Abstract
We have developed an economical, high-throughput nucleic acid amplification test (NAT) for blood-borne viruses, suitable for use in the screening of plasma samples from individual blood donors. This assay system includes a semiautomated procedure, using 96-well glass fiber plates for the extraction of viral nucleic acids from plasma and “universal beacon” technology which permits the detection of all genotypes of highly variable viruses (e.g., human immunodeficiency virus and hepatitis C virus). In this detection system, two fluorescent- detection technologies were employed successfully in a single tube: molecular beacon for West Nile virus (WNV) detection using a 6-carboxyfluorescein fluorophore and TaqMan for internal control detection using a VIC fluorophore. To establish proof of concept, we focused on the development of a robust individual donor NAT for WNV. The assay showed no reactivity to 15 other viruses tested or to 420 blood donor samples from the WNV pre-epidemic season. No cross-contamination was observed on an alternating positive-/negative-well test. The sensitivity (limit of detection, 95%) of the assay for WNV is between 3.79 and 16.3 RNA copies/ml, depending on which material was used as a standard. The assay detected all positive blood donation samples identified by the Roche WNV NAT. The assay can be performed qualitatively for screening and quantitatively for confirmation.
West Nile virus (WNV) is a member of the genus Flavivirus and is part of the Japanese encephalitis virus (JEV) family. WNV was first isolated in Uganda in 1937 and has since been found in Europe, Africa, Asia, and North America. WNV is an arthropod-borne virus which cycles between mosquitoes and vertebrate hosts. The primary vertebrate hosts for WNV are birds (e.g., crows, ravens, jays, etc.). These hosts may harbor very high titers of the virus. Transmission of the virus to other vertebrate hosts (e.g., humans and horses) occurs following mosquito bites. The viral titer in infected immunocompetent humans appears to be quite low relative to that of birds, and the viremic phase of infection appears to be of short duration (1 to 2 weeks) (14).
There have been several recent epidemics of WNV, notably in Israel, Romania, and Russia in the 1990s (14) and in the United States from 1999 to 2004 (1, 13, 15). In 2003, the Centers for Disease Control and Prevention (CDC) reported 9,862 clinical cases from 46 states, including 2,866 cases of meningoencephalitis and 264 deaths (6). Most significantly, the theoretical risk of transfusion-transmitted WNV infection was confirmed (1). Pealer et al. reported at least 21 cases of WNV infection thought to be transmitted by transfusion (13). In addition, cases of transmission via organ donation (7) and through breast milk (4) were reported.
The nucleic acid amplification test (NAT) for WNV RNA in donated blood was implemented in June/July 2003 under an Investigational New Drug exemption issued by the FDA to two U.S. manufacturers. Testing is performed on pools of 6 or 16 samples, depending on the vendor of the test kit. The test algorithm is such that samples in a WNV RNA-positive pool are then tested individually, and the implicated sample is identified. To confirm the presence of WNV RNA, an alternative sample (e.g., from the plasma unit) is also tested. Donors whose samples are found to be positive are invited to enroll in follow-up studies in which the persistence of WNV RNA is tracked. Testing for the appearance of anti-WNV immunoglobulin M is also performed. Donors whose samples are positive for WNV RNA are deferred from donation until 28 to 56 days following the last NAT-reactive sample; products from these donors are discarded.
The results of testing blood donors in 2003 and 2004 have recently been reported (2, 3, 15). Approximately six million donations were tested from June to December 2003. The presence of WNV RNA was confirmed in 818 blood donors nationally. The distribution of cases in blood donors followed the national pattern, with the vast majority of cases in blood donors occurring in the Midwest, West, and Southwest. In 2003, 23 cases of WNV due to transfusion were reported to the CDC (5).
Of specific relevance to our technology, several testing centers in regions that were considered to have high incidence for WNV switched from pool testing to individual donor (ID) testing in 2003. The assumption was that individual testing would be more sensitive than pool testing. This is supported by a recent report of a case of transfusion-transmitted WNV infection in which the six-member donor pool tested negative, while individual donor testing revealed an infected unit (11). The improved sensitivity of single-unit testing has been further confirmed by two recent reports. Stramer et al. observed that of 540 WNV RNA-positive donations, 148 (27%) were detected only by single-unit testing (15). Similarly, Busch et al. reported that, in 2003, 34% of all viremic units detected were detected only by single-unit testing (2). These reports are consistent with the observation that the levels of viremia in infected individuals are very low. An unexplained observation is that the levels of viremia in infected individuals identified in 2003 (0.06 to 0.5 PFU/ml) were lower than those in infected individuals identified in 2002 (0.87 to 75 PFU/ml) (13). The limitations of the conversion from pool tests to individual donation NATs are the availability of reagents, technology, and cost.
With these considerations in mind, it is clear that highly sensitive assays will be required to effectively detect the low levels of virus present in infected individuals. The FDA has recommended that assays for the detection of WNV have a minimum analytical sensitivity of 1,000 RNA copies/ml in a pool test. Current pool testing methods meet this requirement, but, as noted above, sensitivity estimates show that some samples escape detection. The enhanced sensitivity of ID NAT may be required to achieve this goal. Here we describe such a sensitive NAT system, which uses semiautomated extraction from 400 μl of single-donor plasma and uses a highly specific universal beacon probe.
MATERIALS AND METHODS
Positive control reagents.
WNV tissue culture supernatant, provided by Laura Kramer (New York State Department of Health, Albany, NY), was inactivated by heating at 60°C for 1 h, diluted in negative human plasma, and quantitated by reverse transcription (RT)-PCR assay using a panel of samples containing a stated amount of WNV RNA (WNV panel QWN 701, lot no. 104874; Boston Biomedica, Inc. [BBI], Boston, MA). Positive control samples were adjusted to contain 60 and 300 RNA copies/ml, rapidly frozen, and stored as single-use aliquots at −80°C or below. Aliquots were thawed by shaking in a 37°C water bath on the day of use.
Internal control (IC) reagents.
Dengue virus (Hawaii strain) culture supernatant, also provided by Laura Kramer, was inactivated by heating at 60°C for 1 h, diluted to 107 PFU/ml in 10% negative human plasma-phosphate-buffered saline, and aliquoted in amounts sufficient for single-plate reactions. Aliquots were rapidly frozen and stored at −80°C or below and thawed by shaking in a 37°C water bath on the day of use.
Extraction and purification of nucleic acid.
The extraction procedure was a semiautomated method using Tecan Genesis RSP 150 and Genesis Workstation 200. In each run, two WNV-positive plasma controls at 60 and 300 copies/well, six WNV-negative plasma controls, and eight internal control negatives were included. For internal control positives, heat-inactivated dengue virus at 8,000 PFU/ml was mixed into lysis buffer.
Fifty μl of proteinase K (QIAGEN), 400 μl of plasma sample, and 530 μl of AL lysis buffer (QIAGEN) were mixed in a 2-ml deep-well plate (Marsh). The plate was tightly sealed with an adhesive cover and incubated in a shaking water bath for 25 min at 58°C. Following the incubation, 530 μl ethanol was mixed with the lysate.
A total of 630 μl of the mixed lysate was transferred into the wells of an 800-μl 96-well glass fiber filter plate (GF/F; Whatman, Clifton, New Jersey) and filtered at 600 mbar for 4 min. The remaining lysate was transferred into the wells of the glass fiber filter plate and filtered at 600 mbar for 5 min.
A total of 680 μl of wash buffer (AW2; QIAGEN) was transferred into the wells of the glass fiber filter plate and filtered at 600 mbar for 45 seconds. A second aliquot of wash buffer (340 μl, AW2; QIAGEN) was applied to the wells of the glass fiber filter plate and filtered at 600 mbar for 30 seconds.
Droplets from the tip directors of the glass fiber filter plate were removed using clean paper towels. The plate was vacuum dried for 10 min at 600 mbar and air dried for an additional 10 min at room temperature.
To elute the nucleic acids, a 96-well PCR plate (ABI, Foster City, CA) that was prefilled with 30 μl/well of RT master mix was placed under the filter plate on a vacuum manifold. Sixty-five μl of nuclease-free H2O was transferred into the filter plate wells, incubated for 2 min at room temperature, and then vacuum filtered at 500 mbar.
RT reaction.
The RT reaction was carried out in an 80-μl volume containing 50 μl purified nucleic acid, 16 U RNase inhibitor (RNasin; Promega, Madison, WI), 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 5 mM dithiothreitol, 3 mM MgCl2, 0.25 mM each deoxynucleoside triphosphate, 1.25 μM WNV reverse primer (5′-GCTCTTGCCGGGCCCTCCTG-3′, 20mer, positions 110 to 129), 38 nM dengue virus (internal control) reverse primer (13mer, 5′-GCGTTCTGTGCCT-3′), and 80 units Moloney murine leukemia virus reverse transcriptase (Gibco BRL, Rockville, MD). The reaction was run at 42°C for 45 min, followed by 90°C for 2 min.
PCR amplification using universal molecular beacon technology.
As described below, the PCR product generated by amplification of the dengue virus internal control RNA was detected using a fluorescence-labeled TaqMan probe. The PCR product amplified from WNV target RNA was detected using the universal molecular beacon technology described below. Molecular beacons are single-stranded oligonucleotide probes that have a stem-loop structure (17). Because of the sensitivity of molecular beacons to even small numbers of nucleotide mismatches between probe and target sequence (16), we developed a modification of molecular beacon technology (Universal Beacon PCR) to permit their use in detection of even highly variant virus species. The method, which is described in detail in U.S. Patent Application Serial No. 10/399843, relies on the use of forward and reverse primers which are designed to hybridize “nose to nose” on the target nucleic acid sequence to be amplified (i.e., there is no intervening gap between the forward and reverse primers). The molecular beacon probe is designed to hybridize asymmetrically across the junction created by the two primers. For the detection of WNV, primers and probes were targeted to a 47-bp sequence spanning the 5′ untranslated region and the nucleocapsid start site of the WNV genome. The reference sequence used for primer design and nucleotide numbering was the New York 1999 equine isolate reported by Lanciotti et al. (10) (GenBank, AF196835).
At the end of the RT reaction, 40 μl PCR mix was added to each of the cDNA reaction wells. The total of 120 μl reaction mixture contained 33.3 mM Tris-HCl (pH 8.3), 50 mM KCl, 3.33 mM dithiothreitol, 2.63 mM MgCl2, 0.17 mM deoxynucleoside triphosphate, 830 nM WNV reverse primer (5′-GCTCTTGCCGGGCCCTCCTG-3′, 20mer, positions 110 to 129), 83 nM WNV forward primer (5′-GCACGAAGATCTCGATGTCTAAGAAAC-3′, 27mer, positions 83 to 109), 120 nM WNV probe (5′ 6-FAM-cgcacgATCTCGATGTCTAAGAAACCcgtgcg-DABCYL-3′ [probe region is in uppercase and stem nucleotides are in lowercase]), 38 nM dengue virus (internal control) reverse primer (13mer, 5′-GCGTTCTGTGCCT-3′, positions 10686 to 10698), 38 nM dengue virus (internal control) forward primer (20mer, 5′-GCATATTGACGCTGGGAGAGA-3′, positions 10632 to 10652), 10 nM dengue virus probe (19mer, 5′-VIC-AGATCCTGCTGTCTCTACA-MGB-3′, positions 10657 to 10675), and 2.5 units of AmpliTaq gold DNA polymerase (Perkin-Elmer). The reaction was run at 95°C for 10 min to activate AmpliTaq gold and subsequently subjected to 45 cycles at 95°C for 30 seconds, at 58°C for 30 seconds, and at 72°C for 30 seconds on a thermal cycler (ABI 2700; PE-Biosystems, Foster City, CA).
Detection by fluorescence reading and calculations.
A spectrofluorometric thermal cycler (ABI PRISM 7900HT; ABI, Foster City, CA) was used for end point detection at the end of 45 PCR cycles. WNV signal is detected with a FAM-labeled probe, which is read at 522 nm, and IC signal labeled with VIC, which is read at 554 nm. Test runs are considered valid if both negative and positive control values fall within predetermined ranges. Results for individual samples are considered valid if the VIC relative fluorescence unit (RFU) value exceeds the IC cutoff. The IC cutoff is calculated from the mean VIC RFU of IC negative controls plus 3 standard deviations (SD) (n = 8). The WNV-reactive cutoff is calculated from the mean FAM RFU of the WNV negative controls plus 5 SD (n = 4). Samples with lower RFU values than IC cutoff values will be considered IC failures unless they are WNV PCR positive (see Results). A positive sample is one in which the FAM RFU is greater than or equal to the WNV cutoff RFU regardless of IC RFU.
Statistical analysis.
To determine the sensitivity of this assay, 95% and 50% limits of detection (LODs) were obtained by Probit analysis (SPSS 11.5; SPSS Inc., Chicago, IL) at 95% confidence intervals.
RESULTS
The New York Blood Center (NYBC) WNV assay readout system.
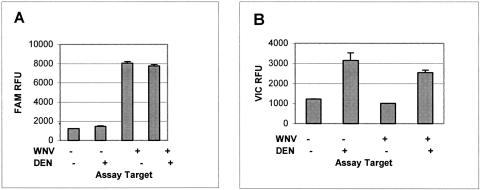
Due to a spectral overlap when both FAM and VIC signals are present at the same time, there is a spectral interaction between the two molecules. To correct this interaction, ABI's assay readout software, SDS 2.2, utilizes spectral compensation. Under our current assay conditions, where the FAM (WNV probe)-to-VIC (IC) ratio is 12:1, VIC signal has little effect on FAM signal; there is a minor increase in FAM background and a slight decrease in WNV signals (Fig. 1A). However, there is a substantial reduction in VIC signals when FAM signals are high enough. This causes IC signal suppression when WNV signals are strong (Fig. 1B). In our assay system, WNV-positive samples will have suppressed IC signals, but that does not interfere with the assay since a confirmed WNV-positive sample indicates the reaction efficacy of that well.
FIG. 1.
WNV and internal control signal interaction. RT-PCR was performed on WNV (100 copies per reaction) or dengue virus RNA (DEN; 2 ng total RNA from culture supernatant per reaction) alone or in combination. The means of four replicates are presented. A, WNV (FAM) fluorescence; B, dengue virus (VIC) fluorescence. Error bars, +ISD.
Evaluation of specificity.
Donor plasma samples from frozen archive plates, collected during the WNV-free season, were screened to assess the specificity, false-positive rate, and IC failure rate of our WNV-NAT reagents. There were no WNV false positives in 420 donor samples collected in March 2000 and October 2000. There was one IC failure. This sample showed a low FAM background as well, due to interfering material on the outer surface of the bottom of the well. Thus, we now place the PCR plate in a PCR plate holder that does not permit the plate to touch any surface before being placed on a thermocycler.
To determine whether the WNV NAT reagents were reactive to other viruses, 15 other viruses were tested in parallel with WNV using the WNV assay as described in Materials and Methods. Extraction and RT-PCR were performed with 400 μl plasma or with 5 μl virus-containing supernatant spiked into 395 μl normal human plasma. As shown in Table 1, the WNV NAT PCR was nonreactive to all 15 other viruses tested in parallel with WNV. The fact that high-titer St. Louis encephalitis virus and Japanese encephalitis virus samples showed no reactivity to our WNV reagent indicates the high specificity of the reagents.
TABLE 1.
Specificity of the WNV assay
| Virusa | Sample type | Sourceb | Strain | Titer of stock tested | No. tested | No. reactive |
|---|---|---|---|---|---|---|
| Dengue virus type 1 | Cell supernatant | NYS | Hawaii | 107.5 PFU/ml | 4 | 0 |
| Dengue virus type 2 | Cell supernatant | NYS | New Guinea | 106.5 PFU/ml | 4 | 0 |
| Dengue virus type 3 | Cell supernatant | NYS | H-87 | 107.3 PFU/ml | 4 | 0 |
| Dengue virus type 4 | Cell supernatant | NYS | H-241 | 106.5 PFU/ml | 4 | 0 |
| SLE | Cell supernatant | NYS | Kern 217 | 108.0 PFU/ml | 4 | 0 |
| Yellow fever virus | Cell supernatant | NYS | 17D | 107.5 PFU/ml | 4 | 0 |
| HTLV-1 | Cell supernatant | NYBC | NYBC donor | NAc | 4 | 0 |
| Rocio | Cell supernatant | NYS | SPH34675 | 106.6 PFU/ml | 4 | 0 |
| CMV | Cell supernatant | ATCC | Towne | NA | 4 | 0 |
| HAV | Cell supernatant | CDC | NA | NA | 4 | 0 |
| Parvovirus B19 | Cell supernatant | NYBC | B19 | 1011 particles/ml | 4 | 0 |
| HCV | Plasma | NYBC | 1b | 106.5 copies/ml | 4 | 0 |
| HIV | Plasma | NYBC | LAV | 105.5 copies/ml | 4 | 0 |
| HBV | Plasma | NYBC | Adw | 104.5 copies/ml | 4 | 0 |
| JEV | Plasmidd | GCCe | Nakayama | 108 copies/ml | 4 | 0 |
| WNV | Plasma | NYS | NYC01035 | 104.5 copies/ml | 4 | 4 |
CMV, cytomegalovirus; HAV, hepatitis A virus; HBV, hepatitis B virus; HTLV-1, human T-cell leukemia virus type 1; SLE, St. Louis encephalitis virus.
NYS, Laura Kramer, New York State Department of Health.
NA, data not provided by supplier.
A plasmid carrying the JEV 5′ untranslated region and core gene (nucleotides 1 to 476) was used for this assay, omitting the RT step.
Mogam Biotechnology Research Institute of the Green Cross Corp., Seoul, Korea.
Cross-contamination study using alternate positive and negative wells.
To assess the possibility of well-to-well cross-contamination and to test donor plasma variability, 80 donor plasma samples from an archive plate were transferred to a deep-well plate and WNV-positive plasma was spiked into 40 alternating wells and assayed. The cutoff (mean RFU from four negatives plus 5 SD) for WNV RFU was 1,233, and mean FAM (WNV) RFU for 40 positive wells (300 WNV copies/ml) was 3,966 ± 963 and for 40 negative wells was 918 ± 79. The cutoff (mean RFU from eight negatives plus 3 SD) for IC RFU was 61; mean IC RFU for negative wells was 264 ± 85. There was no well-to-well cross-contamination. The 40 different plasma sources spiked with WNV were all positive. Although the SD for 40 positives is somewhat high, plasma source variability did not interfere with assay detection.
Analytical sensitivity and reproducibility.
BBI stock (Uganda strain, 7.33 × 104 copies/ml, lot no. 101702C) was diluted in human plasma at 12.5, 6.3, 3.2, 1.6, and 0.8 copies per ml, based on BBI's estimates, and tested to determine the sensitivity of the WNV assay (Table 2). Sixteen replicates per dilution were tested and repeated in five separate experiments (Table 3). These data were subjected to probit analysis to determine the 95% and 50% LOD in each experiment separately (Table 3). Coefficients of variations of these experiments indicated a high degree of reproducibility. As a reliable and accepted WNV standard is not yet available, we used dilutions from the BBI stock for the above-sensitivity determination. However, we found a previous BBI panel (QWN 701, lot no. 104874), with which our quantitation standards were calibrated, to have an average WNV copy number 4.31-fold lower than the stated copy number when estimated with an in-house quantitative PCR method for comparison of the two lots. The data are shown in Table 4. The average difference was 4.31 (n-fold). Thus, our estimate of sensitivity would depend on which panel we use as a standard for the study. With lot no. 101702C, the 95% LOD is 3.79 copies/ml; however, if we used QWN 701 (lot no. 104874), the 95% LOD is 3.79 × 4.31 = 16.3 copies/ml. We await the completion of the evaluation of FDA and CDC panels to resolve this issue.
TABLE 2.
Summary of the data used for determination of WNV assay sensitivity
| WNV dilutionsa (WNV copies/ml) | No. positive/no. tested for run no.
|
||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| 0.8 | 0/16 | 1/16 | 0/16 | 2/16 | 1/16 |
| 1.6 | 7/16 | 3/16 | 6/16 | 6/16 | 1/16 |
| 3.2 | 15/16 | 15/16 | 12/16 | 13/16 | 12/16 |
| 6.3 | 16/16 | 16/16 | 16/16 | 16/16 | 16/16 |
| 12.5 | 16/16 | 16/16 | 16/16 | 16/16 | 16/16 |
Dilutions were made from BBI stock (Uganda, 7.33 × 104 copies/ml, lot no., 101702C) in human plasma.
TABLE 3.
Probit analysis of assay sensitivitya
| LOD (%) | Sensitivity of plates run forb:
|
Meanb | SD | CVc | ||||
|---|---|---|---|---|---|---|---|---|
| Experiment 1 | Experiment 2 | Experiment 3 | Experiment 4 | Experiment 5 | ||||
| 95 | 3.09 | 3.36 | 4.07 | 4.06 | 4.18 | 3.79 | 0.49 | 0.13 |
| 50 | 1.92 | 2.13 | 2.33 | 2.08 | 2.64 | 2.22 | 0.28 | 0.12 |
Results shown are for 16 replicates of each dilution tested on five plates run on different days.
WNV copies/ml.
CV, coefficient of variation.
TABLE 4.
In-house quantitationa estimate for WNV and BBI materials
| Material | Quantitation (copies/ml) for:
|
||
|---|---|---|---|
| Experiment 1 | Experiment 2 | Experiment 3 | |
| WNVb | 100 | 500 | 2,000 |
| BBI panel QWN 701 (lot no. 104874) | 187c | 393 | 1,362 |
| BBI stock (lot no. 101702C) | 816 | 1,360 | 6,998 |
| Difference (n-fold) | 4.35 | 3.45 | 5.14 |
In-house quantitation standards are extracted WNV RNA from cell supernatant calibrated with the BBI material.
Copy numbers in dilutions according to estimates from BBI.
Means of four estimates are shown. Copies/ml based on NYBC quantitative PCR.
Detection of WNV-positive donor samples.
We have analyzed 11 donor plasma samples that were found positive for WNV at the NYBC in screening with the Roche assay during 2003. The results using the NYBC screening assay are summarized in Table 5, and the serological test results on donor follow-up are summarized in Table 6. Of cases successfully followed, eight out of eight seroconverted. The mean titers were log10 2.90 ± 0.93 copies/ml with the NYBC WNV assay and log10 2.70 ± 0.79 copies/ml with the alternate TaqMan assay. The titers of the pool sample would be 6- to 16-fold lower, depending on the commercial kit, further suggesting that single-unit testing would be desirable.
TABLE 5.
Analysis of WNV-positive units detected in 2003 by the Roche pool assay at NYBC: quantitation using NYBC and Taqman quantitative PCR assays
| Donor no. | Sample collection date (mo/day/yr) | Quantitation (log10 copies/ml) forb:
|
|
|---|---|---|---|
| NYBC WNV assay | Taqman alternate assay | ||
| 1 | 7/29/2003 | 3.85a | 3.32 ± 0.22 |
| 2 | 8/21/2003 | 3.84a | 3.72 ± 0.04 |
| 3 | 8/27/2003 | 1.78a | 1.41 ± 0.29 |
| 4 | 9/2/2003 | 3.8a | 3.06 ± 0.15 |
| 5 | 9/9/2003 | 2.7 ± 0.07 | 2.93 ± 0.01 |
| 6 | 9/9/2003 | 1.6 ± 0.32 | 1.88 ± 0.15 |
| 7 | 9/9/2003 | 2.3 ± 0.08 | 2.21 ± 0.10 |
| 8 | 9/11/2003 | 3.9 ± 1.03 | 3.08 ± 0.10 |
| 9 | 9/13/2003 | 1.7 ± 0.29 | Negativec |
| 10 | 10/15/2003 | 3.26 ± 0.07 | Not tested |
| 11 | 10/15/2003 | 3.12 ± 0.08 | Not tested |
| Mean ± SD | 2.90 ± 0.93 | 2.70 ± 0.79 | |
No SD available since only a single reaction was set up for these samples.
Mean ± SD unless otherwise indicated.
Signals were lower than the assay detection limit.
TABLE 6.
Analysis of WNV-positive units detected in 2003 by the Roche pool assay at NYBC: serological test results upon donor follow-upa
| Donor no. | Sample date (mo/day/yr)b | Anti-WNV test results forc:
|
|
|---|---|---|---|
| IgM | IgG | ||
| 1 | 7/29/2003 | Negd | Neg |
| 9/3/2003 | Pose | Pos | |
| 2 | 8/21/2003 | Neg | Neg |
| 9/4/2003 | Pos | Pos | |
| 9/18/2003 | Pos | Pos | |
| 3 | 8/27/2003 | Neg | Neg |
| 9/10/2003 | Pos | Neg | |
| 9/24/2003 | Pos | Pos | |
| 4 | 9/2/2003 | Neg | Neg |
| 9/16/2003 | Pos | Neg | |
| 10/9/2003 | Pos | Pos | |
| 5 | 9/9/2003 | Neg | Neg |
| 9/23/2003 | Pos | Pos | |
| 10/7/2003 | Pos | Pos | |
| 6 | Rf | R | |
| 7 | R | R | |
| 8 | 9/11/2003 | Neg | Pos |
| 9/25/2003 | Pos | Pos | |
| 10/14/2003 | Pos | Pos | |
| 9 | R | R | |
| 10 | 10/15/2003 | Neg | Neg |
| 10/29/2003 | Pos | Pos | |
| 11/12/2003 | Pos | Pos | |
| 11 | 10/15/2003 | Neg | Neg |
| 10/30/2003 | Pos | Pos | |
| 11/13/2003 | Pos | Pos | |
Data tested by the California Department of Health Services for WNV-positive donor samples.
The first sample from each donor is from the day of blood donation.
Neg, negative; Pos, positive; IgG and IgM, immunoglobulins G and M, respectively.
Antibody not detected.
Antibody detected.
R, refused follow-up.
DISCUSSION
In recent years, there have been many WNV detection systems reported using a variety of technologies. Gen-Probe uses transcription-mediated amplification in combination with a chemiluminescent-detection method. Roche uses RT-PCR with a TaqMan detection system, while others reported using the combinations of nucleic acid sequence-based amplification and molecular beacon (9, 18), nucleic acid sequence-based amplification and TaqMan (8), and RT-PCR and SYBR green (12). We have developed a combined RT-PCR, using the universal beacon (modified molecular beacon) probe for WNV detection and the TaqMan probe for IC detection. Use of the ABI 7900 system, which excites fluorescent molecules only at 490 nM, was a challenging task for dual-probe detection. To accommodate a second fluorescent molecule as an internal control in addition to FAM for WNV detection, the most promising fluorophore was VIC, developed by ABI. However, VIC is available exclusively for the TaqMan probe. Therefore, we optimized our assay conditions for a single tube containing FAM-labeled molecular beacon and VIC-labeled TaqMan probes. We intentionally designed a weak VIC internal control signal to prevent FAM signals from being affected by the VIC signal. This was accomplished using a FAM and VIC probe molar ratio of 12:1. Our assay presents a combination of two fluorescent-detection technologies in a single reaction.
The extraction system described here uses proteinase K- and SDS buffer-based lysis, which is capable of isolating viral RNA and DNA simultaneously (W. Pfahler et al., submitted). In combination with the universal beacon, which is capable of detecting all genotypes of highly variable viruses, such as hepatitis C virus (HCV) and human immunodeficiency virus (HIV), this extraction method permits economical multiplex ID NAT screening, since only a single extraction is required for detection of both RNA and DNA viruses. Individual donor NAT screening would benefit the detection of low-titer infections during window periods of WNV, hepatitis B virus, and HIV infections.
The WNV reagents proved to be highly specific for WNV since they were not reactive to JEV, St. Louis encephalitis virus, or 13 other viruses tested. The actual sensitivity of this assay remains to be determined, but our sensitivity should be higher than that with commercial kits using pool testing, as the 6- to 16-fold dilution is avoided. We estimate that this WNV ID NAT system will yield a higher number of WNV-positive samples than those of other commercial kits that use pool testing.
Acknowledgments
We gratefully acknowledge the provision of viral strains for this study by Laura Kramer of the New York State Department of Health, Howard Fields of the CDC, and Doo-Hong Park of the Mogam Biotechnology Research Institute of the Green Cross Corporation, Seoul, Korea.
REFERENCES
- 1.Biggerstaff, B. J., and L. R. Petersen. 2002. Estimated risk of West Nile virus transmission through blood transfusion during an epidemic in Queens, New York City. Transfusion 42:1019-1026. [DOI] [PubMed] [Google Scholar]
- 2.Busch, M. P., S. Caglioti, E. F. Robertson, J. D. McAuley, L. H. Tobler, H. Kamel, J. M. Linnen, V. Shyamala, P. Tomasulo, and S. H. Kleinman. 2005. Screening the blood supply for West Nile virus RNA by nucleic acid amplification testing. N. Engl. J. Med. 353:460-467. [DOI] [PubMed] [Google Scholar]
- 3.Candotti, D., J. Temple, S. Owusu-Ofori, and J. P. Allain. 2004. Multiplex real-time quantitative RT-PCR assay for hepatitis B virus, hepatitis C virus, and human immunodeficiency virus type 1. J. Virol. Methods 118:39-47. [DOI] [PubMed] [Google Scholar]
- 4.CDC. 2002. Possible West Nile virus transmission to an infant through breast-feeding—Michigan, 2002. Morb. Mortal. Wkly. Rep. 51:877-878. [PubMed] [Google Scholar]
- 5.CDC. 2003. Update: detection of West Nile virus in blood donations—United States, 2003. Morb. Mortal. Wkly. Rep. 52:916-919. [PubMed] [Google Scholar]
- 6.CDC. 2004. Update: West Nile virus screening of blood donations and transfusion-associated transmission—United States, 2003. Morb. Mortal. Wkly. Rep. 53:281-284. [PubMed] [Google Scholar]
- 7.Iwamoto, M., D. B. Jernigan, A. Guasch, M. J. Trepka, C. G. Blackmore, W. C. Hellinger, S. M. Pham, S. Zaki, R. S. Lanciotti, S. E. Lance-Parker, C. A. DiazGranados, A. G. Winquist, C. A. Perlino, S. Wiersma, K. L. Hillyer, J. L. Goodman, A. A. Marfin, M. E. Chamberland, and L. R. Petersen. 2003. Transmission of West Nile virus from an organ donor to four transplant recipients. N. Engl. J. Med. 348:2196-2203. [DOI] [PubMed] [Google Scholar]
- 8.Lambert, A. J., D. A. Martin, and R. S. Lanciotti. 2003. Detection of North American eastern and western equine encephalitis viruses by nucleic acid amplification assays. J. Clin. Microbiol. 41:379-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lanciotti, R. S., and A. J. Kerst. 2001. Nucleic acid sequence-based amplification assays for rapid detection of West Nile and St. Louis encephalitis viruses. J. Clin. Microbiol. 39:4506-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lanciotti, R. S., J. T. Roehrig, V. Deubel, J. Smith, M. Parker, K. Steele, B. Crise, K. E. Volpe, M. B. Crabtree, J. H. Scherret, R. A. Hall, J. S. MacKenzie, C. B. Cropp, B. Panigrahy, E. Ostlund, B. Schmitt, M. Malkinson, C. Banet, J. Weissman, N. Komar, H. M. Savage, W. Stone, T. McNamara, and D. J. Gubler. 1999. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 286:2333-2337. [DOI] [PubMed] [Google Scholar]
- 11.Macedo de Oliveira, A., B. D. Beecham, S. P. Montgomery, R. S. Lanciotti, J. M. Linnen, C. Giachetti, L. A. Pietrelli, S. L. Stramer, and T. J. Safranek. 2004. West Nile virus blood transfusion-related infection despite nucleic acid testing. Transfusion 44:1695-1699. [DOI] [PubMed] [Google Scholar]
- 12.Papin, J. F., W. Vahrson, and D. P. Dittmer. 2004. SYBR green-based real-time quantitative PCR assay for detection of West Nile virus circumvents false-negative results due to strain variability. J. Clin. Microbiol. 42:1511-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pealer, L. N., A. A. Marfin, L. R. Petersen, R. S. Lanciotti, P. L. Page, S. L. Stramer, M. G. Stobierski, K. Signs, B. Newman, H. Kapoor, J. L. Goodman, and M. E. Chamberland. 2003. Transmission of West Nile virus through blood transfusion in the United States in 2002. N. Engl. J. Med. 349:1236-1245. [DOI] [PubMed] [Google Scholar]
- 14.Petersen, L. R., and A. A. Marfin. 2002. West Nile virus: a primer for the clinician. Ann. Intern. Med. 137:173-179. [DOI] [PubMed] [Google Scholar]
- 15.Stramer, S. L., C. T. Fang, G. A. Foster, A. G. Wagner, J. P. Brodsky, and R. Y. Dodd. 2005. West Nile virus among blood donors in the United States, 2003 and 2004. N. Engl. J. Med. 353:451-459. [DOI] [PubMed] [Google Scholar]
- 16.Tyagi, S., D. P. Bratu, and F. R. Kramer. 1998. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 16:49-53. [DOI] [PubMed] [Google Scholar]
- 17.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]
- 18.Yates, S., M. Penning, J. Goudsmit, I. Frantzen, B. van de Weijer, D. van Strijp, and B. van Gemen. 2001. Quantitative detection of hepatitis B virus DNA by real-time nucleic acid sequence-based amplification with molecular beacon detection. J. Clin. Microbiol. 39:3656-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]