Abstract
Species-specific sequences were shown to be carried by plasmids of the three main species of Borrelia burgdorferi sensu lato involved in Lyme disease. Libraries of the 16-, 33-, and 25-kb plasmids of B. burgdorferi sensu stricto, Borrelia garinii, and Borrelia afzelii, respectively, were then built and used to isolate species-specific sequences. After sequencing of the cloned inserts, three sets of primers were designed. They were shown to determine species-specific PCR amplification products. The sensitivities of the PCR assay with these primers were 100 spirochetes for B. burgdorferi sensu stricto and 1,000 spirochetes for B. garinii and B. afzelii. The usefulness of these primers for the identification of species in biological samples (tick, serum, and cerebrospinal fluid samples) was ascertained.
Borrelia burgdorferi sensu lato spirochetes, which cause Lyme disease, are transmitted to a wide range of vertebrates by the bite of infected ticks of the genus Ixodes. Genetic and immunological studies (4, 7, 11, 19) have led to the delineation of three distinct species among pathogenic B. burgdorferi sensu lato isolates: B. burgdorferi sensu stricto, Borrelia garinii, and Borrelia afzelii. Recently, Borrelia japonica and Borrelia andersonii were recognized as two novel species of B. burgdorferi sensu lato (16, 20, 25). Up to now, these two species were not found in Lyme disease patients. The identification of the different species of B. burgdorferi sensu lato might be clinically relevant, since they seem to cause different complications (2, 3) by distinct pathogenic mechanisms (13, 14).
B. burgdorferi sensu lato strains have a small linear chromosome of approximately 950 kb. In addition, they harbor several circular and linear plasmids. The contribution of plasmids, considered minichromosomes, to the genetic makeup and variability of Borrelia strains has long been recognized (5, 8, 31, 32). In view of the fact that different clinical outcomes depend on the species of the infecting spirochete, the isolation of species-specific sequences might help provide an understanding of the distinct pathogenic mechanisms. We recently isolated a species-specific sequence of B. burgdorferi sensu stricto repeated on several plasmids of this species (21).
Among the methods able to distinguish the B. burgdorferi sensu lato species, PCRs have the advantage of not requiring previous cultivation of the organisms. In the present study, we extended the isolation of phylogenetically significant plasmid target sequences to the three pathogenic B. burgdorferi sensu lato genospecies. These sequences were exploited for the development of PCR tests. The chosen primers were tested for their ability to amplify DNA from cultured bacteria, biological fluids from patients, and infected ticks in order to facilitate the study of risk and clinical outcome of Lyme disease in areas inhabited by ticks.
Isolation of species-specific B. burgdorferi sensu lato plasmid sequences.
The Borrelia strains used in this study, listed in Table 1, were provided by I. Saint Girons (Institut Pasteur, Paris, France). Their isolation has been described previously (4, 6). We previously showed in each species plasmids with distinct pulsed-field gel electrophoretic profiles, allowing us to identify for each species a plasmid that was present in all tested strains of a given species and that had no prevalent counterpart with an equivalent molecular weight in strains belonging to the other two species (21). These plasmids were chosen as possible carriers of species-specific sequences: a 16-kb plasmid of B. burgdorferi sensu stricto IP1, a 33-kb plasmid of B. garinii N34, and a 25-kb plasmid of B. afzelii UO1. They were excised from the gel, radiolabeled by random priming, and hybridized onto a dot blot with DNAs of different strains of each species (28). As indicated in Fig. 1, the 16-kb plasmid of the B. burgdorferi sensu stricto strain hybridized strongly with DNA of this species and poorly with the DNAs of B. garinii and B. afzelii. The 33-kb plasmid of the B. garinii strain hybridized strongly with the DNAs of the other strains of this species and moderately with the DNAs of B. burgdorferi sensu stricto B31 and B. afzelii VS461 and UMO1. It hybridized poorly with the DNAs of other strains of B. burgdorferi sensu stricto and B. afzelii species. The 25-kb plasmid of the B. afzelii strain hybridized strongly with the DNAs of the other strains of B. afzelii but also hybridized with the DNA of B. garinii 20047. It hybridized poorly with DNAs of other strains of B. burgdorferi sensu stricto and B. garinii. It seemed that these three plasmids contained largely species-specific sequences.
TABLE 1.
Strains of Borrelia used in this study
| Strain | Origin | Geographic location |
|---|---|---|
| B. burgdorferi sensu stricto | ||
| B31 | Ixodes dammini | United States |
| IP1 | Human (CSFa) | France |
| IP2 | Human (CSF) | France |
| IP3 | Human (CSF) | France |
| IRS | Ixodes ricinus | Switzerland |
| B. garinii | ||
| 20047 | Ixodes ricinus | France |
| N34 | Ixodes ricinus | Germany |
| G25 | Ixodes ricinus | Sweden |
| P/Bi | Human (CSF) | Germany |
| B. afzelii | ||
| VS461 | Ixodes ricinus | Switzerland |
| UO1 | Human (skin) | Sweden |
| UMO1 | Human (skin) | Sweden |
| Iper3 | Ixodes persulcatus | Former USSR |
| B. japonica Cow611C | Japan | |
| B. hermsii | ||
| B. parkeri | ||
| B. turicatae |
CSF, cerebrospinal fluid.
FIG. 1.
Hybridization of the 16-kb plasmid of B. burgdorferi sensu stricto, the 33-kb plasmid of B. garinii, and the 25-kb plasmid of B. afzelii with DNAs of the three species of B. burgdorferi sensu lato involved in Lyme disease. DNAs (100 ng) of the B31 (dot 1), IP1 (dot 2), IP2 (dot 3), IP3 (dot 4), and IRS (dot 5) strains of B. burgdorferi sensu stricto, the 20047 (dot 6), N34 (dot 7), G25 (dot 8), and P/Bi (dot 9) strains of B. garinii, and the VS461 (dot 10), UO1 (dot 11), UMO1 (dot 12), and Iper3 (dot 13) strains of B. afzelii were denatured and spotted onto a nylon membrane. The membrane was incubated in the presence of the 16-kb radiolabeled plasmid of B. burgdorferi sensu stricto IP1 (A), the 33-kb radiolabeled plasmid of B. garinii N34 (B), or the 25-kb radiolabeled plasmid of B. afzelii UO1 (C). The membrane was autoradiographed.
For isolation of those specific sequences, the chosen plasmids, excised from the agarose gel, cleaved with Sau3A1, and separately ligated into the BamHI site of the pBluescript SK+ vector. These recombinant plasmids were introduced in Escherichia coli DH5αF′. Recombinant colonies were hybridized in parallel with total cleaved radiolabeled DNA of B. burgdorferi sensu stricto IP1, B. garinii N34, and B. afzelii UO1 (28). Three species-specific recombinants were further studied. Plasmid pMC52, carrying cloned sequences of the 16-kb plasmid of a B. burgdorferi sensu stricto strain, hybridized in a Southern blot analysis to several plasmids of strains belonging to the same species (21). Plasmids pMC159 and pMC18, carrying sequences of the 33-kb plasmid of B. garinii N34 and of the 25-kb plasmid of B. afzelii UO1, respectively, hybridized exclusively to the equivalent plasmid of strains belonging to their respective species (data not shown).
Both strands of the spirochetal inserts were sequenced (29). The insert of plasmid pMC52, specific for B. burgdorferi sensu stricto, contained 1,271 bp. The inserts of plasmid pMC159, specific for B. garinii, and plasmid pMC18, specific for B. afzelii, contained 254 and 347 bp, respectively. These sequences, analyzed by the DNA Strider (17) and oligo4-s (National Biosciences, Inc., Plymouth, Minn.) programs, were analyzed in a database search with the BLAST program for homology with nucleotide and amino acid sequences (1). They did not show homology with any other sequences (BLAST [release, June 1997]).
Selection of primers and specificity of the PCR amplification products.
To select PCR primers suitable for the differentiation of the three species of B. burgdorferi sensu lato involved in Lyme disease, we have chosen a set of primers in each specific sequence derived from each recombinant plasmid (pMC52, pMC159, and pMC18). The nucleotide sequences of the primers and their target species are presented in Table 2. All primers were tested for their ability to amplify DNA from 10 B. burgdorferi sensu lato isolates and 3 relapsing fever agents.
TABLE 2.
PCR primers for detection and identification of B. burgdorferi sensu lato involved in Lyme disease
| Primer set | Oligonucleotide sequence | Target species | Size (bp) of amplification product |
|---|---|---|---|
| MC16 | 5′-TAAAGTTTTGCATAAGC-3′ | B. burgdorferi sensu stricto | 395 |
| 5′-TACTAAAGGTGTTTCTCC-3′ | |||
| MC33 | 5′-CTAACCGCACTAACAGCAGCAAT-3′ | B. garinii | 236 |
| 5′-AGTTTTCATTAGCAGCAA-3′ | |||
| MC25 | 5′-AGAAGGAGATAAAAGAAC-3′ | B. afzelii | 125 |
| 5′-AAAAAGGTATAGCACAGT-3′ | |||
| c/c′a | 5′-CCAACTTTATCAAATTCTGC-3′ | B. burgdorferi sensu lato | 126 |
| 5′-AGGATCTATTCCAAAATC-3′ |
See reference 27.
For PCR, the reaction mixture (50 μl) contained 1.5 mM MgCl2, 175 μM (each) deoxyribonucleotide triphosphates (dATP, dCTP, dTTP, and dGTP), 0.1% Triton X-100, 50 mM KCl, 10 mM Tris-HCl (pH 8.0), 2.5 U of Thermus brockianus DNA polymerase (Dynazyme; Techgen), 50 pmol of each primer, and 10 ng of spirochetal DNA (or volumes of biological samples, as described below). Samples were overlaid with 50 μl of mineral oil (Sigma) and amplified for 35 cycles (with the c/c′ and MC16 primers), 30 cycles (with MC33 primers), or 40 cycles (with MC25 primers) in a thermocycler (New Brunswick Scientific, Benelux b.v.) under the following conditions: 1 min at 96°C; 1 min at 54°C (with the c/c′ and MC16 primers), 60°C (with MC33 primers), or 48°C (with MC25 primers); and 1 min at 72°C. The last cycle was terminated by elongation for 10 min at 72°C. The PCR amplification products were resolved by electrophoresis in a 1.5% agarose gel containing 0.5 μg of ethidium bromide per ml and were visualized under UV light. Possible inhibition of DNA polymerase, yielding a false-negative result for any given sample, was checked by adding 10 ng of DNA of B. burgdorferi sensu lato to the reaction mixture.
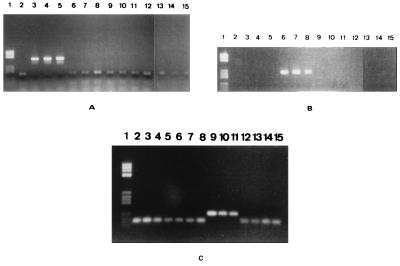
PCR with the MC16 primers amplified an expected 395-bp fragment from the DNA of B. burgdorferi sensu stricto B31, IP1, and IRS but not from the DNA of B. garinii and B. afzelii isolates (Fig. 2A). Fragments of 236 bp from B. garinii 20047, N34, and G25 were amplified in the PCR assay with the MC33 primers but not from the strains of B. burgdorferi sensu stricto or B. afzelii (Fig. 2B). Finally, the MC25 primers amplified an expected 125-bp fragment from the DNA of B. afzelii VS461, UO1, and Iper3 but not from the DNA of B. burgdorferi sensu stricto or B. garinii (Fig. 2C). The MC16, MC33, and MC25 primers did not amplify the DNA of B. japonica Cow611C, B. hermsii, B. parkeri, or B. turicatae. No signal was observed when template DNA was omitted from the amplification reaction mixture (faint signals are nucleotides and oligonucleotides not used in the reaction).
FIG. 2.
Specificity of the chosen primers for amplification of B. burgdorferi sensu stricto, B. garinii, or B. afzelii DNA. DNAs from B. burgdorferi sensu stricto B31 (lanes 3), IP1 (lanes 4), and IRS (lanes 5), B. garinii 20047 (lanes 6), N34 (lanes 7), and G25 (lanes 8), B. afzelii VS461 (lanes 9), UO1 (lanes 10), and Iper3 (lanes 11), B. japonica Cow611C (lanes 12), B. hermsii (lanes 13), B. parkeri (lanes 14), and B. turicatae (lanes 15) were amplified with the MC16 (A), MC33 (B), or MC25 (C) primers. The amplified products were separated on a 1.5% agarose gel and revealed under UV light after the addition of ethidium bromide (10 μg/ml). Lanes 2, control without DNA; lanes 1, DNA molecular mass markers (587, 540, 504, 458, 434, 267, 234, 213, 192, 184, and 124 bp).
The species-specific primers described here recognize different plasmids. Since only strains with high passage numbers were analyzed, these plasmids must be stably maintained and the primers should be able to distinguish most if not all B. burgdorferi sensu lato strains in nature belonging to the three pathogenic species. Other species-specific primers were described on chromosomal (fla, orfX, rRNA) (15, 18, 24, 27) or plasmid (ospA) genes (10, 22). These primers were able to distinguish the species by their ability to recognize specific sequences within a given gene shared by all spirochetes. A current comparison of two different sets of primers for species identification underlines the genetic heterogeneity of spirochetes in nature (20a).
Sensitivity of the PCR amplification assay.
The detection threshold of the PCR assay was determined by performing amplification reactions with serially diluted samples of DNA from B. burgdorferi sensu stricto, B. garinii, or B. afzelii. Amplification reactions were performed with aliquots containing amounts of DNA equivalent to given numbers of spirochetes (from 107 to 1) and with the MC16 (Fig. 3A), MC33 (Fig. 3B), and MC25 (Fig. 3C) primers. It was found that a template DNA input corresponding to 100 spirochetes was sufficient for the detection of the amplified fragment of B. burgdorferi sensu stricto and that 1,000 spirochetes was sufficient for the detection of the amplified fragments of B. garinii and B. afzelii.
FIG. 3.
Sensitivity of the B. burgdorferi sensu stricto, B. garinii, and B. afzelii species-specific primers. DNA corresponding to 107 (lanes 2), 106 (lanes 3), 105 (lane 4), 104 (lanes 5), 103 (lanes 6), 102 (lanes 7), 10 (lanes 8), 1 (lanes 9), or 0 (lanes 10) spirochetes of B. burgdorferi sensu stricto IP1 (A), B. garinii N34 (B), or B. afzelii UO1 (C) was amplified with the MC16 (A), MC33 (B), or MC25 (C) primers. The amplified products were electrophoretically separated in a 1.5% agarose gel and revealed under UV light after the addition of ethidium bromide (10 μg/ml). Lanes 1, DNA molecular mass markers (587, 540, 504, 458, 434, 267, 234, 213, 192, 184, and 124 bp).
The usefulness of these primers for the identification of species in biological samples was ascertained. Biological samples came from patients with Lyme disease diagnosed during the summer of 1996 at the Belgian Reference Center for Borreliosis (G. Bigaignon, Infectious Serology Laboratory, Saint-Luc Hospital, Brussels, Belgium) by positive enzyme-linked immunosorbent assay (Diagast, Lille, France) and PCR (Biocode, Sclessin, Belgium) test results. Sera and cerebrospinal fluids (150 μl) were centrifuged (13,000 × g, 20 min), and the pellets were washed three times with phosphate-buffered saline and resuspended in 60 μl of H2O. After heat denaturation (10 min at 100°C), 20 μl was added to the PCR mixture.
Ticks were collected in July 1996 from vegetation on the ground in the forest near Matagne-la-Petite, Namur Province, Belgium. Ticks were kept at 4°C in alcohol until use. Ticks were dried and incubated in 100 μl of TE (10 mM Tris-HCl [pH 7.8], 1 mM EDTA) containing 200 μg of proteinase K (Boehringer Mannheim, Mannheim, Germany) per ml. After overnight incubation at room temperature, the ticks were crushed with a pipette tip, boiled for 10 min, and then placed on ice for 10 min. The samples were centrifuged at 13,000 × g for 10 min, and supernatants were collected and stored at −20°C. A total of 5 μl of the supernatants was added to the PCR mixture.
Three serum samples, two cerebrospinal fluid samples, and four ticks containing B. burgdorferi sensu lato DNA, as shown by amplification with the c/c′ primers of Rosa et al. (27), were analyzed further to identify the Borrelia species in these samples (Table 3). Only one sample from a human contained DNA from a unique species, whereas the four others contained DNA originating from at least two different species of B. burgdorferi sensu lato. PCR with the MC16 or the MC33 primer amplified DNA from three samples from patients, whereas PCR with the MC25 primer, amplified DNA from four samples. Of the four ticks, one contained the DNA of only one species and the three others contained the DNAs of two or three B. burgdorferi sensu lato species. The three primer sets were thus able to amplify borrelial DNA in biological samples.
TABLE 3.
Presence of different B. burgdorferi sensu lato species in ticks and human biological fluids
| Specimen | PCR results with the species-specific primer setsa
|
||
|---|---|---|---|
| MC16 (B. burgdorferi sensu stricto) | MC33 (B. garinii) | MC25 (B. afzelii) | |
| Tick 1 | + | + | − |
| Tick 2 | − | + | − |
| Tick 3 | + | + | + |
| Tick 4 | − | + | + |
| CSFb 1 | + | + | + |
| CSF 2 | + | − | + |
| Serum 1 | − | + | − |
| Serum 2 | − | + | + |
| Serum 3 | + | − | + |
−, absence of amplification product; +, presence of amplification product.
CSF, cerebrospinal fluid.
The sensitivities of the PCR amplification assays with the primers designed in this work seemed adequate and at least equivalent to those with the general c/c′ primers (27). Indeed, the infecting species could be identified with the primers being studied in all the samples, which were selected by the ability of the c/c′ primers to amplify DNA. The detection of B. burgdorferi sensu stricto seemed more sensitive than the detection of the other two species (Fig. 3). This is probably due to the presence of the target sequence of the corresponding primers (MC16) in several plasmids of this species (21). Interestingly, homologous regions of DNA have been found in different plasmids of Borrelia strains (9, 30, 31, 33). These shared sequences are not specific for any one of the described B. burgdorferi sensu lato genospecies, however.
The PCR data indicated the usefulness of these primers with tick and clinical samples (Table 3). This analysis revealed several cases of infection with multiple Borrelia species, in agreement with observations with ticks (12, 23, 26) and biological fluids from patients with Lyme disease (10).
Nucleotide sequence accession numbers.
The accession numbers for the nucleotide sequences (EMBL Nucleotide Sequence Database) reported here are U12332 for B. burgdorferi sensu stricto, U83998 for B. garinii, and U84145 for B. afzelii.
Acknowledgments
M. C. Misonne received a predoctoral grant from Institut d’Encouragement de la Recherche Scientifique dans l’Industrie et l’Agriculture. P. Hoet is research director of FNRS (National Fund for Scientific Research, Brussels, Belgium).
We thank P. Gilot for critically reading the manuscript.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Anthonissen F M, De Kesel M, Hoet P P, Bigaignon G H. Evidence for the involvement of different genospecies of Borrelia in the clinical outcome of Lyme disease in Belgium. Res Microbiol. 1994;145:327–331. doi: 10.1016/0923-2508(94)90187-2. [DOI] [PubMed] [Google Scholar]
- 3.Assous M V, Postic D, Paul G, Névot P, Baranton G. Western blot analysis of sera from Lyme borreliosis patients according to the genomic species of the Borrelia strains used as antigens. Eur J Clin Microbiol Infect Dis. 1993;12:261–268. doi: 10.1007/BF01967256. [DOI] [PubMed] [Google Scholar]
- 4.Baranton G, Postic D, Saint Girons I, Boerlin P, Piffaretti J C, Assous M, Grimont P A D. Delineation of Borrelia burgdorferi sensu stricto, Borrelia garinii sp. nov., and group VS461 associated with Lyme borreliosis. Int J Syst Bacteriol. 1992;42:378–383. doi: 10.1099/00207713-42-3-378. [DOI] [PubMed] [Google Scholar]
- 5.Barbour A G. Plasmid analysis of Borrelia burgdorferi, the Lyme disease agent. J Clin Microbiol. 1988;26:475–478. doi: 10.1128/jcm.26.3.475-478.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boerlin P, Peter O, Bretz A-G, Postic D, Baranton G, Piffaretti J-C. Population genetic analysis of Borrelia burgdorferi isolates by multilocus enzyme electrophoresis. Infect Immun. 1992;60:1677–1683. doi: 10.1128/iai.60.4.1677-1683.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canica M M, Nato F, du Merle L, Mazie J C, Baranton G, Postic D. Monoclonal antibodies for identification of Borrelia afzelii sp. nov. associated with late cutaneous manifestations of Lyme borreliosis. Scand J Infect Dis. 1993;25:441–448. doi: 10.3109/00365549309008525. [DOI] [PubMed] [Google Scholar]
- 8.Casjens S, Delange M, Ley III H L, Rosa P, Huang W M. Linear chromosomes of Lyme disease agent spirochetes: genetic diversity and conservation of gene order. J Bacteriol. 1995;177:2769–2780. doi: 10.1128/jb.177.10.2769-2780.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casjens S, van Vugt R, Tilly K, Rosa P A, Stevenson B. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J Bacteriol. 1997;179:217–227. doi: 10.1128/jb.179.1.217-227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demaerschalck I, Ben Messaoud A, De Kesel M, Hoyois B, Lobet Y, Hoet P, Bigaignon G, Bollen A, Godfroid E. Simultaneous presence of different Borrelia burgdorferi genospecies in biological fluids of Lyme disease patients. J Clin Microbiol. 1995;33:602–608. doi: 10.1128/jcm.33.3.602-608.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukunaga M, Sohnaka M, Yanagihara Y. Analysis of Borrelia species associated with Lyme disease by rRNA gene restriction fragment length polymorphism. J Gen Microbiol. 1993;139:1141–1146. doi: 10.1099/00221287-139-6-1141. [DOI] [PubMed] [Google Scholar]
- 12.Guttman D S, Wang P W, Wang I-N, Bosler E M, Luft B J, Dykhuizen D E. Multiple infections of Ixodes scapularis ticks by Borrelia burgdorferi as revealed by single-strand conformation polymorphism analysis. J Clin Microbiol. 1996;34:652–656. doi: 10.1128/jcm.34.3.652-656.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isogai E, Isogai H, Kimura K, Hayashi S, Kubota T, Nishikawa T, Nakane A, Fujii N. Cytokines in the serum and brain in mice infected with distinct species of Lyme disease Borrelia. Microb Pathog. 1996;21:413–419. doi: 10.1006/mpat.1996.0072. [DOI] [PubMed] [Google Scholar]
- 14.Isogai E, Kimura K, Fujii N, Nishikawa T, Ishii N, Postic D, Baranton G, Isogai H. Platelet-activating-factor-mediated pathogenesis in Lyme disease. Infect Immun. 1996;64:1026–1029. doi: 10.1128/iai.64.3.1026-1029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson B J B, Happ C M, Mayer L W, Piesman J. Detection of Borrelia burgdorferi in ticks by species-specific amplification of the flagellin gene. Am J Trop Med Hyg. 1992;47:730–741. doi: 10.4269/ajtmh.1992.47.730. [DOI] [PubMed] [Google Scholar]
- 16.Kawabata H, Masuzawa T, Yanagihara Y. Genomic analysis of Borrelia japonica sp. nov. isolated from Ixodes ovatus in Japan. Microbiol Immunol. 1993;37:843–848. doi: 10.1111/j.1348-0421.1993.tb01714.x. [DOI] [PubMed] [Google Scholar]
- 17.Marck C. “DNA strider”: a ’C’ program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marconi R T, Garon C F. Development of polymerase chain reaction primer sets for diagnosis of Lyme disease and for species-specific identification of Lyme disease isolates by 16S rRNA signature nucleotide analysis. J Clin Microbiol. 1992;30:2830–2834. doi: 10.1128/jcm.30.11.2830-2834.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marconi R T, Lubke L, Hauglum W, Garon C F. Species-specific identification of and distinction between Borrelia burgdorferi genomic groups by using 16S rRNA-directed oligonucleotide probes. J Clin Microbiol. 1992;30:628–632. doi: 10.1128/jcm.30.3.628-632.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marconi R T, Liveris D, Schwartz I. Identification of novel insertion elements, restriction fragment length polymorphism patterns, and discontinuous 23S rRNA in Lyme disease spirochetes: phylogenetic analyses of rRNA genes and their intergenic spacers in Borrelia japonica sp. nov. and genomic group 21038 (Borrelia andersonii sp. nov.) isolates. J Clin Microbiol. 1995;33:2427–2434. doi: 10.1128/jcm.33.9.2427-2434.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Misonne, M. C., et al. Unpublished data.
- 21.Misonne M C, Schuttler M, Dernelle J M, De Kesel M, Hoet P. Cloning and sequencing of a species-specific nucleotide fragment of Borrelia burgdorferi sensu stricto, which is repeated in several plasmids of the species. FEMS Microbiol Lett. 1997;150:157–164. doi: 10.1111/j.1574-6968.1997.tb10364.x. [DOI] [PubMed] [Google Scholar]
- 22.Persing D H, Telford III S R, Spielman A, Barthold S W. Detection of Borrelia burgdorferi infection in Ixodes dammini ticks with the polymerase chain reaction. J Clin Microbiol. 1990;28:566–572. doi: 10.1128/jcm.28.3.566-572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pichon B, Godfroid E, Hoyois B, Bollen A, Rodhain F, Pérez-Eid C. Simultaneous infection of Ixodes ricinus nymphs by two Borrelia burgdorferi sensu lato species: possible implications for clinical manifestations. Emerg Infect Dis. 1995;1:89. doi: 10.3201/eid0103.950304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Picken R P. Polymerase chain reaction primers and probes derived from flagellin gene sequences for specific detection of the agents of Lyme disease and North American relapsing fever. J Clin Microbiol. 1992;30:99–114. doi: 10.1128/jcm.30.1.99-114.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Postic D, Belfaiza J, Isogal E, Saint Girons I, Grimont P A D, Baranton G. A new genomic species in B. burgdorferi sensu lato isolated from Japanese ticks. Res Microbiol. 1993;144:467–473. doi: 10.1016/0923-2508(93)90054-6. [DOI] [PubMed] [Google Scholar]
- 26.Rijpkema S G T, Molkenboer M J C H, Schouls L M, Jongejan F, Schellekens J F P. Simultaneous detection and genotyping of three genomic groups of Borrelia burgdorferi sensu lato in Dutch Ixodes ricinus ticks by characterization of the amplified intergenic spacer region between 5S and 23S rRNA genes. J Clin Microbiol. 1995;33:3091–3095. doi: 10.1128/jcm.33.12.3091-3095.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosa P A, Hogan D, Schwan T G. Polymerase chain reaction analyses identify two distinct classes of Borrelia burgdorferi. J Clin Microbiol. 1991;29:524–532. doi: 10.1128/jcm.29.3.524-532.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simpson W J, Garon C F, Schwan T G. Borrelia burgdorferi contains repeated DNA sequences that are species specific and plasmid associated. Infect Immun. 1990;58:847–853. doi: 10.1128/iai.58.4.847-853.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Y, Johnson R C. Analysis and comparison of plasmid profiles of Borrelia burgdorferi sensu lato strains. J Clin Microbiol. 1995;33:2679–2685. doi: 10.1128/jcm.33.10.2679-2685.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zingg B C, Brown R N, Lane R S, LeFebvre R B. Genetic diversity among Borrelia burgdorferi isolates from wood rats and kangaroo rats in California. J Clin Microbiol. 1993;31:3109–3114. doi: 10.1128/jcm.31.12.3109-3114.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zückert W R, Meyer J. Circular and linear plasmids of Lyme disease spirochetes have extensive homology: characterization of a repeated DNA element. J Bacteriol. 1996;178:2287–2298. doi: 10.1128/jb.178.8.2287-2298.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]