Abstract
We compared whole blood dried on filter paper to the standard assay with frozen cell-free plasma for use in the quantitation of the human immunodeficiency virus RNA load in blood. RNA values from filter paper, corrected for the hematocrit, gave results comparable to those of the standard assay in terms of sensitivity and reproducibility.
The measurement of human immunodeficiency virus (HIV) RNA levels in plasma is rapidly becoming the most important laboratory tool for staging HIV infection and for the management of therapy (6, 7). However, quantitation of HIV levels in plasma requires access to certain laboratory equipment which may not be readily available in some settings. By the year 2000 the World Health Organization predicts that 30 million to 40 million new HIV infections will have occurred, with 90% of these occurring in developing countries (5). Even the simple act of centrifuging a tube of blood, aliquoting and freezing the plasma, and subsequently shipping the plasma to a centralized laboratory might prove difficult in some field situations, where many clinical trials of new antiretroviral drugs and potential HIV vaccines as well as transmission intervention studies will be conducted in the future.
The purpose of this research was to develop a simple method for collecting specimens for analysis of the HIV RNA level in plasma that would yield accurate and reproducible results without the need for electricity at the collection site. Our ultimate goal is to obtain blood from a fingerstick or heelstick, dry it on filter paper, and airmail it to a central laboratory, thus avoiding the need for a skilled phlebotomist and laboratory technician on site and also avoiding the need for centrifuges, freezers, and dry ice for shipping. Dried blood spots have been used for many years to screen for several metabolic disorders such as phenylketonuria and sickle cell disease. For HIV they have been used for the anonymous screening of newborns to assess the seroprevalence of HIV among childbearing women (4) and for the diagnosis of perinatal HIV infections by using HIV DNA (2, 3). More recently, it has been reported that DNA obtained from dried blood spots is suitable for gene sequence analysis (1). Consequently, we decided to evaluate the possibility of using whole blood dried onto filter paper for the quantitation of the HIV RNA level in plasma.
One hundred four HIV-positive patients seen in the University of North Carolina’s adult and pediatric infectious disease clinics provided 4 ml of blood collected in EDTA for the purposes of viral load testing. HIV RNA was quantitated from 100 μl of whole blood from each of 76 patients spotted in quadruplicate onto Schleicher & Schuell no. 903 filter paper. A total of 50 μl of whole blood from an additional 28 patients was spotted in eight replicates onto Schleicher & Schuell Isocode filter paper. The blood spots were dried overnight at room temperature in a biohazard hood. We also used the whole blood to determine the hematocrit (mean of two determinations). Cell-free plasma obtained from the same tube of blood was stored at −70°C. HIV type 1 (HIV-1) RNA levels were determined by a commercially available nucleic acid sequence-based amplification assay (NASBA; NASBA HIV-1 QT; Organon Teknika, Durham, N.C.). Two 50-μl aliquots of dried blood spots were placed in 9 ml of NASBA lysis buffer, and the mixture was rocked at room temperature for 2 h to isolate the RNA, after which the filter papers were removed. The HIV-1 RNA from cell-free plasma was isolated by placing 100 μl of plasma in 0.9 ml of NASBA lysis buffer. For the rest of the procedure, the manufacturer’s instructions were followed, including the use of diluted calibrators. The numbers of RNA copies per milliliter of plasma from the spots were calculated as follows: corrected number of spot RNA copies per milliliter of plasma = (number of spot RNA copies per milliliter of blood)/[(100 − hematocrit)/100]. Blood from the first 76 patients was tested singly by using the no. 903 filter paper. Blood from the remaining 28 patients was tested in duplicate by using the Isocode filter paper. Some dried spots were stored at room temperature for various lengths of time before testing.
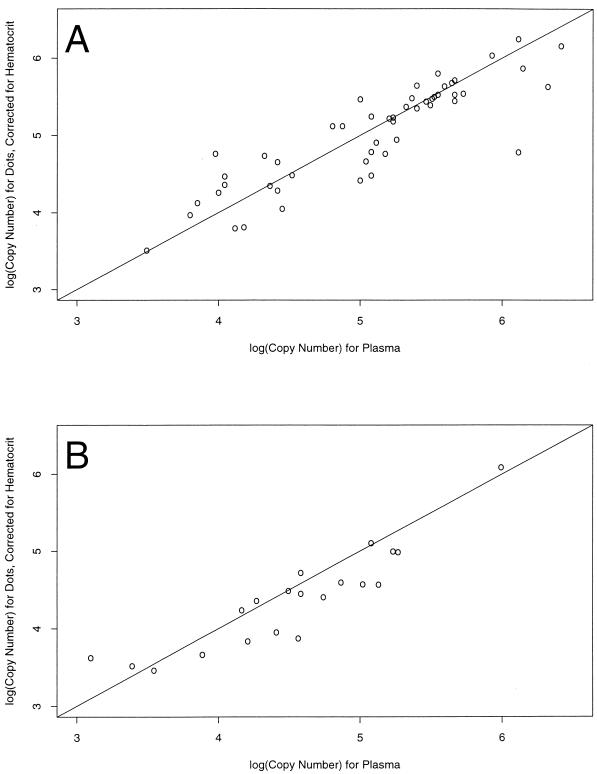
We initially used the no. 903 paper to measure HIV-1 RNA levels from dried blood spots, with encouraging results (Fig. 1A). However, a second lot of paper gave uninterpretable results and we switched to Isocode paper, which is pretreated with guanidinium isothiocyanate. The log10 hematocrit-corrected RNA concentration from the two types of paper is compared with the log10 RNA concentration from plasma in Fig. 1A (no. 903 filter paper) and Fig. 1B (Isocode filter paper). The diagonal line on each graph indicates where the points would fall if the estimates from the dots and the plasma were the same. Correlation coefficients were 0.88 (no. 903 filter paper versus plasma) and 0.90 (Isocode filter paper versus plasma). There is little evidence of a systematic difference between the values from no. 903 filter paper and those from plasma. The mean difference, 0.0478, was not statistically significantly different from zero (P = 0.33). Most (91%) of the differences in RNA values between the no. 903 filter paper dots and plasma were between −0.5 and 0.5 log10. However, the mean difference between values from plasma and Isocode filter paper was significantly different from zero (mean = 0.22; P < 0.01). On average, then, the values from plasma were 1.66 times the values from the Isocode filter paper. The difference did not vary systematically over the range of the data (Fig. 1B), so the use of a correction factor for the Isocode paper might be feasible.
FIG. 1.
(A) Log10 hematocrit-corrected HIV RNA concentrations for 55 patients tested with no. 903 filter paper plotted against the log10 RNA concentration for the matching plasma sample. Samples for which either estimate was below the detection limits were excluded (n = 21). The diagonal line indicates where points would fall if the estimates from the dots and the plasma were the same (correlation coefficient = 0.88). The points in the plot are roughly scattered around this line, indicating that there is little evidence of a systematic difference between the two approaches. For 91% of the hematocrit-corrected dot values, the values were within threefold (0.5 log10) of the value for plasma. (B) Mean log10 hematocrit-corrected HIV RNA concentrations for the Isocode filter paper plotted against the mean log10 RNA concentration for the matching plasma sample (coefficient correlation = 0.90). The points are shifted to the right, indicating that higher values were obtained in the assay with plasma. The mean difference was 0.22 (P = 0.0016), meaning that, on average, the estimates from the plasma were 1.66 times the estimates from the Isocode filter paper. For 95% of the hematocrit-corrected dot values, the values were within threefold (0.5 log10) of the value for plasma.
We tried to correct RNA values from filter paper for the hematocrit in three ways. Initially, we simply adjusted the RNA value obtained from the filter paper for each patient’s hematocrit, producing the data shown in Fig. 1. Then we tried the use of two constant correction factors. The first, 1.82, was derived from the mean hematocrit of 45 for this cohort. The second, 2.0, was based on the assumption that the mean hematocrit was close to 50. Use of a constant factor could be appropriate when hematocrits are not available. The two constants produced very similar results (data not shown).
The sensitivities of filter dot RNA and plasma RNA were compared by classifying each RNA determination as above or below the limit of detection (1,000 HIV RNA copies/ml). For 21 of 76 (28%) samples, values from no. 903 filter paper were below the detection limit, whereas for 17 of 76 (22%) samples, values from plasma were below the detection limit. For 13 samples, values from both no. 903 filter paper and plasma were below the detection limits. For 9 of 28 (32%) samples at least one of two values was below the detection limit on the Isocode filter paper, whereas for 7 (25%) samples at least one of two values from plasma were below the detection limit. For six samples two values from both Isocode filter paper and plasma were below the detection limits. The sensitivities of assays with filter dot RNA and plasma RNA were not significantly different (P > 0.05), but these results should be interpreted with caution. The volume of plasma on the filter paper dots is roughly half the volume in the plasma assay, so a difference in sensitivity should be expected. The absence of such a difference could simply mean that the study included few patients with RNA concentrations in the range needed to show a difference. Alternatively, it may reflect the fact that the HIV-1 RNA from the dried blood spots included both cell-free and cell-associated HIV-1 RNA, while the HIV-1 RNA quantitated from plasma is essentially only cell-free RNA.
The reproducibility of RNA values was examined by using intra-assay standard deviations (SDs) that were estimated from the duplicate day 0 results for 17 patients from both Isocode paper and plasma. The SDs for Isocode filter paper and plasma were 0.19 and 0.15, respectively. However, one pair of Isocode filter paper values differed by almost 0.7 log10. This large difference may reflect a specimen handling error rather than a problem with reproducibility. When this point is excluded, the SD drops to 0.17, which is close to the SD for plasma. For comparison, in a multicenter evaluation of quantitative assays of HIV-1 RNA levels in plasma, the AIDS Clinical Trials Group virology laboratories determined that the intra-assay SD for NASBA was 0.15 (8).
The stability of estimates from Isocode filter paper was examined by using a random effects regression of the log10 RNA level on length of storage (in days). The results for blood from 14 patients, each with storage times of 0, 7, and 28 days, were included. Nine values that were less than the detection limits were set equal to 3.0 (1,000 HIV RNA copies/ml). A statistically significant decline of 0.0261 log10 RNA copies per day was detected (P < 0.01). This slope is equivalent to a loss of approximately 5% per day.
Initial results for the no. 903 filter paper looked very good. However, later results with a different lot of no. 903 filter paper were not reproducible. It may be that this paper should be handled with gloved hands at all stages, including prior to the application of blood, to avoid contamination with RNases present on fingers; this may not be possible in all situations. The Isocode paper is pretreated with guanidinium isothiocyanate, so it does not require special handling prior to application of the specimen.
The preliminary results reported here are extremely encouraging. Using the Isocode filter paper and correcting for hematocrit and even without correcting for the use of the Isocode filter paper, we were able to quantify the amount of HIV RNA in plasma fairly accurately (Fig. 1B). No difference in sensitivity was observed between the assays with Isocode filter paper and the plasma. The reproducibility of the results of assays with Isocode filter paper were comparable to the reproducibility of the results of the standard assay with plasma. However, problems with stability exist. We are not sure whether the RNA is being degraded with time or whether we are simply having increased difficulty in removing the RNA from the filter paper with prolonged storage.
We believe that modifications and refinements of this method can provide an accurate measurement of the plasma HIV RNA level suitable for primitive field conditions. In addition, the nucleic acid recovered from the spots should be amenable to amplification and sequencing and should thus be of value for drug sensitivity and HIV clade testing (1).
Acknowledgments
This study was supported in part by cooperative agreements U01-AI27535 and U01 AI38858 (contract 96VD006) from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Cassol S, Weniger B G, Babu P G, Salminen M O, Zheng X, Htoon M T, Delaney A, O’Shaughnessy M, Ou C Y. Detection of HIV type 1 env subtypes A, B, C, and E in Asia using dried blood spots: a new surveillance tool for molecular epidemiology. AIDS Res Hum Retroviruses. 1996;12:1435–1441. doi: 10.1089/aid.1996.12.1435. [DOI] [PubMed] [Google Scholar]
- 2.Cassol S A, Lapointe N, Salas T, Hankins C, Arella M, Fauvel M, Delage G, Boucher M, Samson J, Charest J. Diagnosis of vertical HIV-1 transmission using the polymerase chain reaction and dried blood spot specimens. J Acquired Immune Defic Synd. 1992;5:113–119. [PubMed] [Google Scholar]
- 3.Comeau A M, Pitt J, Hillyer G V, Landesman S, Bremer J, Chang B H, Lew J, Moye J, Grady G F, McIntosh K. Early detection of human immunodeficiency virus on dried blood spot specimens: sensitivity across serial specimens. J Pediatr. 1996;129:111–118. doi: 10.1016/s0022-3476(96)70197-x. [DOI] [PubMed] [Google Scholar]
- 4.Gwinn M, Pappaioanou M, George J R, Hannon W H, Wasser S C, Redus M A, Hoff R, Grady G F, Willoughby A, Novello A C. Prevalence of HIV infection in childbearing women in the United States: surveillance using newborn blood samples. JAMA. 1991;265:1704–1708. [PubMed] [Google Scholar]
- 5.Joint United Nations Programme on HIV/AIDS. The HIV/AIDS situation in mid 1996: global and regional highlights. UN AIDS Fact Sheet. July 1. New York, N.Y: United Nations; 1996. [Google Scholar]
- 6.Mellors J W, Kingsley L A, Rinaldo C R, Todd J A, Hoo B S, Kokka R P, Gupta P. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann Intern Med. 1995;122:573–579. doi: 10.7326/0003-4819-122-8-199504150-00003. [DOI] [PubMed] [Google Scholar]
- 7.Saag M S, Holodniy M, Kuritzkes D R, O’Brien W A, Coombs R, Poscher M E, Jacobsen D M, Shaw G M, Richman D D, Volberding P A. HIV viral load markers in clinical practice: recommendations of an International AIDS Society-USA Expert Panel. Nature Med. 1996;2:625–629. doi: 10.1038/nm0696-625. [DOI] [PubMed] [Google Scholar]
- 8.Yen-Lieberman B, Brambilla D, Jackson B, Bremer J, Coombs R, Cronin M, Herman S, Katzenstein D, Leung S, Lin H J, Palumbo P, Rasheed S, Todd J, Vahey M, Reichelderfer P. Evaluation of a quality assurance program for quantitation of human immunodeficiency virus type 1 RNA in plasma by the AIDS Clinical Trials Group Virology Laboratories. J Clin Microbiol. 1996;34:2695–2701. doi: 10.1128/jcm.34.11.2695-2701.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]