Abstract
In the summer of 2003 a community-acquired outbreak of Legionella pneumophila occurred in Rome, Italy. Three molecular typing methods, pulse-field gel electrophoresis, amplified fragment length polymorphism analysis, and sequence-based typing (SBT), were used to establish the clonal correlation among the isolates of the epidemic cluster. By comparison of the methods, SBT was the most rapid and the easiest to perform and provided unambiguous results.
Legionella naturally inhabits protozoa aquatic environments as a parasite. The increased frequency of outbreaks of Legionella pneumophila is primarily due to the contamination of artificial aquatic environments. Molecular typing with associated epidemiological investigations can help establish the link between clinical and environmental isolates and identify the source of infection (2, 17, 24). However, the efficiencies of the various typing methods vary, and not all typing methods are standardized (14, 26, 27). The need for extensive comparison among the different molecular typing methods is widely recognized (3, 7). By exploiting the need for molecular characterization of isolates from a recent epidemic of L. pneumophila serogroup 1 infection to establish the source of infection, in the present study we have compared three molecular typing methods. The outbreak occurred in the period from August to October 2003 in a neighborhood of Rome, with 15 notified cases, including one death (21). In accordance with epidemiological investigations, microbiological analyses by incubation of α-buffered charcoal yeast extract (Oxoid, United Kingdom) agar plates at 37°C with 2.5% CO2 showed L. pneumophila contamination of water and air samples from the cooling tower of a shopping center in the area. Monoclonal antibody (MAb) typing, performed by indirect immunofluorescence assay with the “Dresden MAb panel” (14), revealed the common subgroup Philadelphia for the single clinical epidemic-related isolate and the nine environmental epidemic-related isolates.
As serotyping is insufficient as a means of ascertaining the actual source of contamination (3, 15), the genetic relatedness between human and environmental strains was determined both by comparison of the genomic profiles by pulse-field gel electrophoresis (PFGE) and amplified fragment length polymorphism analysis (AFLP) analysis and by comparison of the nucleotide sequences of five genes by sequence-based typing (SBT). Data from the epidemic-related isolates were also compared to data from unrelated strains.
PFGE was performed as described by Castellani Pastoris et al. (5) by using 20 U of NotI (Roche, Italy) for overnight macrorestriction at 37°C of the genomic DNA plugs. Saccharomyces cerevisiae chromosomal DNA (Bio-Rad, Hercules, Calif.) was used as a size marker. As shown in Fig. 1, PFGE produced genomic patterns that were the same for the clinical and the environmental isolates from the epidemic, but these patterns were dissimilar from those for the control strains. The NotI enzyme was chosen because of its rare restriction sites in the genome of Legionella, in which it produces only a few high-molecular-weight fragments. However, comparison of the genomic profiles showed very thick bands that could be constituted by one or more unresolved fragments. In addition, macrorestriction of only one of the epidemic-related environmental samples produced a faint additional band that was absent from the other isolates. This might constitute a confounding factor in the attribution of clonality. Although the results obtained by PFGE were overall in agreement with those obtained by MAb typing, the isolates of L. pneumophila with the same patterns might not come from the same environmental source (18). Therefore, these results, even if they are reliable (16, 22), should be used with caution (7, 25). On the other hand, PFGE is time-consuming, technically difficult, and somewhat cumbersome to perform because of the need to prepare plugs with fresh bacterial cultures, lyse the bacteria, and define the correct parameters for the electrophoretic run.
FIG. 1.
PFGE analysis of genomic DNA from a clinical isolate (sample in lane 1) and environmental isolates (samples in lanes 2 to 10) from the epidemic. The samples in lanes 11 and 12 were unrelated environmental and clinical strains, respectively, and the sample in lane 13 was the L. pneumophila ATCC 33152 type strain. Lanes M, molecular weight marker (S. cerevisiae).
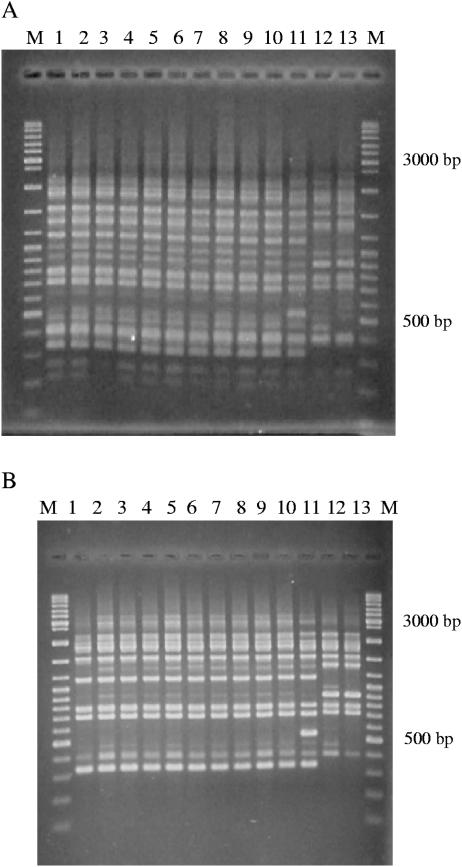
The second method used in this study was AFLP analysis, currently considered the “gold standard” for the typing of L. pneumophila (10, 11). PCR amplification was performed both by using puRe Taq Ready-to-Go PCR beads (Amersham BioSciences, United Kingdom) and by using 2.5 U of Taq polymerase from a commercial PCR kit (Applied Biosystems) with 75 ng selective primer AFLP-PstI-G (5′-GACTGCGTACATGCAGG-3′). The MBI Fermentas Ladder Mix (MBI Fermentas, United Kingdom) was used as a molecular size marker. AFLP analysis is technically easier and faster to perform than PFGE and is less expensive. In our study, it confirmed the high similarity of the DNA fingerprints of the cluster-related clinical and environmental isolates (Fig. 2), but AFLP analysis allowed a more reliable comparison than PFGE, because it showed more numerous smaller fragments. Remarkably, as shown in Fig. 2A, the environmental strain unrelated to the epidemic had a profile very similar to those of the isolates involved in the epidemic, whereas unrelated clinical and type strains had clearly different banding patterns. Figure 2B shows the results of AFLP analysis performed for comparative purposes with Taq polymerase from an Applied Biosystems commercial PCR kit. More bands were detected for the environmental isolates from the epidemic, while unrelated strains had a different genomic pattern. More relevant general profiles were different, depending on the Taq polymerase used; therefore, comparison of the genome types with those in the AFLP database (www.ewgli.org) could give incorrect results. Moreover, as in PFGE, the analysis of the AFLP banding patterns by visual or computational methods proved to be difficult because band discrimination is subjective.
FIG. 2.
AFLP analysis. (A) AFLP performed according to the version 1.2 standard EWGLI AFLP protocol; (B) AFLP performed with Taq polymerase from a commercial PCR kit. The genomic patterns of the clinical isolate (sample in lane 1) and environmental isolates (samples in lanes 2 to 10) from the epidemic are shown. The samples in lanes 11 and 12 are unrelated environmental and clinical strains, respectively. The sample in lane 13 is the L. pneumophila ATCC 33152 type strain. Lanes M, MBI Fermentas ladder mix.
SBT, proposed by Gaia et al. for L. pneumophila serogroup 1 and non-serogroup 1 (12, 13), was also used in this study. We sequenced both strands of the amplicons of five of the six genes (flaA, proA, mip, asd, and pilE) suggested to be used for sequencing with the primers described by Gaia et al. (13). The sequences were determined by using an ABI PRISM BigDye terminator DNA sequencing kit and were analyzed on a 310 ABI DNA sequencer (Applied BioSystems). The sequenced amplicons of each gene from the isolates from the epidemic showed 100% identity. These sequences were compared with those in the European Working Group for Legionella Infections (EWGLI) SBT database, and for every gene, only the sequences of the isolates from the epidemic all matched the same allelic target with the same allelic number (Table 1). The control strains matched different alleles. The results were always clear, reliable, and reproducible. Although all molecular methods demonstrated the clonal relationship of the epidemic cluster, SBT was sufficiently rapid and the easiest to perform by using the same PCR cycle and the same sequencing analysis for all genes. Although AFLP analysis and SBT have similar performance times, for each PCR, SBT requires only 50 ng genomic DNA instead of the 1.5 μg required for AFLP. This means that minipreparations of genomic DNA (1) are sufficient for performance of the SBT assay, which is relevant in typing experiments. Moreover, if we consider that molecular biology laboratories usually have a sequencer, the costs of the equipment required to perform SBT, as well as materials and labor, are similar to those required to perform AFLP analysis, but the results of SBT are unambiguous and reproducible.
TABLE 1.
Allelic profiles of the L. pneumophila serogroup 1 strains used in this study
| Straina | Allele no.
|
||||
|---|---|---|---|---|---|
| flaA | pilE | asd | mip | proA | |
| 1 | 2 | 3 | 9 | 10 | 1 |
| 2 | 2 | 3 | 9 | 10 | 1 |
| 3 | 2 | 3 | 9 | 10 | 1 |
| 4 | 2 | 3 | 9 | 10 | 1 |
| 5 | 2 | 3 | 9 | 10 | 1 |
| 6 | 2 | 3 | 9 | 10 | 1 |
| 7 | 2 | 3 | 9 | 10 | 1 |
| 8 | 2 | 3 | 9 | 10 | 1 |
| 9 | 2 | 3 | 9 | 10 | 1 |
| 10 | 2 | 3 | 9 | 10 | 1 |
| 11 | 1 | 4 | 3 | 1 | 1 |
| 12 | 3 | 4 | 1 | 1 | 1 |
| 13 | 3 | 4 | 1 | 1 | 9 |
Strains 1 to 10 are of the epidemic cluster; strain 11 and 12 are environmental and clinical-unrelated strains, respectively; strain 13 is the L. pneumophila ATCC 33152 type strain.
SBT has been used successfully to type other bacteria for both evolutionary and epidemiological studies (9, 17, 19). Multilocus sequence typing has been developed to determine the allelic variations of multiple housekeeping genes (8, 20, 23). Recently, some studies have explored the suitability of sequencing single genes to differentiate bacterial strains (4, 6). DNA sequence typing has multiple advantages over fingerprinting-based methods: less subjectivity in interpretation of the results, transferability of the data among laboratories, the portability of the data through web-based databases, and the opportunity for use of the data to probe pathogen evolution and to track the spread of clonal groups. Therefore, the use of the SBT method for robust genotyping with high discriminatory power is warranted, and the method is proposed as the new gold standard for the typing of L. pneumophila (12, 13).
Acknowledgments
We are grateful to A. Cassone and G. Orefici for encouragement, support, and useful suggestions throughout this study. We also thank Alessandra Ciervo and Ian Stansfield for critical analysis of the manuscript.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1994. Current protocols in molecular biology. Greene Publishing Associates and Wiley Interscience, New York, N.Y.
- 2.Bartlett, J. G. 1993. Legionnaires' disease: over-treated, under-diagnosed. J. Crit. Illness 8:755-768. [Google Scholar]
- 3.Bernarder, S., K. Jacobson, H. Helbig, P. C. Luck, and M. Lundholm. 2003. A hospital-associated outbreak of Legionnaires' disease caused by Legionella pneumophila serogroup 1 is characterized by stable genetic fingerprinting but variable monoclonal antibody patterns. J. Clin. Microbiol. 41:2503-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai, S., D. Y. Kabuki, A. Y. Kuaye, T. G. Cargioli, M. S. Chung, R. Nielsen, and M. Wiedmann. 2002. Rational design of DNA sequence-based strategies for subtyping Listeria monocytogenes J. Clin. Microbiol. 40:3319-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castellani Pastoris, M., L. Ciceroni, R. Lo Monaco, P. Goldoni, B. Mentore, G. Flego, L. Cattani, S. Ciarrocchi, A. Pinto, and P. Visca. 1997. Molecular epidemiology of an outbreak of Legionnaires' disease associated with a cooling tower in Genova-Sestri Ponente, Italy. Eur. J. Clin. Microbiol. Infect. Dis. 16:883-892. [DOI] [PubMed] [Google Scholar]
- 6.Diggle, M. A., C. M. Bell, and S. C. Clarke. 2003. Nucleotide sequence-based typing of meningococci directly from clinical samples. J. Med. Microbiol. 52:505-508. [DOI] [PubMed] [Google Scholar]
- 7.Drenning, S., J. E. Stout, J. R. Joly, and V. L. Yu. 2001. Unexpected similarity of pulsed-field gel electrophoresis patterns of unrelated clinical isolates of Legionella pneumophila, serogroup 1. J. Infect. Dis. 183:628-632. [DOI] [PubMed] [Google Scholar]
- 8.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feil, E. J., M. C. Enright, and B. G. Spratt. 2000. Estimating the relative contributions of mutation and recombination to clonal diversification: a comparison between Neisseria meningitidis and Streptococcus pneumoniae. Res. Microbiol. 151:465-469. [DOI] [PubMed] [Google Scholar]
- 10.Fry, N. K., J. M. Bangsborg, S. Bernarder, J. Etienne, B. Forsblom, V. Gaia, P. Hasenberger, C. Pelaz, M. Strulens, S. A. Uldum, P. Visca, and T. G. Harrison. 2000. Assessment of intercentre reproducibility and epidemiological concordance of Legionella pneumophila serogroup 1 genotyping by amplified fragment length polymorphism analysis. Eur. J. Clin. Microbiol. Infect. Dis. 19:773-780. [DOI] [PubMed] [Google Scholar]
- 11.Fry, N. K., J. M. Bangsborg, A. Bergmans, S. Bernander, J. Etienne, L. Franzin, V. Gaia, P. Hasenberger, B. Baladron Jimenez, D. Jonas, D. Lindsay, S. Mentula, A. Papoutsi, M. Struelens, S. A. Uldum, P. Visca, W. Wannet, and T. G. Harrison. 2002. Designation of the European Working Group on Legionella Infection (EWGLI) amplified fragment length polymorphism types of Legionella pneumophila serogroup 1 and results of intercentre proficiency testing using a standard protocol. Eur. J. Clin. Microbiol. Infect. Dis. 10:722-728. [DOI] [PubMed] [Google Scholar]
- 12.Gaia, V., N. K. Fry, T. G. Harrison, and R. Peduzzi. 2003. Sequence-based typing of Legionella pneumophila serogroup 1 offers the potential for true portability in legionellosis outbreak investigation. J. Clin. Microbiol. 41:2932-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaia, V., N. K. Fry. B. Afshar, P. C. Luck, H. Meugnier, J. Etienne, R. Peduzzi, and T. G. Harrison. 2005. Consensus sequence-based scheme for epidemiological typing of clinical and environmental isolates of Legionella pneumophila. J. Clin. Microbiol. 43:2047-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haertl, R., and G. Bandlow. 1991. Subtyping of Legionella pneumophila serogroup 1 isolates by small-fragment restriction endonuclease analysis. Eur. J. Clin. Microbiol. Infect. Dis. 10:630-635. [DOI] [PubMed] [Google Scholar]
- 15.Helbig, J. H., S. Arder, M. Castellani Pastoris, J. Etienne, V. Gaia, S. Lauwers, D. Lindsay, P. C. Luck, T. Marques, S. Mentula, M. F. Peeters, C. Pelaz, M. Struelens, S. A. Uldum, G. Wewalka, and T. G. Harrison. 2002. Pan-European study on culture-proven Legionnaires' disease: distribution of Legionella pneumophila serogroups and monoclonal subgroups. Eur. J. Clin. Microbiol. Infect. Dis. 21:710-716. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, W. M., K. Bernarder, T. J. Marrie, and S. D. Tyler. 1994. Discriminatory genomic fingerprinting of Legionella pneumophila by pulsed-field electrophoresis. J. Clin. Microbiol. 32:2620-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotetishvili, M., O. C. Stine, A. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2002. Multilocus sequence typing for characterization of clinical and environmental salmonella strains. J. Clin. Microbiol. 40:1626-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence, C., E. Ronco, S. Dubrou, R. Leclercq, C. Nauciel, and P. Matsiota-Bernard. 1999. Molecular typing of Legionella pneumophila serogroup 1 isolates from patients and the nosocomial environment by arbitrarily primed PCR and pulsed-field gel electrophoresis. J. Med. Microbiol. 48:327-333. [DOI] [PubMed] [Google Scholar]
- 19.Nicolas, P., G. Raphenon, M. Guibourdenche, L. Decousset, R. Stor, and A. B. Gaye. 2000. The 1998 Senegal epidemic of meningitis was due to the clonal expansion of A:4:P1.9, clone III-1, sequence type 5 Neisseria meningitidis strains. J. Clin. Microbiol. 38:198-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noller, A. C., M. C. McEllistrem, O. C. Stine, J. G. Morris, Jr., D. J. Boxrud, B. Dixon, and L. H. Harrison. 2003. Multilocus sequence typing reveals a lack of diversity among Escherichia coli O157:H7 isolates that are distinct by pulsed-field gel electrophoresis. J. Clin. Microbiol. 41:675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rota, M. C., G. Pontrelli, M. Scaturro, A. Bella, A. R. Bellomo, M. O. Trinito, S. Salmaso, and M. L. Ricci. Legionnaires' disease outbreak in Rome, Italy. Epidemiol. Infect., in press. [DOI] [PMC free article] [PubMed]
- 22.Schoonmaker, D., T. Heimberger, and G. Birkhead. 1992. Comparison of ribotyping and restriction enzyme analysis using pulsed-field gel electrophoresis for distinguishing Legionella pneumophila isolates obtained during a nosocomial outbreak. J. Clin. Microbiol. 6:1491-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spratt, B. G. 1999. Multilocus sequence typing: molecular typing of bacterial pathogens in an era of rapid DNA sequencing and the internet. Curr. Opin. Microbiol. 2:312-316. [DOI] [PubMed] [Google Scholar]
- 24.Stout, J. E., and V. L. Yu. 1997. Legionellosis. N. Engl. J. Med. 337:682-687. [DOI] [PubMed] [Google Scholar]
- 25.Thouverez, M., C. Godard, R. Leprat, and D. Talon. 2003. Is pulsed-field gel electrophoresis a valuable tool to identify nosocomial cases of Legionella pneumophila disease? J. Hosp. Infect. 55:254-259. [DOI] [PubMed] [Google Scholar]
- 26.Van Belkum, A., H. Maas, H. Verbrugh, and N. Van Leeuwen. 1996. Serotyping, ribotyping, PCR-mediated ribosomal 16S-23S spacer analysis and arbitrarily primed PCR for epidemiological studies on Legionella pneumophila. Res. Microbiol. 147:405-413. [DOI] [PubMed] [Google Scholar]
- 27.Welsh, J., and M. McClelland. 1990. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 24:7213-7218. [DOI] [PMC free article] [PubMed] [Google Scholar]