Abstract
Real-time quantitative PCR systems (Q-PCR) for the rapid detection and quantification of microorganisms in clinical specimens employ oligodeoxyribonucleotide primers and probes for specificity, which makes them vulnerable to false negatives caused by sequence diversity in the template. Schaade et al. (J. Clin. Microbiol. 39:3809, 2001) reported a sequence variant (C630T) in the cytomegalovirus (CMV) glycoprotein B (gB) gene that, although detectable in their Q-PCR assay, could not be accurately quantified. In an effort to evaluate the impact of CMV sequence variants in our patient population by use of a similar Q-PCR assay, we surveyed 54 isolates of CMV, each from a different patient. We detected evidence for the C630T variant in 4 of 54 (7.4%) patients. Furthermore, isolates from two additional patients were completely negative in the test. Sequencing of these false-negative isolates revealed multiple mutations within the probe hybridization sites. A Q-PCR that targeted the CMV polymerase gene instead of gB detected all 54 isolates. We suggest that Q-PCR assays for viral load be rigorously tested on large panels of viral isolates to assess the impact of sequence diversity on detection as well as quantification.
Real-time quantitative PCR (Q-PCR) is currently the method of choice for the rapid detection and quantification of cytomegalovirus (CMV) in clinical specimens (10, 12). The glycoprotein B gene (gB, UL55) is, overall, one of the less variable regions of the CMV genome (11, 17), except for two variable subregions (4, 16), and has been frequently used as a target for Q-PCR assays (5, 6, 8-10, 19). Schaade et al. (14) described a Q-PCR assay using fluorescence energy transfer technology with two fluorophore-labeled hybridization probes within the gB gene and concluded that their assay was superior in terms of sensitivity, specificity, linear range, and efficiency to the commercially available COBAS Amplicor CMV Monitor assay (Roche), which uses the CMV DNA polymerase gene (pol) as a target. Other groups, using modifications of the assay described by Schaade et al. (14), have reported similar experiences (8, 12).
However, CMV variants that could not be quantified using this Q-PCR have been reported (15). In the initial report (15), 912 clinical specimens were tested, of which 197 were positive for CMV DNA, but for 12 specimens (6.1% of the positive specimens) a viral load could not be determined. The specimens that were detected but not quantified did not produce a crossing point or any evidence of amplification during the PCR. However, in the melting curve analysis following the PCR, a signal was detected with a melting temperature of 53.1°C, which is below the 59.2°C melting temperature found for wild-type CMV. Sequence analysis revealed a single base pair mutation, C630T, in the target sequence of the downstream probe (15). This sequence variant prevented the appearance of a fluorescent signal under the cycling conditions of the assay, which included an annealing temperature of 58°C (15). The melting curve employed a lower annealing temperature (45°C), which allowed hybridization of the probe with the C630T variant (15). Thus, for the C630T gB variant, the assay developed by Schaade et al. (14) was only sufficient to provide qualitative results.
To evaluate the impact of sequence variants in our patient population using a similar Q-PCR assay, we studied 54 CMV isolates, each from a different patient. We found two isolates that were not detectable. Sequence analysis of a portion of the gB gene revealed that the reason for the failure of the assay was a divergent nucleotide sequence in the target region of the probes. This work adds to our knowledge of sequence diversity in the CMV gB gene.
MATERIALS AND METHODS
Specimens.
Culture supernatants, prepared from clinical specimens exhibiting characteristic CMV cytopathic effect in human embryonic lung cells, were thawed from storage at −80°C. A 1:10 dilution of supernatant was used as template without nucleic acid extraction.
gB Q-PCR.
The sequences of the primers and probes were as described previously (2, 14). Ten microliters of 1:10-diluted culture supernatant was added to a 10-μl reaction mixture to give final concentrations of 3 mM MgCl2, 0.5 μM of each primer, 0.25 μM of each probe, and 1× Roche FastStart DNA master hybridization probes mix (Roche Applied Science, Indianapolis, IN) in Roche LC capillaries. The thermal cycle was as follows: an initial 10 min at 95°C to activate the FastStart Taq DNA polymerase, followed by 55 cycles of 5-second denaturation at 95°C, 20-second annealing at 58°C, and 15-second extension at 72°C. After PCR the melting curve thermal profile was 95°C for 10 seconds, 45°C for 1 minute, and then 80°C with a 0.1°C/second temperature transition rate while continuously collecting fluorescence data. Thermal cycling and collection of fluorescence data were done in a Roche LightCycler.
pol gene Q-PCR.
The protocol for pol gene Q-PCR was adapted from Yun et al. (18) for use on the Roche LightCycler. The primers were the same as in the original paper (18); however, we corrected a typographical error in the Yun-3 probe sequence 5′-6-carboxyfluorescein-TGCGCCGTATGCTGCTCGACA-tetramethylcarboxyrhodamine-3′ (Z. Yun, personal communication). Ten microliters of 1:10-diluted culture supernatant was added to a 10-μl reaction mixture to give final concentrations of 2 mM MgCl2, 0.5 μM of each primer, 0.2 μM of dual labeled probe, and 1× LightCycler FastStart DNA master hybridization probes in Roche LC capillaries. Cycling parameters were as follows: an initial 10 min at 95°C for FastStart Taq DNA polymerase activation, 50 cycles of 4-s denaturation at 95°C and 20-s annealing and extension at 58°C. Fluorescence data were collected after every cycle in a Roche LightCycler.
Sequencing.
The isolates that were not detectable in the gB Q-PCR assay were studied by DNA sequence analysis. The I-35 isolate was amplified using the upstream primer 5′-TCCGTCGCACTATATAATGAAACTTTC-3′ and the downstream primer 5′-ATAGGAGGCGCCACGTATTC-3′. The I-60 isolate was amplified using the upstream primer 5′-CTGGTGCCTGGTAGTCTGCGTT-3′ and downstream primer 5′-GGCTGTGCCAYTGATCCTTGAC-3′. The reactions were done in a volume of 25 μl with primers at a concentration of 1 μM, 50 μM each deoxynucleoside triphosphate, 1 U of HotStart Taq DNA polymerase, and 2.5 μl of the 10× buffer provided by the enzyme manufacturer (QIAGEN Inc., Valencia, CA). The reaction was cycled 35 times between 96°C for 20 seconds, 50.3°C for 20 seconds, and 72°C for 2 min, preceded by 10 min at 95°C and followed by 5 min at 72°C in an Eppendorf thermal cycler (Brinkmann Instruments, Westbury, NY). The amplicons were treated with ExoSap (Amersham Biosciences, Piscataway, NJ) to remove the primers and deoxynucleoside triphosphates and sequenced using the PCR primers as sequencing primers and Applied Biosystems (ABI, Foster City, CA) dye terminator chemistry, and then the sequencing reactions were resolved by capillary electrophoresis on the ABI 3100 Prism DNA sequencer.
RESULTS
In an effort to institute CMV viral load testing in our laboratory, we evaluated the assay proposed by Schaade et al. (14) for its ability to detect CMV by retrospectively testing 54 culture supernatants from CMV isolates grown in tissue culture in our clinical virology laboratory. The isolates tested were from 54 different patients and were originally derived from a variety of specimen sources over a 1-year period. Isolates came from patients with the following clinical histories: 22% solid organ transplant, 13% bone marrow transplant, 26% human immunodeficiency virus/AIDS, and 39% with other or unknown diagnoses. Of these 54 isolates, 48 (89%) produced amplification curves as expected for a specimen with an abundance of CMV (data not shown). Four isolates (7.4%) did not produce amplification curves during the PCR but did produce an amplicon that hybridized with the probes under the less stringent conditions used in the annealing step prior to the melting curve. Melting temperature analysis suggested that these four isolates had the C630T mutation which was previously described (15). For this polymorphic CMV variant, the assay developed by Schaade et al. (14) is useful for detection, but not quantification, of the virus.
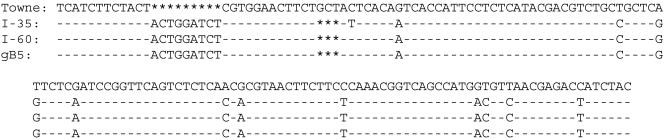
Two additional clinical isolates (I-60 and I-35) could not be quantified using the Q-PCR assay developed by Schaade et al. (14) and also did not produce a melting curve when the probe hybridization was done under less stringent conditions (data not shown). We were able to detect all 54 viral isolates using a Q-PCR assay that targets the viral pol gene (18), thus confirming that all isolates actually did contain CMV DNA. We suspected that the CMV isolates that were not detected by the gB Q-PCR assay might have a more divergent sequence in the target area for the primers and probes. We used the PCR to amplify the target region from the two CMV isolates that produced false-negative results and determined their nucleotide sequences. Both isolates were identical in this region and had two differences within the upstream probe sequence and three differences within the downstream probe sequence (Fig. 1). Further analysis of I-60 and I-35 in the upstream variable region of the gB gene revealed a sequence that was very divergent from standard CMV strains, such as Towne and AD169 (Fig. 2). A BLAST search (1) revealed that these isolates were identical (I-60) or almost identical (I-35) to the gB genotype 5 consensus sequence in the 5′ variable region of the gB gene (16) (Fig. 2).
FIG. 1.
Sequence comparison of the CMV gB gene in the target region for the probes in the Q-PCR assay described by Schaade et al. (14). The reference sequence in the first line is the AD169 strain, accession number X04606, 443 to 493 bp from the translational start codon. The probe sequences from Schaade et al. (14) are shown on the second line, followed by the C630T variant from Schaade et al. (15) on the third line. The last two lines of sequence are from the two isolates that were not detected by the gB Q-PCR assay (I-35 and I-60). —, sequence identity with the AD169 strain.
FIG. 2.
Sequence comparison of the upstream variable region of the CMV gB gene. The reference sequence on the top line is the Towne strain from accession number M22343, nucleotides 73 to 216 from the start of translation. The next two lines show the two isolates sequenced in this study. The bottom line is the consensus gB5 sequence from Shepp et al. (16). —, sequence identity with the Towne strain;  , a deletion.
, a deletion.
Nucleotide sequence accession numbers.
The nucleotide sequence data from these isolates are available in GenBank, accession numbers DQ121372 and DQ121371, for I-35 and I-60, respectively.
DISCUSSION
In our patient population, a commonly used CMV Q-PCR that targets the gB gene (14) failed to quantify the virus in 11% (6/54) of isolates. This was most likely due to a known variant, C630T, in four isolates, which allowed qualitative detection of the virus (15). However, in two isolates sequence variation in the target sequence of the probes made the viral genome invisible to the assay and created a true false-negative result.
The frequency of the gB C630T variant in our small survey was similar (7.4% versus 6.1%) to the frequency reported by Schaade et al. (15). In a population of primarily bone marrow transplant recipients (85%) in Portland, Oregon, the frequency of the gB C630T variant was reported to be 2.3% of patients (8). In a study of 66 solid organ transplant recipients, no C630T variants were found (12). Although the gB Q-PCR assay is unable to quantify CMV with the C630T variant, it still detects the virus.
Far more serious is our finding of two CMV isolates that are more divergent than the C630T variant. These isolates have several additional sequence differences with the probes used in the gB assay (Fig. 1). Due to the additional sequence differences, these CMV isolates are undetectable by this assay.
Additional sequence analysis of the two CMV isolates that produced a false-negative result in the gB Q-PCR (Fig. 2) revealed that both isolates are not new but are most likely the same as the previously discovered gB genotype 5 (gB5) (16). This genotype was defined by sequence analysis of two variable regions in the CMV glycoprotein B gene and was found to be distinct from the other four gB genotypes of CMV (4). The two isolates derived from our patient population are identical or nearly identical to the upstream variable region of the gB5 consensus sequence (Fig. 2) (16). We did not study the downstream variable region of the CMV gB gene in our isolates. The additional differences between the two isolates studied here (GenBank accession numbers DQ121372 and DQ121371) and other published CMV sequences (Fig. 1) reveal that gB5 is likely to be more divergent than previously thought. In particular, the sequence divergence extends outside of the previously known gB variable regions (4, 16). The sequence diversity that we found among our clinical isolates adds to our knowledge of sequence diversity in the CMV gB gene and consequently should be taken into consideration in the design of Q-PCR assays that use this gene as a target.
CMV with the gB5 genotype has not been reported frequently. After its original discovery (16) in five patients with AIDS, another group (3) reported sequence data consistent with a gB5 genotype in five patients with AIDS. This may be due to the actual rarity of the strain and its confinement to AIDS patients, but the apparent rarity of gB5 may also be because it is difficult to detect. The primers used for genotyping by several groups are not able to detect gB5 because of its sequence divergence (4, 13, 16).
In a comparison of sequence diversity among clinical isolates, the CMV DNA polymerase (pol) gene was found to be slightly more conserved than the gene encoding gB, suggesting pol may be a better target, in terms of clinical sensitivity, for determining CMV viral load (17). We found that an adaptation for the LightCycler of the Q-PCR assay described by Yun et al. (18), which targets pol, successfully detected and quantified all 54 of our clinical isolates.
Real-time Q-PCR assays that use both primers and probes are inherently more vulnerable to false-negative results arising from sequence diversity, because they depend on more sequence than a simple PCR to achieve analytical sensitivity and specificity. Based on this principle and our experience, we suggest that laboratories carefully evaluate the clinical sensitivity of a candidate Q-PCR assay in their patient population to avoid errors due to sequence diversity in the target sequences. Consideration should also be given to using more than one target to avoid false negatives due to rare or newly arising variants that escape detection during the initial validation. This approach has been taken by several laboratories (7, 8).
Acknowledgments
We thank David Shepp for helpful advice and Laura Ascroft and the University of Rochester Functional Genomics Center for DNA sequencing.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai, X., B. B. Rogers, P. C. Harkins, J. Sommerauer, R. Squires, K. Rotondo, A. Quan, D. B. Dawson, and R. H. Scheuermann. 2000. Predictive value of quantitative PCR-based viral burden analysis for eight human herpesviruses in pediatric solid organ transplant patients. J. Mol. Diagn. 2:191-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chern, K. C., D. B. Chandler, D. F. Martin, B. D. Kuppermann, R. A. Wolitz, and T. P. Margolis. 1998. Glycoprotein B subtyping of cytomegalovirus (CMV) in the vitreous of patients with AIDS and CMV retinitis. J. Infect. Dis. 178:1149-1153. [DOI] [PubMed] [Google Scholar]
- 4.Chou, S. W., and K. M. Dennison. 1991. Analysis of interstrain variation in cytomegalovirus glycoprotein B sequences encoding neutralization-related epitopes. J. Infect. Dis. 163:1229-1234. [DOI] [PubMed] [Google Scholar]
- 5.Ferreira-Gonzalez, A., R. A. Fisher, L. A. Weymouth, M. R. Langley, L. Wolfe, D. S. Wilkinson, and C. T. Garrett. 1999. Clinical utility of a quantitative polymerase chain reaction for diagnosis of cytomegalovirus disease in solid organ transplant patients. Transplantation 68:991-996. [DOI] [PubMed] [Google Scholar]
- 6.Guiver, M., A. J. Fox, K. Mutton, N. Mogulkoc, and J. Egan. 2001. Evaluation of CMV viral load using TaqMan CMV quantitative PCR and comparison with CMV antigenemia in heart and lung transplant recipients. Transplantation 71:1609-1615. [DOI] [PubMed] [Google Scholar]
- 7.Herrmann, B., V. C. Larsson, C.-J. Rubin, F. Sund, B.-M. Eriksson, J. Arvidson, Z. Yun, K. Bondeson, and J. Blomberg. 2004. Comparision of a duplex quantitative real-time PCR assay and the COBAS Amplicor CMV monitor test for detection of cytomegalovirus. J. Clin. Microbiol. 42:1909-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong, K. M., H. Najjar, M. Hawley, and R. D. Press. 2004. Quantitative real-time PCR with automated sample preparation for diagnosis and monitoring of cytomegalovirus infection in bone marrow transplant patients. Clin. Chem. 50:846-856. [DOI] [PubMed] [Google Scholar]
- 9.Kearns, A. M., M. Guiver, V. James, and J. King. 2001. Development and evaluation of a real-time quantitative PCR for the detection of human cytomegalovirus. J. Virol. Methods 95:121-131. [DOI] [PubMed] [Google Scholar]
- 10.Li, H., J. S. Dummer, W. R. Estes, S. Meng, P. F. Wright, and Y.-W. Tang. 2003. Measurement of human cytomegalovirus loads by quantitative real-time PCR for monitoring clinical intervention in transplant recipients. J. Clin. Microbiol. 41:187-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy, E., D. Yu, J. Grimwood, J. Schmutz, M. Dickson, M. A. Jarvis, G. Hahn, J. A. Nelson, R. M. Myers, and T. E. Shenk. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 100:14976-14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pang, X. L., L. Chui, J. Fenton, B. LeBlanc, and J. K. Preiksaitis. 2003. Comparison of LightCycler-based PCR, COBAS Amplicor CMV Monitor, and pp65 antigenemia assays for quantitative measurement of cytomegalovirus viral load in peripheral blood specimens from patients after solid organ transplantation. J. Clin. Microbiol. 41:3167-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rasmussen, L., C. Hong, D. Zipeto, S. Morris, D. Sherman, S. Chou, R. Miner, W. L. Drew, R. Wolitz, A. Dowling, A. Warford, and T. C. Merigan. 1997. Cytomegalovirus gB genotype distribution differs in human immunodeficiency virus-infected patients and immunocompromised allograft recipients. J. Infect. Dis. 175:179-184. [DOI] [PubMed] [Google Scholar]
- 14.Schaade, L., P. Kockelkorn, K. Ritter, and M. Kleines. 2000. Detection of cytomegalovirus DNA in human specimens by LightCycler PCR. J. Clin. Microbiol. 38:4006-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaade, L., P. Kockelkorn, K. Ritter, and M. Kleines. 2001. Detection of cytomegalovirus DNA in human specimens by LightCycler PCR: melting point analysis is mandatory to detect virus strains with point mutations in the target sequence of the hybridization probes. J. Clin. Microbiol. 39:3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shepp, D. H., M. E. Match, S. M. Lipson, and R. G. Pergolizzi. 1998. A fifth human cytomegalovirus glycoprotein B genotype. Res. Virol. 149:109-114. [DOI] [PubMed] [Google Scholar]
- 17.Wirgart, B. Z., M. Brytting, A. Linde, B. Wahren, and L. Grillner. 1998. Sequence variation within three important cytomegalovirus gene regions in isolates from four different patient populations. J. Clin. Microbiol. 36:3662-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yun, Z., I. Lewensohn-Fuchs, P. Ljungman, L. Ringholm, J. Jonsson, and J. Albert. 2003. A real-time TaqMan PCR for routine quantitation of cytomegalovirus DNA in crude leukocyte lysates from stem cell transplant patients. J. Virol. Methods 110:73-79. [DOI] [PubMed] [Google Scholar]
- 19.Yun, Z., I. Lewensohn-Fuchs, P. Ljungman, and A. Vahlne. 2000. Real-time monitoring of cytomegalovirus infections after stem cell transplantation using the TaqMan polymerase chain reaction assays. Transplantation 69:1733-1736. [DOI] [PubMed] [Google Scholar]