Abstract
In 2004, six puppies and one adult dog from a total of four premises were subjected to necropsy evaluation. For five of the seven dogs, disease caused by canine distemper virus (CDV) infection was suspected based on clinical signs. In all of the dogs, a diagnosis of CDV infection was established by the presence of compatible gross and histologic lesions, immunohistochemical labeling for CDV antigen, and detection of CDV RNA by reverse transcription-PCR. To further characterize the CDV strains detected in the four cases, complete gene sequences were determined for the hemagglutinin (H) and fusion (F) protein genes, while partial gene sequencing was performed for the phosphoprotein gene. A total of 4,508 bases were sequenced for the CDV strains detected from each of the four cases. Two cases were found to have identical sequences except for 2 bases in the intergenic region of the F and H genes. Phylogenetic analysis strongly suggested an evolutionary relationship between sequences detected in these two cases and those of phocine distemper virus 2 and two other strains of CDV not previously detected in the continental United States. Clear phylogenetic relationships were not established for viruses detected in the two additional cases; however, one strain showed similarity to CDV strains detected in a panda from China. Importantly, the three CDV strains detected were demonstrated to be genetically distinct from known vaccine strains and strains previously reported in the continental United States.
Canine distemper virus (CDV) is the etiological agent of one the most important viral diseases of wild and domestic Canidae (dog, dingo, fox, coyote, jackal, and wolf) (2). It occurs worldwide and produces high morbidity and mortality in immunologically naïve populations (22, 29). This virus also infects a broad range of other animals, such as Mustelidae (ferrets, minks, skunks, weasels, and badgers), Procyonidae (raccoons and pandas), Ursidae (bears), Viverridae (civets, genets, and linsangs), hyaenidae (hyenas), and Felidae (lions and tigers) (2, 3, 4, 10, 12, 16, 36). Canine distemper virus is classified in the genus Morbillivirus within the family Paramyxoviridae and has an unsegmented, negative-sense, single-stranded, ∼15.7-kb RNA genome and an enveloped virus particle that is 150 to 300 nm in diameter (29). The pathogenesis of CDV infection in dogs has been well characterized (2, 22, 29, 35). The genome of CDV encodes the following virion proteins: matrix (M), fusion (F), hemagglutinin (H), nucleocapsid (N), polymerase (L), and phosphoprotein (P). The H gene protein is responsible for viral attachment to the cell host (38) and may also play a role in induction of protective immunity (11). The H protein is one of the most variable morbillivirus proteins and thus has often been used to assess genetic changes between CDV isolates (8, 9, 17, 18, 20, 21, 23, 26, 28, 30).
In this study, postmortem findings, immunohistochemical labeling, and reverse transcription (RT)-PCR specific for CDV genomic RNA established a definitive diagnosis of disease caused by CDV infection in four naturally occurring canine cases in Missouri that occurred between June and October 2004. Approximately 4.5 kb of genomic nucleotide sequence was determined for each of these four cases. Phylogenetic analysis performed on complete (F and H genes) and partial (P gene) gene sequences demonstrated that three genetically distinct CDV strains were detected and that these putative isolates were distant from those previously found in North America and most closely related to isolates previously described in either Asia or Europe.
MATERIALS AND METHODS
Cases.
Between June and October 2004, seven dogs were submitted to the University of Missouri's Veterinary Medical Diagnostic Laboratory for routine necropsy examination following death or euthanasia. The dogs originated from four unrelated breeding premises within the state of Missouri and had no history of recent travel. Multiple tissues from individual animals were fixed for 48 to 72 h in 10% neutral buffered formalin, embedded in paraffin, and processed with hematoxylin-eosin stains for routine histologic examination. For immunohistochemical staining, sections of paraffin-embedded tissue blocks were deparaffinized and then mounted on treated slides, which were then steamed in citrate buffer, pH 6.0 (DAKO-Cytomation, Carpenteria, CA). The steamed slides were incubated for 30 min with a 1:800 dilution of anti-CDV monoclonal antibody (DV2-12; Custom Monoclonal, Inc., West Sacramento, CA) targeting the nucleocapsid protein. Envision Plus was used as the detection system with DAB Plus (DAKO-Cytomation, Carpenteria, CA) as the chromogen. Slides were counterstained with Mayer's hematoxylin (Newcomer Supply, Middleton, WI).
Nucleic acid extraction and RT-PCR amplification.
Unfixed tissues were collected at necropsy and homogenized in phosphate-buffered saline, pH 7.4. The RNA extractions were performed with the NucleoSpin RNA II kit (BD Biosciences, Inc., Palo Alto, CA) according to the manufacturer's instructions. For each RNA extraction step, strict protocols were followed to avoid cross-contamination of samples (e.g., sample extraction was performed for samples from only one case at a time, aerosol barrier pipette tips were used for all pipetting steps, gloves were changed between extractions of cases, and a negative extraction control was incorporated with every set of samples). The RNA was stored at −80°C until it was used as the template for RT-PCR amplification.
For the initial diagnosis of CDV infection, amplification of RNA by RT-PCR was performed with oligonucleotide primers previously reported for diagnosis of CDV infection in dogs (15). For nucleotide sequencing, RT-PCR amplification of RNA purified directly from tissues collected at necropsy was performed with the oligonucleotide primers shown in Table 1 and targeted to the P, F, or H gene. All RT-PCR amplifications were performed using each primer at a final concentration of 0.6 μM in a 20-μl reaction mixture with 0.8 μl of QIAGEN one-step RT-PCR enzyme mix in the manufacturer's buffer containing deoxynucleoside triphosphates (0.4 μM each, final concentration). The thermocycling conditions for amplification were 50°C (40 min) and 95°C (12 min), followed by 10 cycles of denaturation (95°C; 30 s), annealing (68°C; 20 s), and extension (72°C; 90 s), with the annealing temperature in these cycles reduced by 1°C each cycle. An additional 35 cycles of denaturation (95°C; 30 s), annealing (54°C; 20 s), and extension (72°C; 90 s) were performed, followed by a final extension (72°C; 7 min). Amplification products were separated in a 1.5% agarose, 1× Tris-acetate-EDTA gel and visualized by ethidium bromide staining and UV transillumination (33). For each step in RT-PCR amplification and analysis, strict protocols were followed to prevent cross-contamination of samples (e.g., use of dedicated pipettes for each step, use of aerosol-barrier pipette tips for all pipetting steps, changing gloves between steps, and performing all postamplification procedures in a room that was physically separate from that used for RT-PCR assembly). Negative control reactions (no template added) were demonstrated to be negative before subsequent analysis of amplification products.
TABLE 1.
Oligonucleotide primers used for RT-PCR amplification and subsequent nucleotide sequencing
| Primer no. | Orientation | Sequence (5′ to 3′) | Nucleotide positiona | Target region |
|---|---|---|---|---|
| 1 | Sense | ACCAGGACCTGGAATACG | 2106-2123 | P gene |
| 2 | Antisense | GAGAAAAGCTCATCATCG | 2721-2738 | |
| 3 | Sense | ACAGGTCAACCAGGTCCA | 4873-4890 | F gene |
| 4 | Sense | GCATCGGAATAGCCAGTC | 5200-5217 | |
| 5 | Antisense | CAGTTTTATGACCAAGTA | 5427-5444 | |
| 6 | Sense | TCAACAACGAACTCGTCC | 5815-5832 | |
| 7 | Antisense | GGGCCAAATATTGACAAC | 5909-5926 | |
| 8 | Sense | CATCTGTAGCCAGAACTCC | 6293-6311 | |
| 9 | Sense | TATTGCCTCCGATACCTG | 6410-6427 | |
| 10 | Antisense | GCAGGTATCGGAGGCAAT | 6411-6428 | |
| 11 | Sense | GTCTCCTCAGTGTTCCTA | 6757-6774 | |
| 12 | Antisense | AATGTCCGTTGGTAGCGTC | 6826-6844 | |
| 13 | Antisense | AATGCCGGATCGACCTTA | 6857-6874 | |
| 14 | Sense | GTCCTTCTCATCCTACTGG | 7199-7217 | H gene |
| 15 | Sense | ACTTCCGCGATCTCCACT | 7372-7389 | |
| 16 | Antisense | AGTGGAGATCGCGGAAGT | 7372-7389 | |
| 17 | Antisense | ACACTCCGTCTGAGATAGC | 7742-7760 | |
| 18 | Sense | TCTCAGACGGCGTGTATG | 7746-7763 | |
| 19 | Sense | TCGACACTCGAGAGATTC | 7806-7823 | |
| 20 | Antisense | GCATGTCATTCAGCCACC | 7851-7868 | |
| 21 | Sense | CATCTTATGGGCGGTTGA | 8289-8306 | |
| 22 | Antisense | GTGAACTGGTCTCCTCTA | 8378-8395 | |
| 23 | Antisense | TGCCTAAGGCCAATTGAG | 8921-8938 | |
| 24 | Antisense | CTGTAAGGGATTTCTCAC | 8950-8967 |
Nucleotide position based on the genome of strain A75/17 (GenBank accession number AF164967).
For nucleotide sequencing, amplification products were excised from agarose gels and purified with the Wizard SV kit (Promega, Inc., Madison, WI) according to the manufacturer's instructions. Purified products were sequenced directly with the oligonucleotide primers used for RT-PCR amplification. Nucleotide sequencing was performed by the DNA core facility, University of Missouri—Columbia. Overlapping sequences, excluding the oligonucleotide primer sequences for the respective fragments, were assembled into contiguous sequences of 540 bp (P gene) and 3,968 bp (F and H genes plus the intergenic region) for each of the four cases studied. For all sequences obtained, a minimum of four overlapping sequencing reactions were performed and subsequently analyzed. Nucleotide sequence assembly and alignments were performed with DNAStar software (DNAStar, Inc., Madison, WI), and BLAST analysis (1) was used to search the public domain database.
Alignments and phylogenetic analysis.
Nucleotide and deduced amino acid sequences were aligned with CLUSTAL X (version 1.8) software. Sequences for comparison were obtained from GenBank and corresponded to nucleotide sequences of the CDV P, F, and H genes and deduced protein sequences. Phylogenetic analyses of nucleotide and amino acid alignments were performed using distance matrix methods (DNADIST or PRODIST, followed by NEIGHBOR) and maximum-parsimony methods (DNAPARS or PROTPARS) within the PHYLIP software package (13, 14). Data sets were subjected to bootstrap analysis, based on 100 resamplings of the original data set, using the SEQBOOT program within the PHYLIP software package (13, 14) to produce a majority rule consensus tree. Completed tree files were visualized using TreeView 1.5 (31).
Nucleotide sequence accession numbers.
The CDV nucleotide sequences that were obtained in this study have been deposited in the National Center for Biotechnology Information database and assigned GenBank accession numbers AY964107 (case 18133; P gene), AY964108 (case 18133; F and H genes), AY964109 (case 19876; P gene), AY964110 (case 19876; F and H genes), AY964111 (case 21261; P gene), AY964112 (case 21261; F and H genes), AY964113 (case 25259; P gene), and AY964114 (case 25259; F and H genes).
RESULTS
Clinical, gross, histologic, and immunohistochemical findings.
Mild to marked upper respiratory disease and neurological dysfunction were the most significant clinical signs observed in five of the seven animals. In two puppies (case 21261), upper respiratory disease was the only clinical sign observed before they were found dead by the owner. Common gross lesions observed included mild to severe conjunctivitis and rhinitis, pulmonary congestion, and pneumonia plus severe thymic atrophy. No gross lesions were noted for one animal (case 25259). Histologic findings (data not shown) of encephalitis, often with demyelination of the white matter of the cerebellum and necrosis, and lymphoid depletion were observed in five of the seven animals examined. Within the areas of necrosis in neurological tissues, astrocytes and glial cells contained droplet-like 2- to 5-μm eosinophilic intranuclear and/or intracytoplasmic viral inclusions. All seven dogs had mild to severe interstitial pneumonia. Viral inclusions were present in numerous bronchiolar epithelial cells, alveolar macrophages, and type II pneumocytes. Mild to marked lymphoid depletion or necrosis was present in sections of thymus, spleen, and lymph nodes of all dogs examined. Viral inclusions were observed in a variety of other tissues, such as the urinary bladder, stomach, and conjunctiva. Positive staining for CDV antigen was present in tissue sections of the brain, lung, spleen, urinary bladder, and stomach in all cases following immunohistochemical staining with an anti-CDV monoclonal antibody. Antemortem serology for detection of anti-CDV antibody was performed for case 19876 using serum dilutions in an indirect immunofluorescence assay. The presence of CDV-specific immunoglobulin M (IgM) and IgG antibodies was detected at dilutions of 1:320 and 1:80, respectively.
RT-PCR and sequence analysis.
Extracted RNA from necropsy tissue samples of all seven dogs from the four cases (which represented four independent premises) were positive for CDV, yielding an amplification product of the expected size (287 bp) using oligonucleotide primers previously described for RT-PCR detection of CDV infection in dogs (15). Samples of the same RNA from a single dog representing each of the four cases (and thus each of the four premises) were subsequently subjected to additional RT-PCR amplification using oligonucleotide primers (Table 1) designed to amplify the F and H genes, plus the intergenic space, and the central region of the P gene. Overlapping amplification products ranging in size from 562 to 1,229 bp were produced, using multiple combinations of the oligonucleotide primers shown in Table 1. The nucleotide sequence of each fragment was determined by direct sequencing of the RT-PCR products. Assembly of the nucleotide sequence data yielded full gene sequences for the F and H genes plus the intergenic region (3,968 bp) and partial sequence (540 bp) for the P gene from each of the four cases.
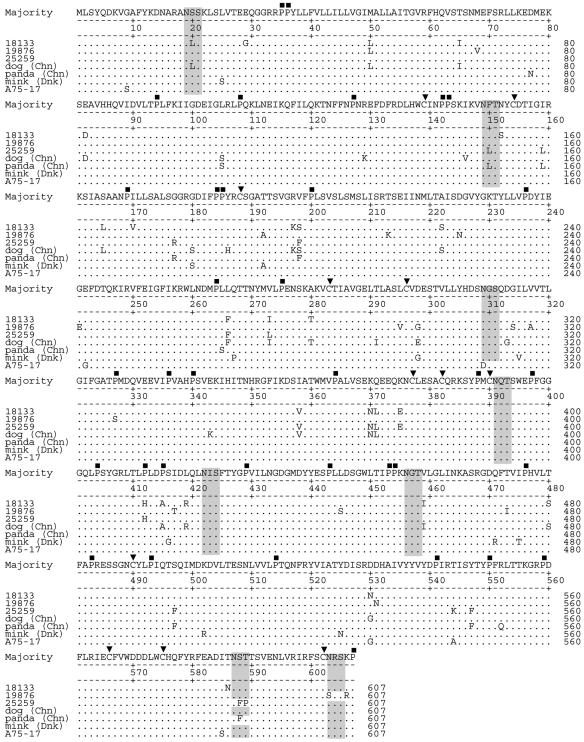
Alignment of the H gene nucleotide and predicted amino acid sequences demonstrated that the nucleotide sequences of the CDVs detected in cases 18133 and 21261 were 100% identical. In contrast, predicted H gene amino acid sequences from these cases were 93.4% and 95.6% identical to those from cases 19876 and 25259, respectively, while case 19876 was 94.6% identical to 25259 (Table 2). Following alignment, similar levels of nucleotide sequence identity were observed between the CDVs detected in these cases (Table 2). Alignment of sequences to CDV sequences from the public databases (Fig. 1) demonstrated identity levels ranging from 93.1% to 97.9% for the predicted amino acid sequences, with similar levels of identity for the nucleotide sequences (Table 2). A total of eight potential N-linked glycosylation sites were identified in the predicted H gene amino acid sequences with either seven (cases 19876 and 25259) or eight (cases 18133 and 21261) of these sites conserved (Fig. 1). Additionally 12 of 12 cysteine residues, 33 or 34 of 35 proline residues, and 33 or 34 of 35 glycine residues were conserved among the viruses detected in the cases in this study when compared by amino acid alignment to other CDV isolates (Fig. 1). Phylogenetic analysis of the H gene nucleotide sequences plus others from the databases using distance matrix methods (Fig. 2) demonstrated that cases 18133 and 21261 were placed into a distinct clade with phocine distemper virus 2 (PDV-2) and CDVs detected in dogs from China and Greenland. A bootstrap value of 100 for this clade suggests robust phylogenetic grouping. Phylogenetic analysis of the H gene nucleotide sequences of case 25259 placed the virus detected closest to that from a lesser panda, which was located in China. Phylogenetic analysis using maximum-parsimony analysis of H gene nucleotide sequences yielded equivalent results for CDV strains detected in cases 18133, 21261, and 25259 (data not shown). By distance matrix methods, analysis of the H gene sequence from case 19876 weakly grouped the virus detected from this case with that detected in a mink from Denmark (GenBank accession number Z47759) (Fig. 2). Maximum-parsimony analysis (data not shown) also implied a weak relationship between H gene amino acid sequences from case 19876 and the same CDV strain from a mink.
TABLE 2.
Nucleotide and deduced amino acid sequence identities of CDV H protein genes
| Virus | % Identitya with:
|
||||||
|---|---|---|---|---|---|---|---|
| 18133b | 19876 | 25259 | Dog (Chn) | Panda (Chn) | Mink (Dnk) | A75/17 | |
| 18133b | 93.4 | 95.6 | 97.4 | 94.2 | 94.1 | 95.1 | |
| 19876 | 93.5 | 94.6 | 93.1 | 95.4 | 96.0 | 96.2 | |
| 25259 | 96.7 | 95.3 | 94.4 | 97.9 | 95.6 | 96.5 | |
| Dog (Chn) | 98.1 | 93.2 | 95.8 | 94.1 | 94.1 | 94.7 | |
| Panda (Chn) | 95.7 | 95.8 | 98.6 | 95.8 | 96.4 | 97.2 | |
| Mink (Dnk) | 95.0 | 95.8 | 96.8 | 94.8 | 97.5 | 97.2 | |
| A75/17 | 95.5 | 95.8 | 97.3 | 95.2 | 98.0 | 97.4 | |
Values for deduced amino acid sequence identities are in boldface. Chn, China; Dnk, Denmark.
Values shown are identical for cases 18133 and 21261.
FIG. 1.
Alignment of deduced amino acid sequences from hemagglutinin genes of CDV strains. Only amino acids that differ from the majority sequence are shown. Identical residues are shown by dots. Potential N-linked glycosylation sites (N-X-S/T) are shaded. Cysteine (▾) and proline (▪) residues are indicated. The predicted amino acid sequences of case 21261 were identical to those shown for case 18133. Chn, China; Dnk, Denmark.
FIG. 2.
Phylogenetic tree for the complete H gene sequences of representative CDVs plus those detected in cases 18133, 21261, 19876, and 25259 (shown in boxes). The unrooted tree was generated using the distance matrix program NEIGHBOR with the jumble option invoked. Distance values were calculated by the DNADIST program within the PHYLIP software package using the Kimura two parameter. Only bootstrap values greater than 70 are shown, and the branch lengths are proportionate to genetic distances. The country of origin of each CDV is indicated by a two- or three-letter abbreviation following the isolate designation; Chn, China; Jpn, Japan; Ger, Germany; SA, South Africa; USA, United States of America; Dnk, Denmark; Trk, Turkey; Twn, Taiwan; Gnd, Greenland; Sib, Siberia. Canine distemper viruses used for comparison with the country of origin and year of isolation (if known), plus the accession numbers, are as follows: A75/17 (USA, 1975), AF164967; black leopard (USA, 1991), Z47763; black panther, strain A92-6 (USA, 1992), Z54166; Chinese leopard, strain A92-27/4 (USA, 1992), Z54156; dog (USA, 1989), Z47762; dog 4513 (Ger), Z77673; dog (Taiwan), AY378091; dog Hamamatsu (Japan, 1992 to 1994), D85754; dog Yanaka (Japan, 1992 to 1994), D85755; dog 5804 (Germany, 1990), AY386315; dog 91A (Denmark, 1991), AF478544; dog 91B (Denmark, 1991), AF478546; dog 2544 (Germany, 1995), Z77672; dog (Turkey), AY093674; dog (China), AF172411; dog GR88 (Greenland, 1988), Z47760; dog 26D (Japan, 1999), AB040766; dog 5B (Japan, 1999), AY297453; dog 98-002 (Japan, 1998), AB025270; dog HM3 (Japan, 1999), AB040767; dog KDK-1 strain (Japan, 1991), AB025271; dog Ueno (Japan, 1992 to 1994), D85753; ferret (German, 1989), X84999; giant panda (China), AF178038; javelina (USA, 1989), Z47764; lesser panda (China), AF178039; mink (Denmark, 1986), Z47759; Onderstpoort (South Africa), AF378705; PDV-2 Siberian seal (Siberia, 1988), X8499; raccoon 98-2646 (USA, 1988), AY542312 (also represents raccoon 98-2655 [USA, 1998], AY548109, and raccoon 98-2666 [USA, 1998], AY548110); raccoon 01-2689 (USA, 2001), AY465925 (also represents raccoon 01-2690 [USA, 2001]); raccoon 00-2601 (USA, 2000), AY443350; raccoon 01-2641(USA, 2001), AY526496; raccoon 01-2676 (USA, 2001), AY498692; raccoon 98-2654 (USA, 1998), AY466011; raccoon (USA, 1989), Z47765; raccoon dog (Japan, 1996), AB016776; Snyder Hill (Germany), AF259552.
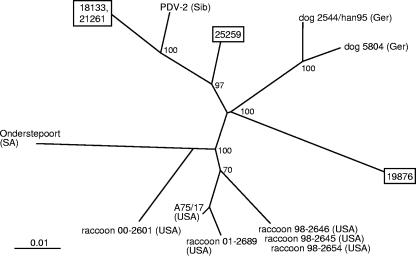
As observed for the H gene sequences, alignment of F gene sequences from cases 18133 and 21261 demonstrated identical nucleotide sequences through the coding region. Of the 4,508 bases sequenced for these two isolates, only two nucleotide changes were noted, and both were in the intergenic region between the F and H gene sequences. Predicted F gene amino acid sequences from cases 18133 and 21261 were 91.2% and 94.9% identical to those from cases 19876 and 25259, respectively, while case 19876 was 94.1% identical to 25259 (Table 3). Following nucleotide sequence alignment, similar levels of identity were observed between the CDV isolates detected in these cases. Alignment of sequences with those in the public databases (data not shown) demonstrated a maximum identity level of 96.7% to PDV-2 for the predicted amino acid sequences and 97.3% for the nucleotide sequences (Table 3). Considerably fewer full-length CDV F gene sequences were available for phylogenetic analysis compared to the number available for the H gene. However, for cases 18133 and 21261, phylogenetic analysis of F gene nucleotide sequences using distance matrix methods supported the results for the H gene by grouping these sequences with those of PDV-2, again with a bootstrap value of 100 (Fig. 3). However, clear phylogenetic relationships were not established for F gene nucleotide sequences from cases 19876 and 25259 with those available for comparison.
TABLE 3.
Nucleotide and deduced amino acid sequence identities of CDV F protein genes
| Virus | % Identitya with:
|
||||
|---|---|---|---|---|---|
| 18133b | 19876 | 25259 | PDV-2 | A75/17 | |
| 18133b | 91.2 | 94.9 | 96.7 | 92.9 | |
| 19876 | 92.7 | 94.1 | 91.5 | 93.7 | |
| 25259 | 96.4 | 94.6 | 95.2 | 96.1 | |
| PDV-2 | 97.3 | 93.2 | 96.4 | 94.4 | |
| A75/17 | 94.0 | 94.3 | 96.4 | 95.0 | |
Values for deduced amino acid sequence identities are in boldface.
Values shown are identical for cases 18133 and 21261.
FIG. 3.
Phylogenetic tree for the complete F gene sequences of representative CDVs plus those detected in cases 18133, 21261, 19876, and 25259 (shown in boxes). The unrooted tree was generated using the distance matrix program NEIGHBOR with the jumble option invoked. Distance values were calculated by the DNADIST program within the PHYLIP software package using the Kimura two parameter. Only bootstrap values greater than 70 are shown, and the branch lengths are proportionate to genetic distances. The country of origin of each CDV is indicated by a two- or three-letter abbreviation following the isolate designation: Ger, Germany; SA, South Africa; USA, United States of America; Sib, Siberia. Canine distemper viruses used for comparison with the country of origin and year of isolation (if known), plus the accession numbers, are as follows: A75/17 (USA, 1975), AF164967; dog 5804 (Germany, 1990), AY386315; dog 2544 (Germany, 1995), AJ007711; raccoon 00-2601 (USA, 2000), AY443350; raccoon 01-2689 (USA, 2001), AY649446; raccoon 98-2646 (USA, 1998), AY542312; raccoon 98-2654 (USA, 1998), AY466011; raccoon 98-2645 (USA, 1998), AY445077; Onderstepoort (South Africa), AF014953; PDV-2 (Siberia, 1988), L07075.
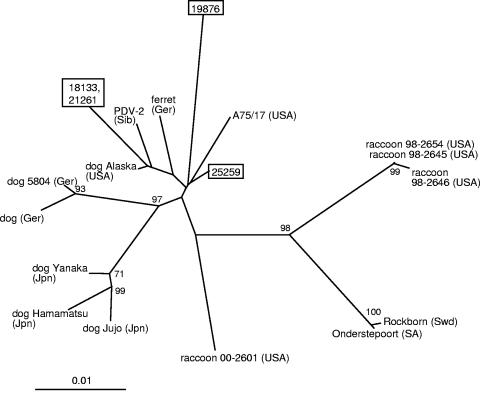
Alignment of the P gene nucleotide and predicted amino acid sequences demonstrated that CDVs detected in cases 18133 and 21261 were again 100% identical in nucleotide sequence for a 540-base fragment of the gene. Predicted P gene amino acid sequences from these cases were 96.1% and 97.2% identical to those from cases 19876 and 25259, respectively, while case 19876 was 97.8% identical to 25259 (Table 4). Following alignment, similar levels of nucleotide sequence identity were observed between the CDVs detected in these cases. Alignment of sequences with those in the public databases (data not shown) demonstrated identity levels ranging from 96.7% to 99.4% for the predicted amino acid sequences, with similar levels of identity for the nucleotide sequences (Table 4). Phylogenetic analysis of the P gene nucleotide sequences, plus others from the database, using distance matrix methods (Fig. 4) supported a relationship between cases 18133 and 21261 and PDV-2. In analysis of the P gene sequence, cases 25259 and 19876 were most closely related to cases 18133 and 21261 plus those of PDV-2, A75/17, and strains detected in a dog from Alaska and a German ferret. However for sequences from each of the cases, low bootstrap values were observed for phylogenetic analysis of the P gene amino acid sequences, suggesting less robust relationships.
TABLE 4.
Nucleotide and deduced amino acid sequence identities of CDV P gene fragments
| Virus | % Identitya with:
|
||||||
|---|---|---|---|---|---|---|---|
| 18133b | 19876 | 25259 | PDV-2 | Alaskan dog | German ferret | A75/17 | |
| 18133b | 96.1 | 97.2 | 96.7 | 97.8 | 96.1 | 96.1 | |
| 19876 | 96.8 | 97.8 | 97.2 | 98.3 | 96.7 | 96.7 | |
| 25259 | 98.4 | 97.6 | 98.3 | 99.4 | 98.9 | 98.9 | |
| PDV-2 | 98.6 | 97.0 | 98.6 | 98.9 | 98.3 | 97.2 | |
| Alaskan dog | 99.2 | 97.6 | 99.2 | 99.4 | 98.3 | 98.3 | |
| Germ ferret | 98.0 | 96.8 | 98.8 | 98.6 | 98.8 | 97.8 | |
| A75/17 | 97.8 | 97.2 | 99.0 | 98.0 | 98.6 | 98.2 | |
Values for deduced amino acid sequence identities are in boldface.
Values shown are identical for cases 18133 and 21261.
FIG. 4.
Phylogenetic tree for partial P gene sequences of representative CDVs plus those detected in cases 18133, 21261, 19876, and 25259 (shown in boxes). The unrooted tree was generated using the distance matrix program NEIGHBOR with the jumble option invoked. Distance values were calculated by the DNADIST program within the PHYLIP software package using the Kimura two parameter. Only bootstrap values greater than 70 are shown, and the branch lengths are proportionate to genetic distances. The country of origin of each CDV is indicated by a two- or three-letter abbreviation following the isolate designation: Bul, Bulgaria; Jpn, Japan; Ger, Germany; SA, South Africa; USA, United States of America; Sib, Siberia; Swd, Sweden. Canine distemper viruses used for comparison with the country of origin and year of isolation (if known), plus the accession numbers, are as follows: A75/17 (USA, 1975), AF164967; dog Alaska (USA, 2003) (25); dog Hamamatsu (Japan, 1992 to 1994), AB028915; dog Jujo (Japan), AB028916; dog Yanaka (Japan, 1992 to 1994), AB028914; dog (Germany, 1993), AF259549; dog 5804 (Germany, 1990), AY386315; ferret (Germany, 1989), AF259550; PDV-2 (Siberia, 1988), AF259551; raccoon 98-2645 (USA, 1998), AY445077; raccoon 98-2646 (USA, 1998), AY542312; raccoon 98-2654 (USA, 1998), AY466011; raccoon 00-2601 (USA, 2000), AY443350; Rockborn (Sweden), AF181446.
DISCUSSION
Over a period of 5 months (June to October 2004), six puppies 6 to 8 weeks of age and one adult dog were subjected to necropsy evaluation. For five of the seven dogs, disease caused by CDV infection was suspected by the referring veterinarian based on clinical signs. Gross and histologic examinations were performed, and all findings were consistent with active, clinical disease caused by CDV infection. Although serologic detection of anti-CDV antibodies was attempted for only one of the seven dogs studied, the presence of anti-CDV IgM antibody suggested recent infection with CDV in that dog. The presence of CDV was confirmed by RT-PCR and immunohistochemical staining for each dog from all four cases. Unfortunately, due to delays between case submission and identification for further study, samples for virus isolation attempts were not available. To further characterize the CDV strains in these clinical cases, nucleotide sequencing of the complete F and H genes plus a portion of the P gene was performed following RT-PCR amplification of RNA purified directly from tissues collected at necropsy. A total of 4,508 bp was sequenced for each of the four cases. Cases 18133 and 21261 were identical except for two base differences in the intergenic region of the F and H genes. However, no known epidemiological link or common source of viral exposure was known between the two premises from which these cases originated. Comparison of 18133 and 21261 to cases 19876 and 25259 demonstrated that in the conserved P gene, identities ranged from 96.1 to 97.8%, while in the more divergent F and H genes, identity values were lower. Phylogenetic analysis of the P, F, and H gene nucleotide and predicted amino acid sequences by both distance and parsimony methods demonstrated that three genetically distinct strains had been detected among the four cases examined in this study. Furthermore, phylogenetic analysis of complete H and F gene sequences suggested that none of the strains detected were closely related to either known vaccine strains or lineages previously detected in the continental United States. Although CDV isolates detected in naturally occurring cases often group by geographical location following phylogenetic analysis (8, 9, 17, 18, 20, 21, 23, 28), other studies have provided examples in which CDV strains do not group by broad geographical location (25, 26, 28), as shown for the cases studied here.
Phylogenetic analyses of CDV P, F, and H gene nucleotide and amino acid sequences have been performed to study evolutionary relationships between CDV isolates and to find genetic variation among wild-type and CDV vaccine strains. The P gene is highly conserved among members of CDV strains and has been used for phylogenetic analysis by others (7, 9, 25, 32, 36). However, relatively few CDV P gene sequences of the length sequenced in this study were available in the public databases for phylogenetic comparison. The F gene is conserved within morbillivirus species (6, 24) and has also been used to determine phylogenetic relationships among these viruses (37). The H gene is more variable among CDV isolates, perhaps due to the role the protein plays in the host immune reaction (11), and thus has been widely used for phylogenetic analysis (8, 17, 18, 20, 28). Additionally, many more complete H gene sequences than complete F gene sequences are available in public databases for phylogenetic comparisons.
In the present study, phylogenetic analysis of the complete H gene sequences provided the most robust, definitive results of the three genes examined due to both the observed sequence divergence and the large number of gene sequences available for comparison relative to the F and P genes. Analysis of the H gene sequences implied an evolutionary relationship of cases 18133 and 21261 to PDV-2, as well as H gene sequences of CDV strains detected in dogs from China and Greenland. The relationship of 18133 and 21261 to PDV-2 was further supported by phylogenetic analysis of complete F gene sequences and partial P gene sequences. Analysis of H gene sequences also suggested that case 25259 may be related to CDV isolates previously detected in a lesser panda from China. Unfortunately, sequence from this panda strain was not available for F or P gene sequence analysis, and thus, the relationship could not be further supported by phylogenetic analyses. Analysis of H gene sequences by both distance and maximum-parsimony methods only weakly implied a relationship for CDV detected in case 19876 to a CDV strain detected in mink from Denmark, suggesting that the strain detected in case 19876 was the most divergent of the those studied.
Given the relationships suggested by phylogenetic analysis, it appears possible that the CDV strains infecting dogs in this study may have originated from noncanine species or, alternatively, may have been transmitted from dogs to the other species. Interspecies transmission has been widely suspected for CDV (10, 16, 19). For example, CDV isolated from captive large felids in the United States was thought to have originated from feral, nonfelid carnivores (18). In a study examining free-ranging carnivores from the Serengeti ecosystem of Africa, closely related CDV isolates were detected from four different species (9), suggesting extensive interspecies transmission. Analysis of PDV-2, which was isolated from Siberian seals, demonstrated a relationship to CDV isolates from a German dog and ferret (26), suggesting that transmission from one of these hosts may have been responsible for the widespread infection seen in these freshwater seals (27).
Recent studies have characterized CDV strains in both dogs and noncanine species in North America. The phylogenetic relationship of CDV strains isolated from raccoons concluded that these viruses were distinct from, yet strongly related to, known American-lineage CDV isolates (23). A CDV outbreak that killed several hundred sled dogs was recently described in Alaska. Sequence analysis of 540 bp of the P gene suggested a close relationship of the strain in the sled dogs to a strain of distemper virus isolated from a Siberian seal (PDV-2) (25). Pairwise alignment of CDV partial P gene sequences from the cases in the present study also demonstrated that the P gene sequences, as well as the H and F gene sequences, of 18133 and 21261 were potentially related to PDV-2 and thus may also be related to the virus detected in the Alaskan dogs in 2003. Unfortunately, only partial sequence of the P gene (but no H or F gene sequences) was available from the CDV outbreak in the Alaskan dogs, and thus, a more definitive link between these cases could not be further explored.
In dog populations with a high rate of vaccination against canine diseases, clinical cases of CDV typically occur sporadically. Speculations put forth to explain these cases are varied but often include vaccine failures, reversion of attenuated CDV vaccine strains to virulence (5), or the emergence of new strains that are sufficiently divergent to evade immune protection elicited by the vaccines used. In the cases studied here, vaccination with modified live vaccine was performed recently in three of the four cases. Importantly, the CDV strains detected in these clinical cases were clearly distinct from known vaccine strains, as has been demonstrated in previous studies of other CDV field strains (8, 17, 21, 23, 25, 28). The CDV strain Lederle, which was used as a vaccine strain in the past (19) and may still be circulating in wildlife, is phylogenetically related to the CDV strain Snyder Hill (23) and thus genetically distant from the strains detected in the present study. The widely used Galaxy-D CDV vaccine (Schering-Plough, Kenilworth, NJ) originates from the Onderstepoort strain, which is also genetically distant from the CDV strains detected in each of the four cases described here. To further explore the possibility of vaccine virus reverting to virulence, partial sequencing (979 bases; data not shown) of the H gene was performed from the same production lot of the CDV vaccine (Vanguard 5; Pfizer, Inc., Exton, PA) used in dogs from case 18133. The nucleotide sequence detected from this vaccine was 97.7% identical to that of strain A75/17 (34), which was the closest match in GenBank over the region sequenced, but only 95.0% identical to the virus detected in cases 18133 and 21261. Together, these observations suggest that a recent reversion of vaccine virus to virulence was not likely to be the cause of the clinical CDV disease seen in the dogs for which vaccine had been recently used. However, the presence of a minor though virulent strain as a contaminant of the vaccine virus stock certainly cannot be excluded.
In summary, phylogenetic analysis of H and F genes showed that the CDV strains detected in this study were genetically distinct from viruses previously detected in the continental United States and most closely matched strains from Asia or possibly Europe. In three of the four cases examined, a recent history of CDV vaccination was reported, yet none of the viruses detected were related to known vaccine strains. These data suggest the possible emergence of novel CDV strains in North America, which may require renewed efforts by vaccine manufacturers to ensure adequate protection following immunization of dogs against CDV.
Acknowledgments
I.D.R.P. was supported in part by a Minority Biomedical Research Training Initiative grant (NIH DHHS 2R25 GM056901-06).
We thank Susan K. Schommer and William J. Mitchell, Jr., for insightful discussion and reading of the manuscript.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appel, M. 1987. Canine distemper virus, p. 133-159. In M. J. G. Appel (ed.), Virus infections of carnivores. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 3.Appel, M. J., R. A. Yates, G. L. Foley, J. J. Bernstein, S. Santinelli, L. H. Spelman, L. D. Miller, L. H. Arp, M. Anderson, M. Barr, S. Pearce-Kelling, and B. A. Summers. 1994. Canine distemper epizootic in lions, tigers, and leopards in North America. J. Vet. Diagn. Investig. 6:277-288. [DOI] [PubMed] [Google Scholar]
- 4.Appel, M. J., and B. A. Summers. 1995. Pathogenicity of morbilliviruses for terrestrial carnivores. Vet. Microbiol. 44:187-191. [DOI] [PubMed] [Google Scholar]
- 5.Appel, M. J. G. 1978. Reversion to virulence of attenuated canine distemper virus in vivo and in vitro. J. Gen. Virol. 41:385-393. [Google Scholar]
- 6.Barrett, T., M. S. Subbarao, G. J. Belsham, and B. W. J. Mahy. 1991. The molecular biology of the morbilliviruses, p. 82-102. In D. Kingsbury (ed.), The paramyxoviruses. Plenum Press, New York, N.Y.
- 7.Barrett, T., I. K. Visser, L. Mamaev, L. Goatley, M. F. van Bressem, and A. D. Osterhaus. 1993. Dolphin and porpoise morbilliviruses are genetically distinct from phocine distemper virus. Virology 193:1010-1012. [DOI] [PubMed] [Google Scholar]
- 8.Bolt, G., T. D. Jensen, E. Gottschalck, P. Arctander, M. J. Appel, R. Buckland, and M. Blixenkrone-Moller. 1997. Genetic diversity of the attachment (H) protein gene of current field isolates of canine distemper virus. J. Gen. Virol. 78:367-372. [DOI] [PubMed] [Google Scholar]
- 9.Carpenter, M. A., M. J. Appel, M. E. Roelke-Parker, L. Munson, H. Hofer, M. East, and S. J. O'Brien. 1998. Genetic characterization of canine distemper virus in Serengeti carnivores. Vet. Immunol. Immunopathol. 65:259-266. [DOI] [PubMed] [Google Scholar]
- 10.Deem, S. L., L. H. Spelman, R. A. Yates, and R. J. Montali. 2000. Canine distemper in terrestrial carnivores: a review. J. Zoo Wildl. Med. 31:441-451. [DOI] [PubMed] [Google Scholar]
- 11.Diallo, A. 1990. Morbillivirus group: genome organization and proteins. Vet. Microbiol. 23:155-163. [DOI] [PubMed] [Google Scholar]
- 12.Evermann, J. F., C. W. Leathers, J. R. Gorham, A. J. McKeirnan, and M. J. Appel. 2001. Pathogenesis of two strains of lion (Panthera leo) morbillivirus in ferrets (Mustela putorius furo). Vet. Pathol. 38:311-316. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein, J. 1985. Confidence intervals on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package) version 3.5c. University of Washington, Seattle.
- 15.Frisk, A. L., M. König, A. Moritz, and W. Baumgärtner. 1999. Detection of canine distemper virus nucleoprotein RNA by reverse transcription-PCR using serum, whole blood, and cerebrospinal fluid from dogs with distemper. J. Clin. Microbiol. 37:3634-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haas, L., H. Hofer, M. East, P. Wohlsein, B. Liess, and T. Barrett. 1996. Canine distemper virus infection in Serengeti spotted hyenas. Vet. Microbiol. 49:147-152. [DOI] [PubMed] [Google Scholar]
- 17.Haas, L., W. Martens, I. Greiser-Wilke, L. Mamaev, T. Butina, D. Maack, and T. Barrett. 1997. Analysis of the haemagglutinin gene of current wild-type canine distemper virus isolates from Germany. Virus Res. 48:165-171. [DOI] [PubMed] [Google Scholar]
- 18.Harder, T. C., M. Kenter, H. Vos, K. Siebelink, W. Huisman, G. van Amerongen, C. Orvell, T. Barrett, M. J. Appel, and A. D. Osterhaus. 1996. Canine distemper virus from diseased large felids: biological properties and phylogenetic relationships. J. Gen. Virol. 77:397-405. [DOI] [PubMed] [Google Scholar]
- 19.Harder, T. C., and A. D. Osterhaus. 1997. Canine distemper virus—a morbillivirus in search of new hosts? Trends Microbiol. 5:120-124. [DOI] [PubMed] [Google Scholar]
- 20.Hashimoto, M., Y. Une, and M. Mochizuki. 2001. Hemagglutinin genotype profiles of canine distemper virus from domestic dogs in Japan. Arch. Virol. 146:149-155. [DOI] [PubMed] [Google Scholar]
- 21.Iwatsuki, K., N. Miyashita, E. Yoshida, T. Gemma, Y. S. Shin, T. Mori, N. Hirayama, C. Kai, and T. Mikami. 1997. Molecular and phylogenetic analyses of the haemagglutinin (H) proteins of field isolates of canine distemper virus from naturally infected dogs. J. Gen. Virol. 78:373-380. [DOI] [PubMed] [Google Scholar]
- 22.Jubb, K. V. F., P. C. Kennedy, and N. Palmer. 1993. Pathology of domestic animals, 4th ed., vol. 2, p. 617-622, 626-628. Academic Press, Inc., San Diego, Calif.
- 23.Lednicky, J. A., J. Dubach, M. J. Kinsel, T. P. Meehan, M. Bocchetta, L. L. Hungerford, N. A. Sarich, K. E. Witecki, M. D. Braid, C. Pedrak, and C. M. Houde. 2004. Genetically distant American canine distemper virus lineages have recently caused epizootics with somewhat different characteristics in raccoons living around a large suburban zoo in the USA. Virol. J. 1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liermann, H., T. C. Harder, M. Lochelt, V. von Messling, W. Baumgartner, V. Moennig, and L. Haas. 1998. Genetic analysis of the central untranslated genome region and the proximal coding part of the F gene of wild-type and vaccine canine distemper morbilliviruses. Virus Genes 17:259-270. [DOI] [PubMed] [Google Scholar]
- 25.Maes, R. K., A. G. Wise, S. D. Fitzgerald, A. Ramudo, J. Kline, A. Vilnis, and C. Benson. 2003. A canine distemper outbreak in Alaska: diagnosis and strain characterization using sequence analysis. J. Vet. Diagn. Investig. 15:213-220. [DOI] [PubMed] [Google Scholar]
- 26.Mamaev, L. V., N. N. Denikina, S. I. Belikov, V. E. Volchkov, I. K. Visser, M. Fleming, C. Kai, T. C. Harder, B. Liess, A. D. Osterhaus, and T. Barrett. 1995. Characterization of morbilliviruses isolated from Lake Baikal seals (Phoca sibirica). Vet. Microbiol. 44:251-259. [DOI] [PubMed] [Google Scholar]
- 27.Mamaev, L. V., I. K. Visser, S. I. Belikov, N. N. Denikina, T. Harder, L. Goatley, B. Rima, B. Edginton, A. D. Osterhaus, and T. Barrett. 1996. Canine distemper virus in Lake Baikal seals (Phoca sibirica). Vet. Rec. 138:437-439. [DOI] [PubMed] [Google Scholar]
- 28.Mochizuki, M., M. Hashimoto, S. Hagiwara, Y. Yoshida, and S. Ishiguro. 1999. Genotypes of canine distemper virus determined by analysis of the hemagglutinin genes of recent isolates from dogs in Japan. J. Clin. Microbiol. 37:2936-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy, F. A., E. P. J. Gibbs, M. C. Horzinek, and M. J. Studdert. 1999. Veterinary virology, 3rd ed., p. 411-428. Academic Press, San Diego, Calif.
- 30.Orvell, C., M. Blixenkrone-Møller, V. Svansson, and P. Have. 1990. Immunological relationships between phocid and canine distemper virus studied with monoclonal antibodies. J. Gen. Virol. 71:2085-2092. [DOI] [PubMed] [Google Scholar]
- 31.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comp. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 32.Rzeutka, A., and B. Mizak. 2003. Sequence analysis of the fragment of the phosphoprotein gene of Polish distemper virus isolates. Arch. Virol. 148:1623-1631. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Stettler, M., and A. Zurbriggen. 1995. Nucleotide and deduced amino acid sequences of the nucleocapsid protein of the virulent A75/17-CDV strain of canine distemper virus. Vet. Microbiol. 44:211-217. [DOI] [PubMed] [Google Scholar]
- 35.Summers, B. A., J. F. Cummings, and A. de Lahunta. 1995. Veterinary neuropathology, p. 102-110. Mosby-Year Book, St. Louis, Mo.
- 36.van de Bildt, M. W., T. Kuiken, A. M. Visee, S. Lema, T. R. Fitzjohn, and A. D. Osterhaus. 2002. Distemper outbreak and its effect on African wild dog conservation. Emerg. Infect. Dis. 8:211-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Visser, I. K., R. W. van der Heijden, M. W. van de Bildt, M. J. Kenter, C. Orvell, and A. D. Osterhaus. 1993. Fusion protein gene nucleotide sequence similarities, shared antigenic sites and phylogenetic analysis suggest that phocid distemper virus type 2 and canine distemper virus belong to the same virus entity. J. Gen. Virol. 74:1989-1994. [DOI] [PubMed] [Google Scholar]
- 38.Von Messling, V., G. Zimmer, G. Herrler, L. Haas, and R. Cattaneo. 2001. The hemagglutinin of canine distemper virus determines tropism and cytopathogenicity. J. Virol. 75:6418-6427. [DOI] [PMC free article] [PubMed] [Google Scholar]