Abstract
Genotyping based on variable-number tandem repeats (VNTR) is currently a very promising tool for studying the molecular epidemiology and phylogeny of Mycobacterium tuberculosis. Here we investigate the polymorphisms of 48 loci of direct or tandem repeats in M. tuberculosis previously identified by our group. Thirty-nine loci, including nine novel ones, were polymorphic. Ten VNTR loci had high allelic diversity (Nei's diversity indices ≥ 0.6) and subsequently were used as the representative VNTR typing set for comparison to IS6110-based restriction fragment length polymorphism (RFLP) typing. The 10-locus VNTR set, potentially providing >2 × 109 allele combinations, obviously showed discriminating capacity over the IS6110 RFLP method for M. tuberculosis isolates with fewer than six IS6110-hybridized bands, whereas it had a slightly better resolution than IS6110 RFLP for the isolates having more than five IS6110-hybridized bands. Allelic diversity of many VNTR loci varied in each IS6110 RFLP type. Genetic relationships inferred from the 10-VNTR set supported the notion that M. tuberculosis may have evolved from two different lineages (high and low IS6110 copy number). In addition, we found that the lengths of many VNTR loci had statistically significant relationships to each other. These relationships could cause a restriction of the VNTR typing discriminating capability to some extent. Our results suggest that VNTR-PCR typing is practically useful for application to molecular epidemiological and phylogenetic studies of M. tuberculosis. The discriminating power of the VNTR typing system can still be enhanced by the supplementation of more VNTR loci.
Over a decade, the fingerprinting method based on restriction fragment length polymorphism (RFLP) of IS6110 insertion sequences has been established as the standard for typing strains of Mycobacterium tuberculosis. IS6110 RFLP fingerprinting is very powerful when it is used to classify M. tuberculosis isolates harboring a large number of IS6110 in their chromosomes (33). However, the prevalence of M. tuberculosis strains harboring no, single, or few copies of IS6110 in their chromosomes dramatically lowers the discriminating efficiency of the method (1). In this regard, many alternative RFLP-based fingerprinting methods, e.g., direct repeat (DR) and polymorphic GC-rich repetitive sequence RFLP fingerprinting, are supplementarily used for differentiation (10, 24). In addition, various methods based on PCR, for example, ligation-mediated PCR, mixed-linker PCR, double repetitive element PCR, and DR-based spoligotyping, were developed mainly in order to avoid the technical demand of RFLP (5, 11, 14, 21). However, most PCR-based methods displayed poor discrimination power compared to the standard IS6110 RFLP typing, whereas others were critically confronted with limitations with respect to reproducibility and reliability (16).
Variable-number tandem repeats (VNTR), often referred to as micro- or minisatellite DNA, are ubiquitous in eukaryotes and humans. They have been extensively studied in humans, and the fingerprinting methods based on them are successfully used for paternity and genetic linkage tests between human individuals (13). VNTR can be directly amplified by PCR and analyzed by agarose gel electrophoresis systems commonly available in general laboratories. Therefore, VNTR-based PCR analysis is easy, rapid, and highly specific and can be conducted by investigators worldwide. In addition, VNTR typing generates portable digit-based data, unlike the analog information obtained from RFLP-based fingerprinting methods. In this regard, investigators can easily compare the genotypic data of independent studies between different laboratories. With all of these advantages, the use of VNTR for investigating M. tuberculosis has attracted great attention from investigators in recent years.
VNTR of M. tuberculosis have been initially identified by many investigators (6, 9, 12, 20, 30). Using a homology search (27) we found 49 potential VNTR at 48 loci in the M. tuberculosis genome. Some of these repeats were equivalent to the major interspersed repeated units (MIRU) (31). In that study, we also performed in silico genome comparison and confirmed that at least 22 loci of the repeats in the two sequenced strains of M. tuberculosis, H37Rv and CDC1551, were polymorphic. In the meanwhile, a number of VNTR loci were introduced into epidemiological studies (7). At first, compared to the standard IS6110 RFLP typing method, VNTR typing was less discriminating (16), probably because too few VNTR loci were used. However, many investigators attempted to perform high-resolution VNTR typing as the number of new informative VNTR increased (17, 23, 26, 28, 31). Nowadays, the discrimination capacity of VNTR typing has been much improved, to a level comparable to that of IS6110 RFLP (18). Moreover, VNTR typing has a greater discriminating power for M. tuberculosis with low-copy-number IS6110 than RFLP typing (18).
Herewith, we report the allelic polymorphisms of all 48 VNTR loci previously identified by our group, including 13 novel ones, among M. tuberculosis strains in Thailand. Many of the novel loci were informative and could be useful for future epidemiological studies. We affirmed that VNTR typing had greater discrimination than IS6110 RFLP typing. We also found that the allelic diversity of VNTR loci could greatly vary in the different RFLP types of M. tuberculosis. In addition, this study also demonstrates correlations of the lengths of many VNTR loci.
MATERIALS AND METHODS
Mycobacterial strains.
Ninety-one initial samples of M. tuberculosis were collected from smear-positive pulmonary tuberculosis patients in Amnat Charoen Hospital in the northeastern region of Thailand during 1999 and 2000.
The hospital is a 400-bed provincial hospital of the Amnat Charoen province, which borders the southern part of Laos and is about 570 km from Bangkok, the capital of Thailand. The population in the province is about 330,000, with the majority working in the agricultural sector. There are no trains or flights to the province, but the province is readily accessible by car. The prevalence of tuberculosis in the province is about 105/100,000 population. The human immunodeficiency virus infection rate among pregnant women is 0.6%.
All of the samples were isolated by cultivation on Lowenstein-Jensen medium. M. tuberculosis H37Rv and Mt14323 were originally obtained from the National Reference Center for Tuberculosis, Canada, and Jan D. A. van Embden, respectively.
DNA extraction and IS6110 RFLP fingerprinting.
The bacteria were cultured in Lowenstein-Jensen medium for 3 weeks. The cells were then harvested, and chromosomal DNA was extracted by an enzymatic lysis method (22). Two micrograms of DNA of each isolate, and the Mt14323 strain, which was used as a marker, was then digested with PvuII and Southern blotted to a nylon filter. The filter was then hybridized with digoxigenin-labeled plasmid pDC73, which contained a segment of IS6110. The details of the methods were previously described (22).
The IS6110 hybridization patterns were analyzed by Gelcompar II version 1.5 (Applied Maths, Kortrijk, Belgium). Isolates were classified as members of the Beijing family (n = 21) based on 78% or more similarity to the previously described isolates (8). The rest of the bacteria had a single band (n = 18), a few (two to five) bands (n = 28), or more than five hybridized bands with heterogeneous banding patterns (n = 24).
VNTR polymorphism analysis.
PCR was performed for each of the 48 loci of the tandem and direct repeats that were previously identified by our group (27). The primers (Table 1) were designed from the genome sequence of M. tuberculosis H37Rv. In general, the final PCR mixture was composed of 10 mM Tris-HCl, pH 9.0, 50 mM KCl, 0.1% Triton X-100, 1.5 mM MgCl2, 200 μM concentrations of each deoxynucleoside triphosphate, a 0.5 μM concentration of each primer, 2.5 U of Thermus aquaticus (Taq) DNA polymerase enzyme, and 40 ng of DNA template in a total volume of 50 μl. Thermocycling conditions included a denaturation step at 95°C for 1 min, an annealing step at 55 to 65°C for 1 min, and an extension step at 72°C for 2 min. PCR was done for 30 cycles in a PCR gradient machine (Eppendorf, Germany). In addition, the reaction mixtures of some primer pairs were supplemented with 4% dimethyl sulfoxide for improving amplification. The amplified products were analyzed by electrophoresis in 1 to 2% agarose gels at 100 V. Visualization was done on a UV light illuminator after ethidium bromide staining. To estimate the lengths of the amplified products, the migratory distances of the amplified products were used to compare with those of DNA fragments of the λ DNA cut by PstI. The copy number of the amplified products was inferred from the difference between the molecular weights of the amplified products of the samples and those of the H37Rv strain. If there were doubts regarding the number of copies of VNTR, the amplified DNA was purified by QIAquick PCR purification kit or gel extraction kit (QIAGEN, Germany) and sequenced.
TABLE 1.
Sequences of primers used for amplifying VNTR in this studya
| Locus | Forward sequence (5′-3′) | Reverse sequence (5′-3′) | Expected size of H37Rv's amplified product |
|---|---|---|---|
| DR0272 | AGCGATCCTGCTGGTGG | TGCTGTTAGGGTCAAACG | 305 |
| DR0642 | CCACTAGCAGATGGCCGTT | GCTCCAAGCGTAGTGATCCT | 231 |
| DR2068 | CACGACGTAGACGAATGC | ATGACACGCTTTCTGCCC | 336 |
| DR3074 | GTCACGATTGACACGCGGT | CATGGCCTCCGTTGTACTC | 172 |
| DR3319 | TGGTAGGTCTGGTTCCGC | ATGTGCATCCTCAACGGG | 574 |
| DR3991 | CCAACCTAGGCGTGTTCG | GATGTTCACCCCGAATGG | 534 |
| DR4110 | TTTAGACGATCGCACCGC | AACGGAATCGTGGTCAGC | 531 |
| ETR4087 | CTGATGAGGGTCGGGATT | ACGCTCCATCTCTTATCC | 500 |
| VNTR0154 | GCTCTGTGTCAACAACCTGT | CTGGTGAACGTTACTTCG | 696 |
| VNTR0569 | TCAGCGTGTTTGTTACCC | CTCTCGCCCATACCGGA | 632 |
| VNTR0577 | TGGGGCAGGTGTTAGAGG | TTCTGAGTCCGGAACGG | 424 |
| VNTR0580 | CTGCGGTCAAACAGGTCA | CATACATCGGTACCCGAC | 463 |
| VNTR0595 | GGCGATGTTGATCGGGAT | TCTAGACGCCAATCACGC | 437 |
| VNTR0917 | CTTCGACTGTTCCAGCTGAC | TATCTTCGAGCGCGAGGACT | 436 |
| VNTR0960 | GGTGTGGACCTGTTCGGG | GTGACTTCGCCATCCACC | 357 |
| VNTR1281 | CGGACTGGGACGACTACC | AGCTGGACTCTTGCGGG | 292 |
| VNTR1305 | TGGAGGAGACCATCTCGC | CCTTGTCGAGTCGGTTGC | 344 |
| VNTR1451 | GTAGCCGTCGTCGAGAAGC | CTCGTCGTAGAGACTCAGCG | 370 |
| VNTR1644 | CTCGCACATCTCGACGG | TGATCCTCCTGATTGCCC | 425 |
| VNTR1895 | AGCAGACTGGATTTGCCG | AGTCAGTCCGCAGTTGGG | 642 |
| VNTR1907 | GAACGTTGGAAGAGATCAGCC | TACATCGGTACGCTCTCAACG | 589 |
| VNTR1955 | AGACGTCAGATCCCAGTT | ACCCGACAACAAGCCCA | 267 |
| VNTR2059 | GTTAGTATCGTCGGTTCGTG | GAAACCAATCACCGTTACATC | 371 |
| VNTR2074 | CACTAGCTGCGTCACTGG | GCTGATTCCCGACGAAAG | 533 |
| VNTR2347 | ATAAACCTCGGCGTCGAA | CGATGATGGCACACCGAA | 378 |
| VNTR2372 | ACCTCCGTTCCGATAATC | CAGCTTTCAGCCTCCACA | 298 |
| VNTR2401 | GTGGTCGCTTGAAGCCC | CACTCGGACCAACCAACC | 306 |
| VNTR2461 | GATACCGCTTCAGTATCAG | TGTCGCCCTGATGGACCT | 907 |
| VNTR2531 | CGAATTCTTCGGTGGTCTCGAGT | ACCGTCTGACTCATGGTGTCCAA | 395 |
| VNTR2687 | TATCCGTCGATCGGTTGC | CTCATGCAGCGCTGACAC | 563 |
| VNTR2703 | GTACAAAATCCGACACGG | GAGGTCTTCGACGTGGTA | 409 |
| VNTR2990 | TCAACTGGATGAGATGGG | TTTCGGGTCGAAGAACG | 524 |
| VNTR2996 | TGGAGTTCATCGGGTTCG | CTGTACGGAGCTAGCCG | 439 |
| VNTR3007 | AACGCATAGCGGACGAAAAC | GGAGCGGATCAAGACGTTAC | 338 |
| VNTR3155 | GGTGATCTGGTCCATCGC | TGTACCAGGGCCAAGACG | 599 |
| VNTR3171 | ATAAGCCGTCAGGCCAACCA | AGAGGATACGTGGATTTGAGC | 228 |
| VNTR3192 | CGAGAGAACATGCCAGGG | GTCTTGATGGCCAAAGCG | 533 |
| VNTR3232 | CGGAACTCACCAGCATCC | GCTCACCAAACTGGACGG | 585 |
| VNTR3239 | CCAGCCGTATTGAGTGTC | ACTGGATTGATGGTGATG | 571 |
| VNTR3336 | TGTCAGACGTTATCGACCAC | CGACGATGATCGAGACGAC | 399 |
| VNTR3594 | ACCAGTACGAACCAACCTGC | AACCGTGAGCTGAAGGCG | 498 |
| VNTR3690 | GTCGGGCAACGCGTCCAA | AGCACGACGAACACCTTCGA | 942 |
| VNTR3820 | TGCGCGGTGAATGAGACG | ACCTTCATCCTTGGCGAC | 444 |
| VNTR4052 | GAGCCAAATCAGGTCCGG | GAGGTATCAACGGGCTTGT | 879 |
| VNTR4120 | GTTCACCGGAGCCAACC | GAGGTGGTTTCGTGGTCG | 447 |
| VNTR4155 | GCAGATCGAGTTTTCCCAG | TTCCGGTGATGCCTGAAG | 758 |
| VNTR4156 | ACCGCAAGGCTGATGATCC | GTGCATCTCGTCGACTTCC | 704 |
| VNTR4348 | ACAAGGAGAGCGGTGTCG | CATCCTGTAGATGGCGGC | 516 |
DR, ETR, and VNTR are direct repeats, exact tandem repeats, and variable-number tandem repeats, respectively. The expected sizes of H37Rv's amplified products conform to the genomic sequences reported by Cole et al. (3) except for VNTR3336 and VNTR3690. The repeat units of H37Rv at these two VNTR loci (eight copies at VNTR3336 and five copies at VNTR3690) were found to be different from the ones deducted by the genomic sequence (five and two copies, respectively). The last four digits in the name of the locus denote the position of the locus in the genome of the H37Rv strain in kilobases (3). The exact positions of all repeat loci were given previously (27).
Analysis of VNTR allelic diversity, genetic relationships, and correlations.
The allelic diversity of each VNTR locus was evaluated by Nei's diversity index (polymorphic information content [PIC]), which is equal to 1 − Σ(allele frequencies)2 (15). Genetic relationships among the isolates were estimated by the unweighted pair group method with arithmetic averages in PAUP version 4.0 b1 software using the distance measurement according to total character differences of the copy numbers of VNTR. The correlation coefficient (r) between the numbers of copies of VNTR was calculated for every pair of VNTR loci. A statistical significance of α = 0.01 was used.
RESULTS
VNTR analysis of M. tuberculosis.
Each VNTR locus was separately amplified by a pair of primers specific for the flanking regions of the VNTR. Thirty-nine loci were polymorphic and will therefore be referred to as VNTR instead of DR or exact tandem repeats (ETR) as previously described (27). Two loci were previously described to be polymorphic by our group (VNTR0595 and VNTR4155). The variability of 30 VNTR loci in this study was also affirmed by other investigators (6, 17, 20, 31), despite the differences in the number of alleles at some VNTR loci. VNTR2059 was previously reported to be polymorphic but was not found to be variable in this study (4, 25, 31). The dissimilar result at VNTR2059 as well as the differences in the number of allelic variants at other VNTR loci between our study and the others might be mainly influenced by the total number of M. tuberculosis isolates and the composition of different lineages of the isolates in each study. Nine polymorphic loci in this study were new (Table 2). Three VNTR (VNTR3820, VNTR3232, and VNTR4052) had more than nine allelic variants. VNTR3820 had the highest number of alleles (16), with a variation of 3 to 32 copies of repetitive units (Fig. 1). Among the eight loci of tandem repeats residing inside coding regions, seven were polymorphic. These included VNTR2347 (3 alleles) in the DNA polymerase I gene (polI) and VNTR4155 (10 alleles) in the α-isopropylmalate synthase gene (leuA). The latter was shown to code for the polymorphic α-isopropylmalate synthase in M. tuberculosis (2).
TABLE 2.
PICs of the 48 potential VNTR locia
| Tandem repeat locus | Designation by other researchers | No. of alleles | Copy number | PIC for:
|
||||
|---|---|---|---|---|---|---|---|---|
| All isolates | Beijing isolates | Few-banded (2-5) isolates | Heterogeneous (≥6) isolates | Single-banded isolates | ||||
| DR0272 | MIRU3 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| DR0642 | MIRU6 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| DR2068 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | |
| DR3074 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | |
| DR3319 | MIRU33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| DR3991 | MIRU37 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| DR4110 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | |
| ETR4087 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | |
| VNTR2059 | MIRU20 | 1 | 2 | 0 | 0 | 0 | 0 | 0 |
| VNTR0154 | MIRU2 | 3 | 1-3 | 0.041 | 0 | 0.074 | 0 | 0.133 |
| VNTR0569 | 6 | 1-6 | 0.666 | 0.100 | 0.764 | 0.604 | 0.180 | |
| VNTR0577 | ETR-C | 3 | 1-3 | 0.118 | 0 | 0 | 0.269 | 0.245 |
| VNTR0580 | ETR-D, MIRU4 | 9 | 1*, 2*, 3, 4*, 5*, 6*, 7*, 9*, 10* | 0.795 | 0.172 | 0.681 | 0.753 | 0.724 |
| VNTR0595 | msx.4 | 9 | 1, 3-4, 6-7, 9-12 | 0.728 | 0 | 0.743 | 0.570 | 0.722 |
| VNTR0917 | 2 | 2-3 | 0.061 | 0 | 0.077 | 0 | 0.245 | |
| VNTR0960 | MIRU10 | 7 | 0-5, 7 | 0.619 | 0.177 | 0.334 | 0.461 | 0.449 |
| VNTR1281 | QUB-1281 | 2 | 2-3 | 0.020 | 0 | 0 | 0 | 0.133 |
| VNTR1305 | 4 | 0-3 | 0.100 | 0 | 0.074 | 0.074 | 0.133 | |
| VNTR1451 | QUB-1451 | 3 | 1-3 | 0.410 | 0 | 0.142 | 0.074 | 0.133 |
| VNTR1644 | MIRU16 | 4 | 1-4 | 0.491 | 0 | 0.269 | 0.260 | 0.521 |
| VNTR1895 | QUB-1895 | 6 | 2-7 | 0.419 | 0.529 | 0.351 | 0.346 | 0.545 |
| VNTR1907 | 2 | 1-2 | 0.021 | 0 | 0 | 0 | 0.133 | |
| VNTR1955 | Mtub21 | 8 | 0-6, 9 | 0.688 | 0.694 | 0.589 | 0.747 | 0.459 |
| VNTR2074 | Mtub24 | 4 | 1-4 | 0.291 | 0.308 | 0.210 | 0.342 | 0.245 |
| VNTR2347 | Mtub29 | 3 | 1-3 | 0.495 | 0.172 | 0.071 | 0.370 | 0.337 |
| VNTR2372 | 4 | 1-4 | 0.464 | 0.463 | 0.074 | 0.423 | 0.133 | |
| VNTR2401 | Mtub30 | 5 | 0-3, 7 | 0.387 | 0.100 | 0 | 0.074 | 0.272 |
| VNTR2461 | ETR-B | 6 | 1-6 | 0.689 | 0 | 0.611 | 0.717 | 0.314 |
| VNTR2531 | MIRU23 | 4 | 0-1, 4-5 | 0.236 | 0 | 0.254 | 0.091 | 0.142 |
| VNTR2687 | MIRU24 | 3 | 0-2 | 0.481 | 0 | 0.074 | 0.426 | 0.449 |
| VNTR2703 | 2 | 2-3 | 0.041 | 0.091 | 0.074 | 0 | 0 | |
| VNTR2990 | Mtub31 | 4 | 2-4, 17 | 0.511 | 0 | 0.077 | 0.448 | 0.439 |
| VNTR2996 | MIRU26 | 7 | 1-7 | 0.460 | 0.188 | 0 | 0.334 | 0.272 |
| VNTR3007 | MIRU27, QUB-5 | 2 | 2-3 | 0.020 | 0 | 0 | 0 | 0.133 |
| VNTR3155 | QUB-15 | 4 | 1-3, 6 | 0.159 | 0.188 | 0.150 | 0.083 | 0.219 |
| VNTR3171 | Mtub34 | 3 | 1-3 | 0.119 | 0.095 | 0.260 | 0 | 0.142 |
| VNTR3192 | ETR-E, MIRU31 | 5 | 1-5 | 0.450 | 0.180 | 0.434 | 0.603 | 0.417 |
| VNTR3232 | QUB-3232 | 11 | 0, 3, 6, 9-15, 17 | 0.425 | 0.844 | 0 | 0.310 | 0.180 |
| VNTR3239 | ETR-F | 5 | 1+1, 2+1, 1+2, 2+2, 3+2 | 0.247 | 0.331 | 0 | 0.150 | 0.520 |
| VNTR3336 | QUB-3336 | 8 | 3-8, 10-11 | 0.407 | 0.283 | 0.480 | 0.345 | 0.403 |
| VNTR3594 | 3 | 2-4 | 0.041 | 0 | 0 | 0.074 | 0.133 | |
| VNTR3690 | Mtub39 | 9 | 1-9 | 0.758 | 0 | 0.723 | 0.785 | 0.198 |
| VNTR3820 | 16 | 3-10, 12-15, 17-18, 31-32 | 0.889 | 0.642 | 0.781 | 0.720 | 0.786 | |
| VNTR4052 | QUB-26 | 10 | 1-8, 10, 4* | 0.684 | 0.449 | 0.503 | 0.633 | 0.367 |
| VNTR4120 | 9 | 1-5, 7, 10-12 | 0.755 | 0.580 | 0.556 | 0.396 | 0.531 | |
| VNTR4155 | mpp.8 | 10 | 2-3, 10-11, 14, 16-19, 21 | 0.440 | 0 | 0.701 | 0.350 | 0.656 |
| VNTR4156 | QUB-4156 | 4 | 1-4 | 0.441 | 0.472 | 0 | 0.145 | 0.255 |
| VNTR4348 | MIRU39 | 3 | 1-3 | 0.265 | 0.346 | 0.083 | 0.226 | 0.444 |
ETR-B to F are tandem repeats previously identified by Frothingham and Meeker-O'Connell (6). MIRU, QUB, and Mtub prefixes are designations of tandem repeats by Supply et al. (31), Roring et al. (23), and Le Flèche et al. (17), respectively. msx.4 and mpp.8 are terms used by Namwat et al. (20). Allelic diversity indices (PICs) were calculated for all M. tuberculosis isolates and for each IS6110 RFLP type: Beijing, few-banded (two to five bands), heterogeneous (at least six bands), and single-banded strains. The PICs of the 10 most polymorphic loci in each IS6110 type are in bold. Asterisks denote the allelic variants found to contain internal deletions of the VNTR sequence. A zero in the copy number column indicates the allelic variants possessing only the incomplete sequence. VNTR3239 is a complex locus composed of 55-bp and 79-bp repeat units of tandem repeats.
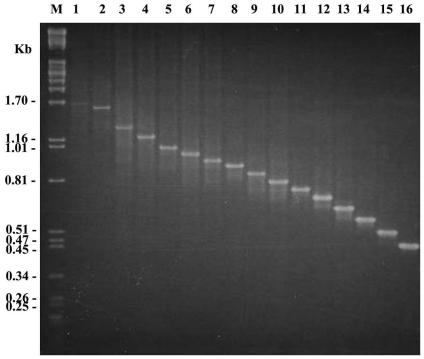
FIG. 1.
PCR products of various clinical isolates using primers that amplify VNTR3820. VNTR3820 has from 32 copies (lane 1) to 3 copies (lane 16) of the repeat unit. Lane M denotes the λ DNA cut by PstI.
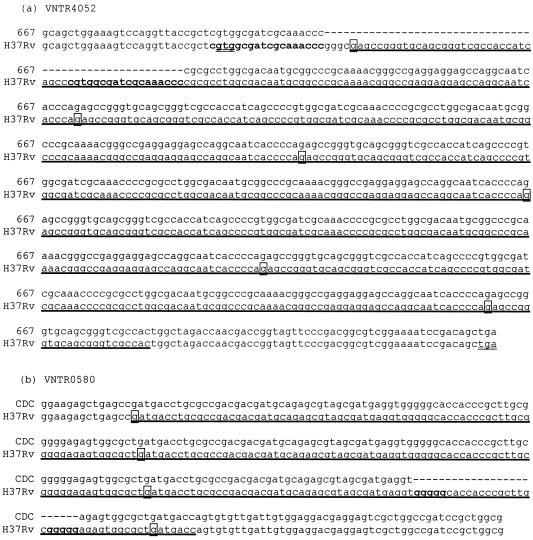
The differences between the lengths of the PCR products of the different alleles were approximately multiples of a unit length, indicating insertion or deletion of complete repetitive units, as similarly found by others (31). The numbers of copies of complete repeated units were calculated. Seven alleles of the polymorphic loci contained only incomplete repeated units. All were confirmed by sequencing (GenBank accession numbers DQ114228 to DQ114236 and DQ116947 to DQ116948). Alleles of two VNTR loci (VNTR0580 and VNTR4052) contained partial deletions. At VNTR0580, most alleles lacked a 24-bp segment from the last copy of the repeats, as also found by other researchers (31). In an allele of VNTR4052, a 54-bp segment was absent. The segment contained the first 50-bp segment of the first copy of the VNTR and the adjacent 4-bp segment (Fig. 2). The allele had a frequency of 4% in this study but was not previously reported (26). The presence of this allele was not related to any particular IS6110 type of M. tuberculosis.
FIG. 2.
Comparison of the sequences of M. tuberculosis H37Rv and the isolates containing a partial deletion of the VNTR sequence. (a) A 54-bp-long fragment of VNTR4052 and its flanking sequence are absent from isolate 667. There are two perfect 18-bp direct repeats (bold) at the border of the deleted fragment. VNTR4052 resides in a possible coding region (Rv3611). The start and stop codons of Rv3611 are double underlined. (b) A 24-bp-long fragment is absent from VNTR0580. Two pentaguanine runs are found at the border of the deleted sequence. VNTR regions are marked by thick single underlines. Boxed nucleotides and dashes denote the first nucleotide of a VNTR copy and the deleted regions, respectively.
It was noteworthy that in most VNTR loci, the found alleles distributed across the range of the possible alleles. However, at VNTR2990 and VNTR3820 there were isolates of which the number of repeats appeared to be very different from the others. At VNTR2990, while most isolates carried 2 to 4 copies of the repeat unit, one isolate carried 17 copies. There were no isolates with 5 to 16 copies. At VNTR3820, while most isolates harbored 3 to 18 copies of the repeat unit, two isolates carried 31 and 32 copies and no isolates harbored 19 to 30 copies.
Allelic diversity of VNTR loci.
The PIC of each VNTR locus was calculated (Table 2). We categorized the polymorphism arbitrarily into three levels: high (≥0.6), moderate (0.4 to <0.6), and low (<0.4). Ten loci (three novel: VNTR3820, VNTR4120, and VNTR0569) were considered high discriminants. Thirteen loci (one novel: VNTR2372) were moderate discriminants, and the remaining loci (five novel) were low discriminants. The PIC values of some VNTR loci of various IS6110 RFLP types were different. For examples, 3 of the 10 most polymorphic loci were not polymorphic at all among the Beijing strains, while the other 3 loci were lowly polymorphic. Similarly, among the single-banded isolates, the polymorphisms of 4 of the 10 most polymorphic loci were much lower than average.
Phylogenetic analysis of M. tuberculosis.
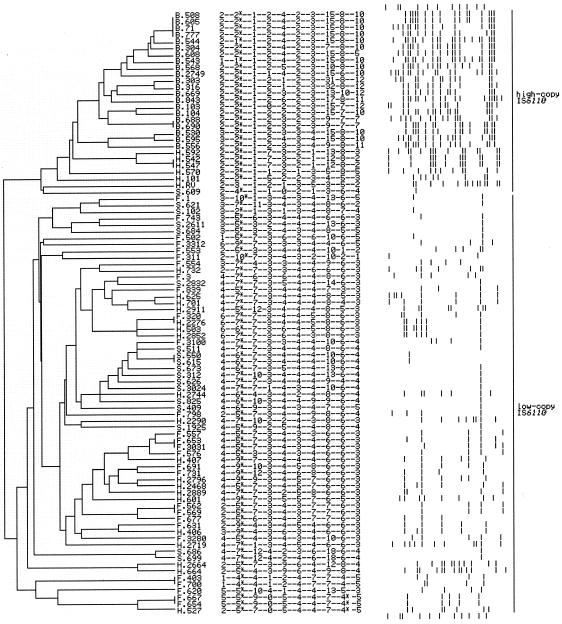
In assessing the genetic relationships between M. tuberculosis strains and the discriminating power of VNTR typing for M. tuberculosis, the 10 VNTR loci with the highest average PICs, including the three new VNTR loci, were used as the representative VNTR typing set. The set could potentially provide >2 × 109 allele combinations.
Two major separations on the phylogenetic tree were obtained. The larger group (64 isolates) did not contain any Beijing strains. Virtually all of the isolates in this group had one to eight copies of IS6110, with the exceptions of the three heterogeneous strains, which had more than eight copies. The number of copies of IS6110 appeared not to associate with any segregation in the phylogenetic tree of this group. In contrast, the smaller group contained all of the 21 Beijing strains as well as the 4 other isolates possessing >10 copies of IS6110, an 8-IS6110-copy isolate, a single-IS6110-copy isolate, and the H37Rv strain. All Beijing isolates were closely formed into a subgroup (Fig. 3).
FIG. 3.
Dendrogram of genetic relationships of 92 isolates of M. tuberculosis (denoted by numbers) based on the 10 VNTR loci. The consensus tree is built using the unweighted pair group method with arithmetic averages algorithm in the PAUP program. The isolates are classified into two separate lineages: high IS6110 copy number (more than eight IS6110 copies, except for the single-banded isolate S.609 and the eight-banded isolate H.101) and low IS6110 copy number (one to eight IS6110 copies, except for the three heterogeneous isolates H.664, H.2664, and H.2719, which have more than eight IS6110 copies). Letters B, F, H, and S denote the different IS6110 types: Beijing, few-banded, heterogeneous, and single-banded isolates, respectively. The VNTR profiles are shown in the 10-digit column, representing the number of copies of the repeat units in VNTR0569, VNTR0580, VNTR0595, VNTR0960, VNTR1955, VNTR2461, VNTR3690, VNTR3820, VNTR4052, and VNTR4120, respectively. Asterisks at VNTR0580 and VNTR4052 indicate partial deletions in the VNTR sequences. The IS6110 RFLP profiles are shown, with high to low molecular weight oriented from left to right. The top and bottom rows represent the IS6110 RFLP of the Mt14323 strain.
Discriminatory power of VNTR typing.
In total, 10-locus VNTR typing showed greater discriminating capacity than IS6110 RFLP typing. Among the 92 isolates, there were 82 VNTR and 66 RFLP patterns.
Excluding the isolates with five copies of IS6110 or less, VNTR typing gave a slightly better discrimination than IS6110 RFLP typing (of 41 distinct VNTR and 40 RFLP patterns) among the 46 isolates, whereas both methods could differentiate the isolates into 44 distinct patterns. There were 18 unique RFLP and 17 distinct VNTR patterns among the 21 Beijing strains. The isolates in the Beijing group were differentiated into 20 distinct patterns by both methods. In the heterogeneous group, VNTR typing showed a slightly better discrimination than the IS6110 method, with 24 and 22 unique patterns, respectively. VNTR typing for the 28 few-banded isolates also rendered a slightly better resolution than IS6110 RFLP typing (25 and 23 patterns, respectively). The few-banded isolates were dissolved into 26 patterns by both methods. For the 18 single-banded strains, VNTR typing clearly generated a greater number (17) of patterns than IS6110 typing (3 patterns; 1.4-, 5.0-, and 5.5-kb-long bands). Two isolates (one heterogeneous and one few-banded strain) had identical VNTR patterns.
Relationships of results of VNTR typing at different loci.
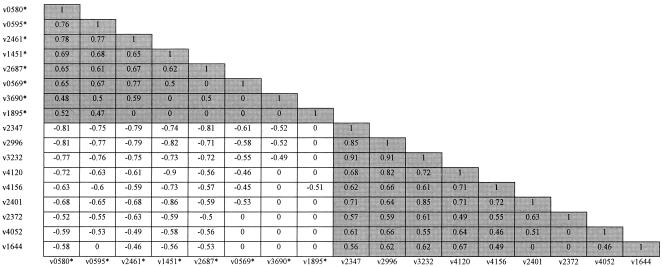
Based on the lengths of all 39 VNTR loci, a correlation coefficient of each pair of VNTR loci was calculated. Among all possible relationships, 125 pairs of correlated VNTR loci were found across 32 VNTR loci, 65 being positive and 60 being negative (for α = 0.01). Among these 32 VNTR loci, 4 correlating pairs did not correlate with any other VNTR loci; VNTR0514 and VNTR2703, VNTR0917 and VNTR1907, VNTR1305 and VNTR2531, and VNTR3007 and VNTR3336. Twenty-three loci had different degrees of either positive or negative correlations to more than one other locus. It was interesting that there were 17 loci forming two major correlating groups. Each group was composed of several VNTR loci that had positive correlations to the others in the same group (Fig. 4). However, members of these two groups were negatively correlated. It was interesting that 7 of the 10 highest discriminants were within these two groups as well. The last locus (VNTR2074) had a correlation with only VNTR0960, although the latter correlated with others. Seven loci did not correlate to any other loci at all: VNTR0577, VNTR1955, VNTR3155, VNTR3171, VNTR3192, VNTR3239, and VNTR4155.
FIG. 4.
The lengths of 17 VNTR loci appear to correlate, and these loci can be classified into two groups. Correlation coefficients (r; in boxes) were calculated for each pair of VNTR loci based on the lengths of the alleles. Negative and positive r values indicate negative and positive correlations, respectively. Zeros denote statistically insignificant correlations. Two major groups of VNTR loci, one marked with asterisks and the other not marked, represent the loci having positive correlations to one another in the same group (shaded cells). However, VNTR loci of these two groups are negatively correlated to each other (unshaded cells).
DISCUSSION
The polymorphisms of 48 potential VNTR loci previously identified by us (27) were simultaneously investigated and characterized here. It was demonstrated in this study that among M. tuberculosis isolates, there were variations of the repeat units in the majority of the repeats (39 loci), including VNTR in 9 novel loci. Most VNTR studied here were present in the noncoding or intergenic regions of the M. tuberculosis chromosome. Intriguingly, almost all of the coding tandem repeats were also variable and therefore likely to code polymorphic proteins. These polymorphic proteins may play a role in the adaptive mechanism of M. tuberculosis, for instance, in protecting the bacterium from the defensive barriers of the human host. Examples of the VNTR found in the possible surface protein coding regions of M. tuberculosis were recently reported (26). The polymorphic proteins caused by these VNTR may be potential sources of antigenic variation allowing the bacteria to evade the immune response.
Short mutations in the different copies of the VNTR sequence, which consist of few substitutions, insertions, and deletions (indels), could be seen in many VNTR loci of M. tuberculosis (23, 31). In contrast, long indels in the VNTR sequence were rarely found. To our knowledge, up to now, only one VNTR locus (VNTR0580 or MIRU4) has been reported to have such a long mutation, which was confirmed in this study (27, 31). This study revealed an additional locus (VNTR4052) having a 54-nucleotide deletion in the VNTR and the nearby flanking sequence. Sequence analysis revealed that the deletion at VNTR4052 was most likely caused by the homologous recombination of the perfect 18-bp direct repeats lying exactly at the boundaries of the deleted sequence (Fig. 2). The proportion of the isolates containing the deletion in the allele of VNTR4052 was relatively low and had no correlation with IS6110 patterns, suggesting that deletion in this region may not have resulted from a single mutational event. Although VNTR4052 resided in a possible coding unit (Rv3611), such internal deletion did not affect the translational frame of the remaining coding sequence. Internal deletion was also observed in alleles of VNTR0580 in this study, possibly associating with the notable pentaguanine (G5) direct repeats lying at the border of the deleted sequence (Fig. 2). However, the underlying cause of this deletion could not be explained by the general recombination process.
The deletion in VNTR4052 also suggests that homologous recombination between different repeat units of the VNTR sequence can occur and contribute to the allelic variation of VNTR. The recombination can either reduce or increase the copy number. The latter case occurs if a double crossover between two DNA strands occurs after the chromosome replication fork has passed the site but before the two daughter chromosomes have been partitioned into the two daughter cells. Alternatively, the addition and deletion of VNTR copies could be generated from the DNA polymerase slippage process, as also suggested from a previous study (32). This mechanism is typically characterized by the stepwise alteration of the complete repeat unit in the variable alleles. In this study, many alleles of VNTR loci had only the incomplete copy and lacked a complete repeat unit. The isolates without the complete repeat unit should not be able to regain the variability property, and therefore VNTR in these loci are practically lost.
The polymorphism of each tandem repeat locus was found to be different. The degree of polymorphism may relate inversely with the selective pressure acting on each locus. However, there appeared to be no differences between the degrees of polymorphism of VNTR residing inside and outside coding sequences. In the absence of selective constraints, one may assume that the diversity of a particular VNTR is correlated with its evolutionary rate. Therefore, a VNTR locus with a high level of polymorphism would be assumed to have a faster evolutionary rate than a locus with a lower level. If so, the rate seemed not to relate to the length of the repeat unit or the position in the genome.
Many VNTR loci in this study had moderate or high allelic diversity (PIC ≥ 0.4). These VNTR loci are useful for the differentiation of M. tuberculosis strains. It was shown in this study that the 10-VNTR set had a resolution comparable to standard IS6110 RFLP typing when tested with the panel of M. tuberculosis isolates containing high copy numbers (more than five) of IS6110; meanwhile, this VNTR set could remarkably differentiate the isolates having only one IS6110 copy. These findings are similar to those of other studies using different VNTR typing sets and M. tuberculosis isolates from various geographical origins (18, 29), which also suggests that VNTR typing is suitable for use in the global epidemiological study of tuberculosis.
Molecular genotyping based on VNTR-PCR analysis has several advantages over standard IS6110 RFLP and other typing methods (16, 19). Also, an individual VNTR locus can be examined independently, giving the investigators the flexibility of using it to modify and improve their own typing format. We found that many VNTR had biased diversity for each IS6110 RFLP type; for instance, VNTR3232 gave the maximal diversity for Beijing strains, whereas this locus showed very low or no discriminating value for the remaining RFLP types. This strain-dependent property of VNTR can be beneficial to the investigation of an outbreak caused by a particular IS6110 type of M. tuberculosis.
VNTR-based analysis can be used to infer the phylogenetic relationships of M. tuberculosis strains. A dendrogram constructed from the 10-VNTR set agrees with the hypothesis that M. tuberculosis may have evolved from two separate lineages, the high- and low-IS6110-copy-number isolates. In this study, the high-IS6110-copy-number group was mostly composed of the isolates having >10 IS6110 copies (Fig. 3). All Beijing isolates were included in this high-IS6110-copy-number group, which conforms to the notion that the Beijing strains constitute a homogeneous M. tuberculosis family. At the same time, the isolates in the low-copy group comprised those possessing one to four copies of IS6110 as well as those having five to eight IS6110 copies. The latter were classified in the high-copy lineage in a previous analysis (18).
In this study, many VNTR loci exhibited significant copy number correlations. These correlations may be caused by several factors. We observed that correlations of VNTR loci could occur between loci with different degrees of allelic diversity. It is probable that the correlations between VNTR loci having low allelic diversity may represent false correlations occurring from the combined effect of the small number of allele combinations between the VNTR loci and the biased allele frequencies of the VNTR loci. In contrast, significant relationships of the VNTR loci having relatively high allelic diversity were unlikely to occur artificially due to a large number of allele combinations between the VNTR loci and should represent the actual correlations. This may happen because the bacterial populations in the study were of multiple clonal origins and M. tuberculosis possesses few mechanisms for horizontal genetic exchanges. If the number of copies of the repeats varies gradually, the correlation of the lengths between some sites may just be the result of the fact that the ancestors originally had VNTR of different lengths at those sites. Alternatively, the correlated VNTR may be subject to the same biological constraint, such as binding to the same proteins, and therefore tend to evolve in the same way. Similar associations between different VNTR loci were also previously recognized in M. tuberculosis (32).
Correlations of VNTR could limit the number of possible patterns among M. tuberculosis isolates. As a consequence, these relationships would reduce to some extent the potential discriminating power of the VNTR typing system composed of the correlating VNTR loci.
The present VNTR typing systems could not define all unique isolates and still require the complementation of other typing methods, such as IS6110 RFLP. The power of the VNTR typing system can be improved by the supplementation of an extra number of VNTR loci. It was shown in this study that our 10-VNTR set was powerful for distinguishing M. tuberculosis isolates. This VNTR set has already been developed in our laboratory for application in the molecular epidemiological study of M. tuberculosis. We adopted multiplex PCR with three separate sets of primers to target those 10 VNTR loci, in conjunction with analysis by agarose gel electrophoresis. Without the requirement of the automated gel analysis equipment, our system is inexpensive and so can be exploited by general researchers. We are now applying this 10-VNTR typing format to examining different M. tuberculosis populations in Thailand.
Supplementary Material
Acknowledgments
We thank Tada Juthayothin and Chaiporn Thangthong for their help in software utilization.
This work was supported by the National Science and Technology Development Agency (NSTDA), Thailand.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Bauer, J., A. B. Andersen, K. Kremer, and H. Miorner. 1999. Usefulness of spoligotyping to discriminate IS6110 low-copy-number Mycobacterium tuberculosis complex strains cultured in Denmark. J. Clin. Microbiol. 37:2602-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chanchaem, W., and P. Palittapongarnpim. 2002. A variable number of tandem repeats result in polymorphic α-isopropylmalate synthase in Mycobacterium tuberculosis. Tuberculosis 81:1-6. [DOI] [PubMed] [Google Scholar]
- 3.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McClean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. Al Rajandream, J. Rodgers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 4.Cowan, L. S., L. Mosher, L. Diem, J. P. Massey, and J. T. Crawford. 2002. Variable-number tandem repeat typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110 by using mycobacterial interspersed repetitive units. J. Clin. Microbiol. 40:1592-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman, C. R., M. Y. Stoeckle, W. D. Johnson, Jr., and L. W. Riley. 1995. Double-repetitive-element PCR method for subtyping Mycobacterium tuberculosis clinical isolates. J. Clin. Microbiol. 33:1383-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable number of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 7.Frothingham, R. 1995. Differentiation of strains in Mycobacterium tuberculosis complex by DNA sequence polymorphisms, including rapid identification of M. bovis BCG. J. Clin. Microbiol. 33:840-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gillespie, S. H., N. Kennedy, F. I. Ngowi, N. G. Fomukong, S. al-Maamary, and J. W. Dale. 1995. Restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolated from patients with pulmonary tuberculosis in northern Tanzania. Trans. R. Soc. Trop. Med. Hyg. 89:335-338. [DOI] [PubMed] [Google Scholar]
- 9.Goyal, M., D. Young, Y. Zhang, P. A. Jenkins, and R. J. Shaw. 1994. PCR amplification of variable sequence upstream of katG gene to subdivide strains of Mycobacterium tuberculosis complex. J. Clin. Microbiol. 32:3070-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groenen, P. M., A. E. Bunschoten, D. van Soolingen, and J. D. van Embden. 1993. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis: application for strain differentiation by a novel typing method. Mol. Microbiol. 10:1057-1065. [DOI] [PubMed] [Google Scholar]
- 11.Haas, W. H., W. R. Butler, C. L. Woodley, and J. Crawford. 1993. Mixed-linker polymerase chain reaction: a new method for rapid fingerprinting of isolates of the Mycobacterium tuberculosis complex. J. Clin. Microbiol. 31:1293-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hermans, P. W. M., D. van Soolingen, and J. D. A. van Embden. 1992. Characterization of a major polymorphic tandem repeat in Mycobacterium tuberculosis and its potential use in the epidemiology of Mycobacterium kansasii and Mycobacterium gordonae. J. Bacteriol. 174:4157-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeffreys, A. J., V. Wilson, and S. L. Thein. 1985. Hypervariable “minisatellite” regions in human DNA. Nature 314:67-73. [DOI] [PubMed] [Google Scholar]
- 14.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Sooligen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. D. A. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 1997. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. M. Hermans, C. Martin, P. Palittappongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. A. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Flèche, P., M. Fabre, F. Denoeud, J. L. Koeck, and G. Vergnaud. 2002. High resolution, on-line identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. BMC Microbiol. 2:37-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazars, E., S. Lesjean, A. U. Banuls, M. Gillbert, V. Vincent, B. Gicquel, M. Tibayrenc, C. Locht, and P. Supply. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. USA 98:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moström, P., M. Gordon, C. Sola, M. Ridell, and N. Rastogi. 2002. Methods used in the molecular epidemiology of tuberculosis. Clin. Microbiol. Infect. 8:694-704. [DOI] [PubMed] [Google Scholar]
- 20.Namwat, W., P. Luangsook, and P. Palittapongarnpim. 1998. The genetic diversity of Mycobacterium tuberculosis strains in Thailand studied by amplification of DNA segments containing direct repetitive sequences. Int. J. Tuberc. Lung Dis. 2:153-159. [PubMed] [Google Scholar]
- 21.Palittapongarnpim, P., S. Chomyc, A. Fanning, and D. Kunimoto. 1993. DNA fingerprinting of Mycobacterium tuberculosis isolates by ligation-mediated polymerase chain reaction. Nucleic Acids Res. 21:761-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palittapongarnpim, P., P. Luangsook, S. Tansuphaswadikul, C. Chuchottaworn, R. Prachaktam, and B. Sathapatayavongs. 1997. Restriction fragment length polymorphism study of Mycobacterium tuberculosis in Thailand using IS6110 as probe. Int. J. Tuberc. Lung Dis. 1:370-376. [PubMed] [Google Scholar]
- 23.Roring, S., A. Scott, D. Brittain, I. Walker, G. Hewinson, S. Neill, and R. Skuce. 2002. Development of variable-number tandem repeat typing of Mycobacterium bovis: comparison of results with those obtained by using existing exact tandem repeats and spoligotyping. J. Clin. Microbiol. 40:2126-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross, B. C., K. Raios, K. Jackson, and B. Dwyer. 1992. Molecular cloning of a highly repeated DNA element from Mycobacterium tuberculosis and its use as an epidemiological tool. J. Clin. Microbiol. 30:942-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savine, E., R. M. Warren, G. D. van der Spuy, N. Beyers, P. D. van Haden, C. Locht, and P. Supply. 2002. Stability of variable-number tandem repeats of mycobacterial interspersed repetitive units from 12 loci in serial isolates of Mycobacterium tuberculosis. J. Clin. Microbiol. 40:4561-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skuce, R. A., T. P. McCorry, J. F. McCarroll, S. M. M. Roring, A. N. Scott, D. Brittain, S. L. Hughes, R. G. Hewinson, and S. D. Neill. 2002. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology 148:519-528. [DOI] [PubMed] [Google Scholar]
- 27.Smittipat, N., and P. Palittapongarnpim. 2000. Identification of possible loci of variable number of tandem repeats in Mycobacterium tuberculosis. Tuberc. Lung Dis. 80:69-74. [DOI] [PubMed] [Google Scholar]
- 28.Spurgiesz, R. S., T. N. Quitugua, K. L. Smith, J. Schupp, E. G. Palmer, R. A. Cox, and P. Keim. 2003. Molecular typing of Mycobacterium tuberculosis by using nine novel variable-number tandem repeats across the Beijing family and low-copy-number IS6110 isolates. J. Clin. Microbiol. 41:4224-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Supply, P., J. Magdalena, S. Himpens, and C. Locht. 1997. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol. Microbiol. 26:991-1003. [DOI] [PubMed] [Google Scholar]
- 31.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36:762-771. [DOI] [PubMed] [Google Scholar]
- 32.Supply, P., R. M. Warren, A. L. Banuls, S. Lesjean, G. D. van der Spuy, L. A. Lewis, M. Tibayrenc, P. D. van Helden, and C. Locht. 2003. Linkage disequilibrium between minisatellite loci supports clonal evolution of Mycobacterium tuberculosis in a high tuberculosis incidence area. Mol. Microbiol. 47:529-538. [DOI] [PubMed] [Google Scholar]
- 33.van Soolingen, D., P. E. W. de Haas, P. W. M. Hermans, P. M. A. Groenen, and J. D. A. van Embden. 1993. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J. Clin. Microbiol. 31:1987-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.