nodes. Type strains are indicated with boldface type. P. africana and S. prolificans were used as outgroups. Pb, P. boydii; Pe, P. ellipsoidea; Pf, P. fusoidea; Pa, P. angusta; Pm, P. minutispora; Sa, S. aurantiacum; Paf, P. africana; Sp, S. prolificans; S, source; T, teleomorph; G, Graphium anamorph; CI, consistency index; RI, retention index; HI, homoplasy index; c, clinical; e, environmental; +, presence; −, absence.
Morphology.
All the 60 isolates that constituted the ingroups in the different trees obtained in our phylogenetic analyses were clearly identified by the presence of a characteristic Scedosporium anamorph. Although no relevant morphological differences were observed among them, the isolates that constitute clade 1 showed narrower conidia (2 to 5 μm wide) than the rest (3 to 6 μm wide). In addition, the members of this clade produced a yellow diffusible pigment, absent in the rest, and showed a maximum growth at 45°C compared to 40°C for the rest of isolates. Practically all isolates also developed conidia growing directly on vegetative hyphae, which were usually brown, thick-walled, and ellipsoidal to obovoidal. This latter feature was an important characteristic in differentiating the members of the subclade where the type strain of P. boydii is included, in which such conidia were globose to subglobose (Table 2). Another typical feature, common to the half of the isolates included in the study, was the development of a second type of anamorph, namely, Graphium sp. Graphium is characterized by the production of synnemata terminated in a slimy head of conidia. The size of the synnemata was very variable (80 to 750 μm long) and depended on the culture medium used and the age of the culture. The production of Graphium was not exclusive of any clade since the isolates that produced them were distributed in all the five clades (Fig. 5). However, it was most common in clade 4 (in 14 of the 22 isolates) and in the two isolates of clade 2. The presence of the teleomorph (the sexual state) was less frequent than the above-mentioned structures; it was produced only by the members of clades 5 (in 18 of the 27 isolates) and 2 (in the two isolates). Interestingly, the teleomorph of the isolates of the latter clade showed ascospores clearly different from those of the other species of Pseudallescheria included in the study. On the basis of the morphological differences, supported by the molecular data, we think that clades 1 and 2 represent two different species from those up to now accepted in Scedosporium and Pseudallescheria, respectively, and are consequently here proposed as new.
TABLE 2.
Relevant features to differentiate the clinical species of Pseudallescheria/Scedosporium spp.
| Species | Diffusible pigment on PDA at 25°C | Maximum growth temp (°C) | Teleomorph
|
Anamorph
|
||
|---|---|---|---|---|---|---|
| Development of ascomata | Shape of ascospores | Shape of conidiogenous cells | Most common shape of conidia borne on vegetative hyphae | |||
| P. boydii | − | 40 | ± | Broadly fusiform | Cylindrical | Globose |
| P. ellipsoidea | − | 40 | + | Ellipsoidal | Cylindrical | Ellipsoidal |
| S. aurantiacum | + (yellow) | 45 | − | Cylindrical or slightly flask-shaped | Obovoid | |
| S. prolificans | − | 40 | − | Flask-shaped | Globose | |
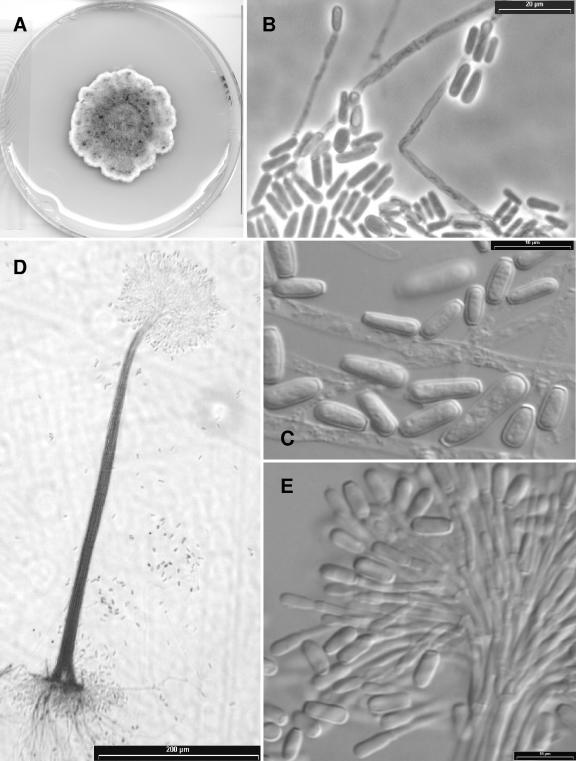
Scedosporium aurantiacum.
Gilgado, Cano, Gené, et Guarro, sp. nov. (Fig. 6).
FIG. 6.
Scedosporium aurantiacum isolates (A, D, E) IHEM 15458 and (B, C) FMR 8630. (A) Colony growing on PDA after 14 days of incubation at 25°C. (B, C) A conidiogenous cell and conidia from solitary conidiophores. (D) A synnema of the Graphium anamorph. (E) Apical part of a synnema producing conidia.
Etym.: referred to the yellow color of the diffusible pigment of the colonies.
Coloniae in agaro dense, plerumque gossypinae, luteolus vel brunneolus griseae; reversum brunneolus aurantiacum. Pigmentis flavis in culturis formantis. Conidiophora solitaria vel synnematica. Synnemata erecta, 330 to 750 μm alta, cum caule atro griseo, cylindrico, 7.5 to 17.5 μm lato, et capitulo mucoso usque ad 60 μm alto, 70 to 140 μm lato. Conidia obovoidea, subcylindrica vel claviformia, 5 to 14 by 2 to 5 μm. Conidia sessilis copiosae, laterales, unicellularia, brunnea, plerumquam obovoidea, 6 to 10 by 3 to 5 μm. Teleomorphosis ignota. Holotypus, IMI 392886, ex mycosis humanum (cultura viva, FMR 8630, IHEM 21147, CBS 116910).
The colonies on PDA attained a diameter of 40 to 50 mm after 14 days at 25°C. They were dense and usually cottony, but in some isolates they were lanose, especially at the center, frequently showing a concentric growth of aerial mycelium of different colors, yellowish gray combined with brownish gray areas, usually with a whitish, irregularly lobate and fimbriate margin, and the reverse was brownish orange at the center and brown to colorless towards the periphery. All isolates produced a light yellow diffusible pigment on PDA and OA after a few days of incubation. Conidiophores were solitary on aerial mycelium or grouped to form synnemata (Graphium) mainly on the agar surface. Solitary conidiophores were often reduced to a conidiogenous cell growing laterally on undifferentiated mycelium or branched, usually bearing verticils of two to three conidiogenous cells. Synnemata were present only in the isolates IHEM 15458 and RKI 2782/95. They were erect, 330 to 750 μm long, consisting of a cylindrical stipe 7.5 to 17.5 μm wide, dark gray, smooth-walled, and slightly roughened apically, and they terminated in a slimy head of conidia, up to 60 μm long by 70 to 140 μm wide. The conidiogenous cells were percurrent, lateral, or terminal, subhyaline, smooth-walled, cylindrical, or slightly flask shaped, 10 to 37 μm long by 1.5 to 2.5 μm wide, less frequent intercalary as a lateral projection on hyphae, and up to 5 μm long by 2 μm wide. There were three types of conidia: (i) those produced on solitary conidiophores were subhyaline, smooth-walled, obovoid, or subcylindrical, and 5 to 14 μm by 2 to 5 μm; (ii) those produced on synnemata were predominantly cylindrical or claviform, 6 to 12 μm by 3 to 5 μm with a wide truncate base; (iii) those developed mainly from the undifferentiated hyphae of the substrate were sessile or on short protrusions, solitary, lateral, brown, smooth, and thick-walled, usually obovoid, 6 to 10 μm long by 3 to 5 μm wide. The latter were abundantly produced by all isolates. Teleomorph was unknown.
The optimum growth temperature was from 37°C to 40°C with colonies on PDA attaining a diameter up to 60 to 67 mm after 14 days. Maximum growth was at 45°C. The fungus did not grow at 50°C.
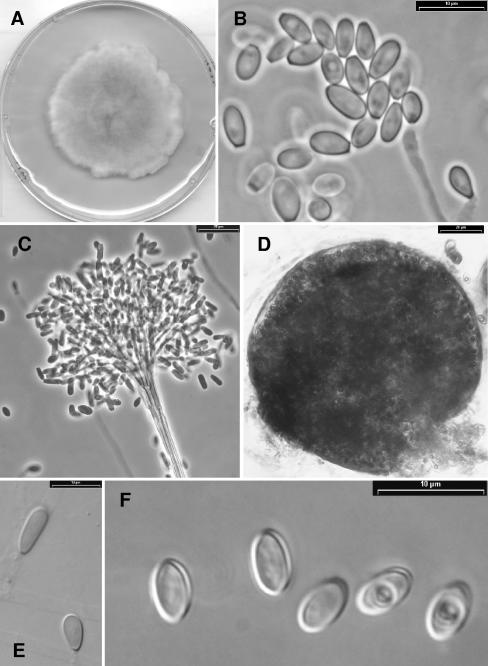
Pseudallescheria minutispora.
Gilgado, Gené, Cano, et Guarro, sp. nov. (Fig. 7).
FIG. 7.
Pseudallescheria minutispora strain FMR 4072. (A) Colony growing on PDA after 14 days of incubation at 25°C. (B) Conidiogenous cells and conidia of the Scedosporium anamorph. (C) Apical part of a synnema of the Graphium anamorph producing conidia. (D) Ascoma. (E) Conidia borne on undifferentiated hyphae. (F) Ascospores.
Etym.: referred to the small size of the ascospores.
Coloniae in agaro dense, gossypinae vel lanosae, aurantium griseae vel brunneolus griseae; reversum incoloratum. Pigmentis in culturis non formantis. Ascomata solitaria, non ostiolata, globosa vel subglobosa, 50 to 150 μm, luteolus vel brunneolus grisea, cum peridium ex textura epidermoidea. Asci octospori, globosi vel subglobosi, 12 to 15 by 10 to 13 μm, evanescentes. Ascosporae unicellulares, subhyalinae vel dilute brunneae, laeves, tenuitunicateae, ellipsoideae, 5 to 7 by 3 to 4 μm. Conidiophora solitaria vel synnematica. Synnemata erecta, 180 to 300 μm alta, cum caule atrobrunneo, cylindrico, 7.5 to 17.5 μm lato, et capitulo mucoso, 60 to 100 μm alto et 80 to 170 μm lato. Conidia obovoidea, ellipsoidea, cylindrica vel claviformia, 5 to 14 by 2.5 to 4.5 μm. Conidia sessilis parcusae, laterales, unicellularia, subhyalina, plerumquam obvoidea, 7 to 10 by 2.5 to 5 μm. Holotypus, IMI 392887, ex sedimentis fluvialibus (cultura viva, FMR 4072, IHEM 21148, CBS 11691).
The colonies on PDA attained a diameter of 50 to 57 mm after 14 days at 25°C. They were dense, cottony to lanose, orange gray combined with brownish gray areas, with a whitish, lobate or irregular, and fimbriate margin; the reverse was colorless. Diffusible pigment was absent. All isolates developed abundant ascomata on OA. The ascomata were solitary, nonostiolate, globose to subglobose, and 50 to 150 μm in diameter, with a peridium of textura epidermoidea, yellowish gray to brownish gray, and often covered with brown, thick-walled septate, 2.2 to 3 μm wide. The asci were eight-spored, globose to subglobose, and 12 to 15 μm long by 10 to 13 μm wide with evanescent walls. Ascospores were unicellular, subhyaline to light brown, smooth and thin-walled, ellipsoidal, and 5 to 7 μm long by 3 to 4 μm wide, with a germ pore at each pole and usually with oil drops. Both isolates developed the two typical anamorph simply or scarcely branched conidiophores, up to 35 μm long. Its conidia were subhyaline to light brown, smooth-walled, obovoid, ellipsoidal or subclaviform, and 6 to 11 μm long by 3 to 4 μm wide. The Graphium anamorph produced synnemata which were erect and 180 to 300 μm long, with a cylindrical stipe that was 7.5 to 17.5 μm wide, and they were smoke brown, smooth-walled, slightly roughened apically, slightly inflated at the base, and up to 25 μm wide and terminated in a slimy head of conidia that was 60 to 100 μm long by 80 to 170 μm wide. The conidia were predominantly cylindrical or claviform, 5 to 14 μm long by 2 to 4.5 μm wide, with a wide truncate base. The conidiogenous cells were percurrent, lateral or terminal, subhyaline, smooth-walled, usually cylindrical, 10 to 35 μm long by 1.5 to 2 μm wide, and less frequently intercalary as a lateral projection on hyphae, up to 6 μm long by 2 μm wide. Conidia from undifferentiated hyphae were scarcely produced. They were lateral, usually sessile, subhyaline, smooth and thick-walled, ellipsoidal to obovoid, and 7 to 10 μm long by 3.5 to 5 μm wide.
The optimum growth temperature was from 25°C to 30°C with colonies on PDA attaining a diameter up to 50 to 61 mm after 14 days. Maximum growth was at 40°C. The fungus was unable to grow at 42°C.
DISCUSSION
DNA sequences from four loci were analyzed to investigate phylogenetic relationships and species limits within the P. boydii species complex. Until recently, the genus Pseudallescheria was considered to comprise the following seven species (20): P. africana, P. angusta, P. boydii, P. desertorum, P. ellipsoidea, P. fimeti, and P. fusoidea. All of these species are morphologically very similar, and the main distinction among them is based on the size of the cleistothecia and ascospores (20). Recently, Rainer et al. (16), using different molecular techniques, concluded that P. ellipsoidea, P. fusoidea, and P. angusta are probable synonyms of P. boydii. In our study, we have included the type strains of these three species, and the results confirmed that all of them are genetically and morphologically different from P. boydii. We have also detected a high number of phylogenetic species, but only two of them can be clearly recognized morphologically.
The information provided by the four loci analyzed was very similar, which proved to be excellent phylogenetic markers for species level systematics within Pseudallescheria. The less informative locus was ITS, which only resolved 9 phylogenetically distinct species, whereas CAL, BT2, and TUB resolved 12, 14, and 15 species, respectively. Apart from P. africana, which was the outgroup of the present analysis, the two phylogenetic species most clearly separated were the two proposed as new, i.e., S. aurantiacum, represented by four clinical isolates, and P. minutispora, represented by two environmental isolates. However, we have recently studied another isolate of clinical origin from Germany (RKI 866/94) that genetically and morphologically matches the features of the latter species. The two clades formed by these species were highly supported in all the phylogenetic trees. Up to know, practically all the described species of Pseudallescheria, with the exception of P. boydii, P. fusoidea, and P. ellipsoidea, were monotypic; i.e., they are known by only one isolate. In the present study, the type strain of P. angusta nested with another soil isolate (FMR 8541) from Argentina. Both isolates showed identical ITS sequences, and those of the other loci studied were only different in 1 or 2 nucleotides (BT2, 1 nucleotide; TUB, 1 nucleotide; CAL, 2 nucleotides). The type strain of P. fusoidea nested with an environmental isolate from Zaire in three of the four loci analyzed. Both isolates also showed identical ITS sequences and differed in a few nucleotides in the other genes (BT2, 8 nucleotides; TUB, 4 nucleotides; CAL, 2 nucleotides). Our phylogenetic study revealed that P. fusoidea and P. angusta were phylogenetically very close. They only differed in 22 bp in the combined data set. However, P. angusta showed smaller ascomata (up to 110 μm in diameter versus up to 160 μm in diameter for P. fusoidea) and narrower ascospores (3 to 3.5 μm wide versus 4 to 4.5 μm wide for P. fusoidea). In addition, the isolates of P. fusoidea grew faster than those of P. angusta (69 to 70 mm versus 52 to 54 mm at 14 days on PDA at 25°C). The teleomorphs developed by the isolates of P. angusta, P. ellipsoidea, and P. fusoidea were consistent with the morphological features for the species described previously by von Arx et al. (20). However, the isolates that nested with the type strain of P. boydii (CBS 101.22) showed larger ascospores (6 to 9 by 5 to 6 μm) than those described previously by von Arx et al. (20) for such species (6 to 7 by 3.5 to 4 μm). Unfortunately, in our study, the type strain of P. boydii only produced the Scedosporium anamorph. It is an old strain that has probably lost the ability to develop the sexual state. P. ellipsoidea was the species that was genetically and morphologically closest to the group of isolates that nested in the same branch as the type strain of P. boydii. However, P. ellipsoidea can be distinguished by its ellipsoidal ascospores, while the ascospores of the members of such groups of isolates are broadly fusiform. Moreover, in the former, the conidia from vegetative hyphae are ellipsoidal to obovoid and scarce, while those of the P. boydii branch are abundant and predominantly globose to subglobose (Table 2). Further studies of this group of isolates are required in order to define the morphological features of P. boydii sensu stricto.
Up to now, P. boydii had been considered the only pathogenic species of the genus Pseudallescheria, but this study has demonstrated that other phylogenetic species of the P. boydii complex also included clinical isolates. However, the clinical strains are not homogeneously distributed in the different clades, and some correlation between the clades and the clinical origin of the strains could be observed instead. Clinical strains were mainly concentrated in three of the five clades. Scedosporium aurantiacum (clade 1) grouped European clinical strains exclusively. Clades 4 and 5 were the biggest ones and included numerous strains each. However, 72% of the isolates of clade 5 were clinical, while only 50% of the isolates of clade 4 had such origin. It is noteworthy that two of the three strains included in the P. ellipsoidea group (CBS 418.73, FMR 7884, and FMR 8623) had caused disseminated infections, which emphasizes the clinical relevance of this species.
Most of the terminal branches that grouped more than one isolate included clinical and environmental isolates. This seems to demonstrate that any environmental strain can cause infection under the appropriate conditions. Using a murine model of invasive infection by S. prolificans, Ortoneda et al. (15) proved that there are no virulence differences between environmental strains and those that caused colonization or infection.
As expected, these results have demonstrated that P. boydii does not represent a single species. It encompasses a high number of phylogenetic species, although only a few of them can be recognized morphologically. One of the most important findings of this work is to provide phenotypic features useful for the distinction of some of these species (Table 2). Considering that not all the hospitals have facilities for molecular diagnosis and that not all these species are equally involved in human infections, these results can be especially useful for clinical microbiologists or laboratorians in order to identify these fungi. Judging by the high clinical relevance of this fungal group, further investigation is expected in the near future. It is especially important to determine if these species, and perhaps others, that could be identified in the future using similar approaches and involving more isolates from different sources and geographical regions are equally pathogenic to humans. Knowledge of the degree of virulence of these species and their response to the antifungal drugs may also be very useful in order to choose the appropriate treatment of the severe and refractory infections attributed to P. boydii sensu lato. Furthermore, taking into account that many of these species can only be reliably separated through molecular phylogenetics of DNA sequences, finding morphological apomorphies for their laboratory identification would also be very valuable.
Acknowledgments
We are indebted to the curators of the Centraalbureau voor Schimmelcultures (Utrecht, The Netherlands), BCCM/IHEM Biomedical Fungi and Yeasts Collection (Brussels, Belgium), Mycotheque de l'Universite Catholique de Louvain (Belgium), the Institute for Tropical Medicine (Antwerp, Belgium), and the Robert Koch Institute (Berlin, Germany) and to P. Godoy (Escola Paulista de Medicina, Universidade Federal de São Paulo, São Paulo, Brazil), J. M. Torres (IMIM, Hospital del Mar, Barcelona, Spain), A. Rezusta (Hospital Universitario Miguel Servet, Zaragoza, Spain), R. Negroni (Hospital de Infecciosas Francisco Javier Muñiz, Buenos Aires, Argentina), J. Llovo (Complejo Hospitalario Universitario de Santiago de Compostela, Santiago de Compostela, Spain), and A. del Palacio (Hospital Universitario Doce de Octubre, Madrid, Spain) for supplying many of the strains used in the study.
This study was supported by the Spanish Ministerio de Ciencia y Tecnología, grant CGL 2004-00425/BOS.
Footnotes
A publication of the ECMM Working Group on Pseudallescheriasis.
REFERENCES
- 1.Capilla, J., C. Serena, F. J. Pastor, M. Ortoneda, and J. Guarro. 2004. Efficacy of voriconazole in treatment of systemic scedosporiosis in neutropenic mice. Antimicrob. Agents Chemother. 48:4009-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carrillo, A., and J. Guarro. 2001. In vitro activities of four novel triazoles against Scedosporium spp. Antimicrob. Agents Chemother. 45:2151-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castiglioni, B., D. A. Sutton, M. G. Rinaldi, J. Fung, and S. Kusne. 2002. Pseudallescheria boydii (anamorph Scedosporium apiospermum) infection in solid organ transplant recipients in a tertiary medical center and review of the literature. Medicine 81:333-348. [DOI] [PubMed] [Google Scholar]
- 4.Cazin, J., Jr., and D. W. Decker. 1964. Carbohydrate nutrition and sporulation of Allescheria boydii. J. Bacteriol. 88:1624-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cazin, J., Jr., and D. W. Decker. 1965. Growth of Allescheria boydii in antibiotic-containing media. J. Bacteriol. 90:1308-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cruse, M., R. Telerant, T. Gallagher, T. Lee, and J. Taylor. 2002. Cryptic species in Stachybotrys chartarum. Mycologia 94:814-822. [PubMed] [Google Scholar]
- 7.Defontaine, A., R. Zouhair, B. Cimon, J. Carrère, E. Bailly, F. Symoens, M. Diouri, J. N. Hallet, and J. P. Bouchara. 2002. Genotyping study of Scedosporium apiospermum isolates from patients with cystic fibrosis. J. Clin. Microbiol. 40:2108-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Hoog, G. S., J. Guarro, J. Gené, and M. J. Figueras. 2000. Atlas of clinical fungi, 2nd. ed. Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands, and University Rovira i Virgili, Reus, Spain.
- 9.Gordon, M. A. 1957. Nutrition and sporulation of Allescheria boydii. J. Bacteriol. 73:199-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gueho, E., and G. S. de Hoog. 1991. Taxonomy of the medical species of Pseudallescheria and Scedosporium. J. Mycol. Med. 1:3-9. [Google Scholar]
- 11.Kishino, H., and M. Hasegawa. 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J. Mol. Evol. 29:170-179. [DOI] [PubMed] [Google Scholar]
- 12.Koufopanou, V., A. Burt, T. Szaro, and J. W. Taylor. 2001. Gene genealogies, cryptic species, and molecular evolution in the human pathogen Coccidioides immitis and relatives (Ascomycota, Onygenales). Mol. Biol. Evol. 18:1246-1258. [DOI] [PubMed] [Google Scholar]
- 13.O'Donnell, K. 2000. Molecular phylogeny of the Nectria haematococca-Fusarium solani species complex. Mycologia 92:919-938. [Google Scholar]
- 14.O'Donnell, K., H. I. Nirenberg, T. Aoki, and E. Cigelnik. 2000. A multigene phylogeny of the Giberella fujikuroi species complex: detection of additional phylogenetically distinct species. Mycoscience 41:61-78. [Google Scholar]
- 15.Ortoneda, M., F. J. Pastor, E. Mayayo, and J. Guarro. 2002. Comparison of the virulence of Scedosporium prolificans strains from different origins in a murine model. J. Med. Microbiol. 51:924-928. [DOI] [PubMed] [Google Scholar]
- 16.Rainer, J., G. S. de Hoog, M. Wedde, I. Graser, and S. Gilges. 2000. Molecular variability of Pseudallescheria boydii, a neurotropic opportunist. J. Clin. Microbiol. 38:3267-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steinbach, W. J., and J. R. Perfect. 2003. Scedosporium species infections and treatments. J. Chemother. 15:16-27. [DOI] [PubMed] [Google Scholar]
- 18.Swofford, D. L. 2001. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods) (version 4.0). Sinauer Associates, Sunderland, Mass.
- 19.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Arx, J. A., M. J. Figueras, and J. Guarro. 1988. Sordariaceous ascomycetes without ascospore ejaculation. Beihefte Nova Hedwigia 94:1-104. [Google Scholar]
- 21.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to the methods and applications. Academic Press, New York, N.Y.