Abstract
Cefepime is a potentially useful antibiotic for treatment of infections with Enterobacter cloacae. However, in our institution the MIC90 for E. cloacae bloodstream isolates is 16 μg/ml. PCR amplification of bla genes revealed that one-third (15/45) of E. cloacae bloodstream isolates produced SHV-type extended-spectrum beta-lactamases (ESBLs) in addition to hyperproduction of AmpC-type beta-lactamases. The majority (11/15) of ESBL producers also produced the TEM-1 beta-lactamase. The SHV types included SHV-2, -5, -7, -12, -14, and -30. All but two of the ESBL-producing E. cloacae isolates, but none of the non-ESBL-producing strains, had MICs of cefepime of ≥2 μg/ml. The MIC90 for cefepime for ESBL-producing strains was 64 μg/ml, while for non-ESBL producers it was 0.5 μg/ml. Using current Clinical and Laboratory Standards Institute breakpoints for cefepime, two thirds (10/15) of ESBL-producing isolates would have been regarded as susceptible to cefepime. Phenotypic ESBL detection methods were generally unreliable with these E. cloacae isolates. Based on these results, pharmacokinetic, pharmacodynamic, and clinical reevaluation of cefepime breakpoints for E. cloacae may be prudent.
Enterobacter cloacae is a leading cause of ventilator-associated pneumonia, bloodstream infections, and urinary tract infections in hospitalized patients. A chromosomal gene in E. cloacae characteristically encodes the AmpC beta-lactamase (9). Mutations that increase the expression of the gene encoding AmpC are responsible for the emergence of resistance of the organism to cephalosporins such as ceftriaxone, cefotaxime, and ceftaxidime during therapy (4, 11). Approximately 30% of E. cloacae isolates from patients in intensive care units in the United States are resistant to cephalosporins such as ceftriaxone, cefotaxime, and ceftaxidime (15).
Cefepime is potentially a very useful antibiotic for the treatment of serious infections with E. cloacae, even in the presence of increased production of the AmpC beta-lactamase (23). In vitro studies performed prior to the commercial availability of cefepime showed that the MIC90 ranged from 0.06 to 0.5 μg/ml (12, 25, 26). In contrast, an assessment of isolates from North American intensive care units in 2003 showed that the MIC90 was 2 μg/ml, with MICs of some isolates as high as >16 μg/ml (20). In our institution in 2003, the MIC90 of cefepime for E. cloacae bloodstream isolates was 16 μg/ml. For this reason, we sought to investigate the mechanisms of reduced cefepime susceptibility in bloodstream isolates occurring at our institution.
MATERIALS AND METHODS
Bacterial strains.
Consecutive bloodstream isolates of E. cloacae from patients at the University of Pittsburgh Medical Center (UPMC) were studied. The isolates were collected from March 2003 through July 2004. Species identification was done by standard biochemical tests. The strains were supplied by the hospital's clinical microbiology laboratory as part of an Institutional Review Board approved study on mechanisms of antibiotic resistance in hospital pathogens.
Antibiotic susceptibility.
The clinical microbiology laboratory at UPMC routinely assesses the MICs of cefepime and other injectable antibiotics commonly used in the treatment of infections with gram-negative bacteria by broth microdilution methods using Clinical and Laboratory Standards Institute (CLSI) criteria, current as of 1 January 2005. Additionally, the MICs of piperacillin-tazobactam, cefotaxime, ceftazidime, ceftazidime-clavulanic acid, cefotaxime, cefotaxime-clavulanic acid, cefepime, cefepime-clavulanic acid, cefoxitin, gentamicin, ertapenem, imipenem, meropenem, and ciprofloxacin were determined by Etest for the strains (AB Biodisk, Solna, Sweden). Escherichia coli ATCC 25922 and Klebsiella pneumoniae ATCC 700603 were used as the reference strains for antimicrobial susceptibility testing.
Analytical isoelectric focusing.
Analytical isoelectric focusing was performed on isolates with cefepime MICs of 0.5 μg/ml or more, as described previously (19). After removal of whole cells and debris by centrifugation, the supernatant was used to determine the isoelectric point (pI). Electrophoresis was performed using precast polyacrylamide gels, pH 3 to 10 (Bio-Rad, Hercules, CA). Enzyme activity was detected by placing filter paper soaked in nitrocefin (500 μg/ml) (Becton Dickinson, Sparks, MD) over the focused gel. Standards from Bio-Rad (Bio-Rad, Hercules, CA) were used, with the following isoelectric points: 4.45, 4.65, 4.75, 5.1, 6.0, 6.5, 6.8, 7.0, 7.1, 7.5, 7.8, 8.00, 8.20, and 9.6.
Plasmid profiles.
The plasmid DNA of the extended-spectrum beta-lactamase (ESBL)-producing clinical isolates was extracted with a plasmid extraction kit (Wizard Plus Minipreps DNA purification system; Promega, Madison, WI), according to the manufacturer's instructions. Plasmid DNA electrophoresis was performed with 0.8% agarose gel and visualized with ethidium bromide under UV light. λ HindIII (Promega, Madison, WI) was used as a molecular size marker.
PFGE.
Genomic DNA was isolated and digested with XbaI (New England Biolabs, Beverly, Mass.). Pulsed-field gel electrophoresis (PFGE) was performed with the CHEF III system (Bio-Rad, Hercules, CA) with the following run parameters: block I, with a switch time of 3 to 65 s and a run time of 17 h, and block II, with a switch time of 15 to 30 s and a run time of 6 h. Dendrograms were created with BioNumerics (Bio-Rad, Hercules CA) by using the Dice coefficient, unweighted-pair group method with arithmetic means, and a position tolerance of 1.3%. Relatedness of the isolates was determined by the criteria of Tenover et al. (24).
Detection of ESBL genes by PCR.
Detection of genes encoding ESBLs was attempted for all isolates with cefepime MICs of 0.5 μg/ml or more. A single colony of each test isolate was resuspended in 400 μl water and boiled for 15 min. The resulting supernatant was used as a bacterial template DNA in PCR assays. The primers for detection of the blaTEM, blaSHV, and blaCTX-M genes are as follows: 5′-ATGAGTATTCAACATTTCCGTG-3′ and 5′-TTACCAATGCTTAATCAGTGAG-3′ for blaTEM (6), 5′-ATTTGTCGCTTCTTTACTCGC-3′ and 5′-TTTATGGCGTTACCTTTGACC-3′ for blaSHV (6), and 5′-CGCTTTGCGATGTGCAG-3′ and 5′-ACCGCGATATCGTTGGT-3′ for blaCTX-M (2). PCRs were performed with RedTaq DNA polymerase (Sigma, St Louis, MO), according to the instructions of the manufacturer, in the presence of 2 μl of the template DNA preparation in a total volume of 30 μl. The DNA amplification programs consisted of an initial denaturation step (96°C, 5 min) followed by 30 cycles of denaturation (96°C, 30 s), annealing (annealing temperature designed for each primer set, 30 s), and extension (72°C, 1 min), and a final extension of 5 min at 72°C. Ten microliters of reaction mixture containing the PCR product was analyzed by electrophoresis in 0.8% (wt/vol) agarose (Bio-Rad, Hercules, CA).
Sequence analysis.
The amplified products were sequenced using ABI 4500 and ABI 3100 genetic analyzers according to the manufacturer's instructions. Sequencing reactions were performed with corresponding primers specific for the blaTEM-1 and blaSHV-1 genes used for the previous amplification. Sequence analysis was performed using Lasergene DNASTAR sequencing analysis software (DNAStar, Madison, WI). Each sequence of blaTEM and blaSHV genes was identified by comparison with known ESBL sequences available in the GenBank and EMBL databases by multiple-sequence alignment using the BLAST program.
Phenotypic detection of beta-lactamase production.
Disk diffusion testing was performed using ceftazidime, cefotaxime, and cefepime alone (30 μg each; Remel, Lenexa, KS), and in combination with clavulanic acid: ceftazidime-clavulanic acid, cefotaxime-clavulanic acid (30/10 μg each; Becton Dickinson, Sparks, MD), and cefepime-clavulanic acid (30/10 μg; Rosco, Prolab) disks were used. The disk tests were performed using confluent growth on Mueller-Hinton agar (Becton Dickinson, Sparks, MD), incubated at 35°C for 24 h. The sensitivities and specificities of phenotypic confirmatory tests for ESBL detection in Klebsiella spp. and E. coli, but applied to E. cloacae, were determined. No interpretative criteria for use of cefepime versus cefepime-clavulanic acid exist for any species.
Additionally, double-disk synergy tests were performed by placing disks of ceftazidime, cefotaxime, and cefepime at distances of 20 and 30 mm (center to center) from a disk containing amoxicillin plus clavulanic acid (20/10 μg; Remel, Lenexa, KS). A “keyhole” phenomenon was regarded as positive for ESBL production.
MICs for cefotaxime, ceftazidime, and cefepime were determined by using Etest strips. These MICs were compared to those obtained from the same isolates by using Etest strips containing ceftazidime-clavulanic acid, cefotaxime-clavulanic acid, and cefepime-clavulanic acid.
Classification of E. cloacae strains as having inducible, partially derepressed, or derepressed AmpC production was determined by the methods of Sanders et al. (22), in which the cefoxitin-cefotaxime antagonist test was performed (16, 22). The E. cloacae isolates were categorized as follows: derepressed AmpC mutants had a cefoxitin MIC of ≥32 μg/ml, a cefotaxime MIC of ≥16 μg/ml, and a negative cefoxitin-cefotaxime antagonist test; partially derepressed AmpC mutants had the same characteristics as derepressed AmpC mutants but with a positive cefoxitin-cefotaxime antagonist test; and inducible AmpC-producing strains had a cefotaxime MIC of ≤8 μg/ml and a positive cefoxitin-cefotaxime antagonist test (16).
RESULTS
PFGE.
There was no relatedness between 41/45 isolates. All 30 isolates which were found not to be ESBL producers were unrelated. Two of 15 ESBL producers (ES1 and ES22) were closely related, and two additional ESBL-producing strains (ES31 and ES43) were possibly related (data not shown).
Antibiotic susceptibility.
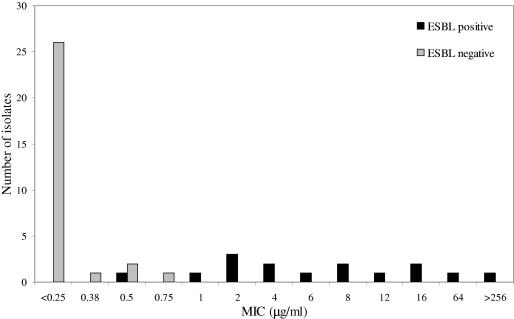
The antibiotic susceptibilities of the non-ESBL- and ESBL-producing E. cloacae strains are seen in Tables 1 and 2, respectively. Forty percent (18/45) of isolates had cefepime MICs of 0.5 μg/ml or more. The distribution of the cefepime MICs is shown in Fig. 1. The MIC90 for cefepime for ESBL-producing strains was 64 μg/ml, while for non-ESBL producers it was 0.5 μg/ml.
TABLE 1.
Antibiotic susceptibilities (μg/ml) of E. cloacae strains without ESBL production
| Antibiotic(s) | Susceptibility (μg/ml) ofa:
|
||
|---|---|---|---|
| Inducible strains | Partially derepressed strains | Derepressed mutants | |
| Piperacillin-tazobactam | 0.75-3 | 0.38-8 | 16->256 |
| Piperacillin | 0.5-64 | 0.5-24 | 32->256 |
| Cefoxitin | 0.19-6 | 12-128 | 128->256 |
| Ceftazidime | 0.125-0.5 | 0.094-12 | 12->256 |
| Ceftazidime-clavulanic acid | 0.38->4 | 0.064-1.5 | >4 |
| Cefotaxime | 0.045-1 | 0.125-12 | 24-128 |
| Cefotaxime-clavulanic acid | 0.125-1 | 0.125->1 | >1 |
| Cefepime | 0.032-0.094 | 0.064-0.5 | 0.125-0.75 |
| Cefepime-clavulanic acid | 0.064-0.094 | 0.016-0.5 | 0.016-0.5 |
| Ciprofloxacin | 0.006-0.012 | 0.006-0.094 | 0.016->32 |
| Imipenem | 0.19-0.25 | 0.19-0.5 | 0.25-0.38 |
| Gentamicin | 0.25-0.5 | 0.094-0.75 | 0.25-1 |
Results are shown for 4 inducible strains, 20 partially derepressed strains, and 6 derepressed mutants.
TABLE 2.
MIC values of the ESBL-producing E. cloacae isolates
| Isolate | MIC (μg/ml) for:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Piperacillin-tazobactam | Ceftazidime | Ceftazidime-clavulanic acid | Cefotaxime | Cefotaxime-clavulanic acid | Cefepime | Cefepime-clavulanic acid | Ciprofloxacin | Ertapenem | Imipenem | Meropenem | Gentamicin | |
| ES1 | 16 | >256 | >4 | 128 | >1 | 4 | 0.125 | 0.38 | 0.125 | 0.25 | 0.032 | 24 |
| ES6 | >256 | >256 | >4 | >256 | >1 | 8 | >4 | >32 | 2 | 0.25 | 0.19 | 256 |
| ES7 | >256 | >256 | >4 | >256 | >1 | >256 | >4 | 0.064 | 0.75 | 0.19 | 0.064 | 0.75 |
| ES11 | 8 | 48 | >4 | 16 | >1 | 2 | 0.5 | 0.5 | 3 | 0.19 | 0.125 | 8 |
| ES15 | >256 | >256 | >4 | >256 | >1 | 6 | 4 | 0.012 | 0.75 | 0.25 | 0.094 | 4 |
| ES18 | >256 | >256 | >4 | >256 | >1 | 64 | >4 | 8 | 4 | 0.38 | 0.38 | 48 |
| ES20 | >256 | >256 | >4 | >256 | >1 | 8 | >4 | 0.125 | 2 | 0.25 | 0.094 | 4 |
| ES22 | 8 | 256 | >4 | 192 | >1 | 2 | 0.094 | 0.38 | 0.5 | 0.5 | 0.032 | 128 |
| ES24 | >256 | >256 | >4 | 12 | >1 | 16 | 2 | >32 | 2 | 0.125 | 0.19 | >256 |
| ES31 | 1.5 | 128 | >4 | 8 | >1 | 1 | <0.016 | 0.094 | 0.125 | 0.25 | 0.032 | 256 |
| ES37 | >256 | >256 | >4 | >256 | >1 | 12 | >4 | 8 | 2 | 0.25 | 0.19 | 48 |
| ES40 | 12 | >256 | >4 | 96 | >1 | 4 | 1.5 | 8 | 1 | 0.38 | 0.064 | 96 |
| ES43 | 8 | >256 | 1 | 64 | >1 | 2 | 0.38 | 4 | 0.125 | 0.125 | 0.016 | 96 |
| ES44 | >256 | >256 | >4 | >256 | >1 | 16 | >4 | 4 | 1 | 0.25 | 0.125 | 64 |
| ES45 | 16 | 2 | 0.5 | 1 | >1 | 0.5 | 0.19 | 6 | 0.38 | 0.125 | 0.032 | 8 |
FIG. 1.
Distribution of cefepime MICs for E. cloacae strains with and without ESBL.
Analytical isoelectric focusing and genotypic detection of ESBLs.
Fifteen of 18 isolates with a cefepime MIC of 0.5 μg/ml or more carried ESBL genes. All of the ESBLs were of the SHV type (Table 3). Eleven of the 15 ESBL-producing isolates harbored blaTEM-1 in addition to blaSHV. blaCTX-M genes were not amplified from any isolates by using our primers. The ESBLs encoded by the E. cloacae strains included SHV-2 (one isolate), SHV-5 (one isolate), SHV-7 (eight isolates), SHV-12 (two isolates), SHV-14 (three isolates), and SHV-30 (one isolate). One isolate produced both SHV-7 and SHV-30. The isoelectric points of the isolates are presented in Table 3. The three non-ESBL producers with cefepime MICs of 0.5 μg/ml or more produced only one beta-lactamase each, with an isoelectric point consistent with that for AmpC.
TABLE 3.
Characterization of the ESBL-producing E. cloacae isolates
| Isolate | Beta-lactamase characterization | Genotypic evaluation
|
pI | Plasmid profile | |
|---|---|---|---|---|---|
| SHV | TEM-1 | ||||
| ES1 | Derepressed AmpC + ESBL | SHV-7 | Present | 5.4+7.6+9.0 | P1 |
| ES6 | Derepressed AmpC + ESBL | SHV-7 | Present | 5.4+7.6+9.0 | P2 |
| ES7 | Derepressed AmpC + ESBL | SHV-5 | Absent | 8.2+9.0 | P3 |
| ES11 | Derepressed AmpC + ESBL | SHV-14 | Present | 5.4+7.0+9.0 | P4 |
| ES15 | Derepressed AmpC + ESBL | SHV-14 | Absent | 7.0+9.0 | P5 |
| ES18 | Derepressed AmpC + ESBL | SHV-7 | Present | 5.4+7.6+9.0 | P6 |
| ES20 | Derepressed AmpC + ESBL | SHV-2 | Absent | 7.6+9.0 | P7 |
| ES22 | Derepressed AmpC + ESBL | SHV-7 | Present | 5.4+7.6+9.0 | P1 |
| ES24 | Derepressed AmpC + ESBL | SHV-7, SHV-30 | Present | 5.4+7.0+7.6+9.0 | P8 |
| ES31 | Derepressed AmpC + ESBL | SHV-12 | Present | 5.4+8.2+9.0 | P9 |
| ES37 | Derepressed AmpC + ESBL | SHV-7 | Present | 5.4+7.6+9.0 | P10 |
| ES40 | Derepressed AmpC + ESBL | SHV-7 | Present | 5.4+7.6+9.0 | P11 |
| ES43 | Partially derepressed + ESBL | SHV-12 | Present | 5.4+8.2+9.0 | P9 |
| ES44 | Derepressed AmpC + ESBL | SHV-7 | Present | 5.4+7.6+9.0 | P10 |
| ES45 | Derepressed AmpC + ESBL | SHV-14 | Absent | 7.0+9.0 | P12 |
Plasmid profile analysis.
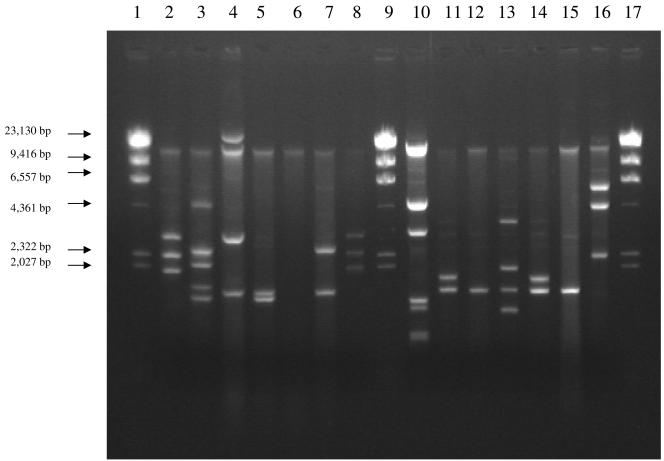
Selected ESBL-producing E. cloacae strains isolated during this period were analyzed for their plasmid content (Fig. 2; Table 3). All ESBL-producing E. cloacae isolates harbored plasmids, although there was diversity in the sizes of these plasmids (Fig. 2).
FIG. 2.
Plasmids from representative ESBL-producing E. cloacae strains. Lanes 1, 9, and 17 contain λ HindIII size markers. Lanes 2 to 8 contain ESBL-producing E. cloacae strains ES1, ES6, ES7, ES11, ES15, ES18, and ES22, respectively. Lanes 10 to 16 contain E. cloacae strains ES24, ES31, ES37, ES40, ES43, ES44, and ES45, respectively.
Antibiotic susceptibility testing of ESBL-producing strains.
Antibiotic susceptibility testing results are shown in Table 2. The cefepime MIC distributions of ESBL-producing isolates were 0.5 μg/ml (one isolate), 1 μg/ml (one isolate), 2 μg/ml (three isolates), 4 μg/ml (two isolates), 6 μg/ml (one isolate), 8 μg/ml (two isolates), 12 μg/ml (one isolate), 16 μg/ml (two isolates), 64 μg/ml (one isolate), and >256 μg/ml (one isolate). According to current CLSI breakpoints for Enterobacteriaceae, 10/15 isolates would have been regarded as cefepime susceptible, 3/15 as cefepime intermediate, and 2/15 as cefepime resistant. The majority of ESBL-producing isolates were nonsusceptible to numerous antibiotics: 14/15 ESBL-producing isolates were nonsusceptible to ceftazidime, 8/15 were nonsusceptible to piperacillin-tazobactam, 12/15 were nonsusceptible to gentamicin, 8/15 were nonsusceptible to ciprofloxacin, and 2/15 were nonsusceptible to ertapenem. All 15 isolates were susceptible to imipenem and meropenem.
Phenotypic detection of ESBL production.
Applying CLSI ESBL screening disk diffusion criteria to E. cloacae showed that 23 isolates had a “positive” result, but 9 of these isolates were non-ESBL producers (Table 4). Confirmatory results were positive for 7/15 ESBL-producing strains but for no non-ESBL-producing isolate. Applying CLSI ESBL screening MIC criteria gave similar results (Table 5), although confirmatory tests were positive for just 2/15 ESBL-producing strains. The conventional double disk diffusion tests utilizing ceftazidime and cefotaxime disks were rarely positive (Table 4).
TABLE 4.
Distribution of the results of different disk diffusion techniques
| Test and drug | Test result | Category (no. of isolates)
|
Sensitivity | Specificity | |
|---|---|---|---|---|---|
| ESBL | Non-ESBL | ||||
| CLSI ESBL initial screena | |||||
| Ceftazidime | Positive | 14 | 9 | 0.93 | 0.70 |
| Negative | 1 | 21 | |||
| Cefotaxime | Positive | 14 | 9 | 0.93 | 0.70 |
| Negative | 1 | 21 | |||
| CLSI ESBL phenotypic confirmatory testb | |||||
| Ceftazidime | Positive | 7 | 0 | 0.47 | 1 |
| Negative | 8 | 30 | |||
| Cefotaxime | Positive | 5 | 0 | 0.33 | 1 |
| Negative | 10 | 30 | |||
| Double disk diffusion test (20 mm) | |||||
| Ceftazidime | Positive | 6 | 0 | 0.40 | 1 |
| Negative | 9 | 30 | |||
| Cefotaxime | Positive | 7 | 0 | 0.47 | 1 |
| Negative | 8 | 30 | |||
| Double disk diffusion test (30 mm) | |||||
| Ceftazidime | Positive | 1 | 0 | 0.07 | 1 |
| Negative | 14 | 30 | |||
| Cefotaxime | Positive | 7 | 0 | 0.47 | 1 |
| Negative | 8 | 30 | |||
The CLSI disk inhibition break points for ESBL initial screen of E. coli and Klebsiella spp. were <22 mm for ceftazidime (30 μg) and <27 mm for cefotaxime (30 μg).
A >5-mm increase in zone diameter in combination with clavulanic acid versus when tested alone.
TABLE 5.
Distribution of the results of MICs obtained by Etest as a means of ESBL detection and phenotypic confirmation
| Test stage and drug | Test result | Category (no. of isolates)
|
Sensitivity | Specificity | |
|---|---|---|---|---|---|
| ESBL | Non-ESBL | ||||
| Initial screeninga | |||||
| Ceftazidime | Positive | 15 | 8 | 1 | 0.73 |
| Negative | 0 | 22 | |||
| Cefotaxime | Positive | 15 | 8 | 1 | 0.73 |
| Negative | 0 | 22 | |||
| Phenotypic confirmationb | |||||
| Ceftazidime | Positive | 2 | 0 | 0.13 | 1 |
| Negative | 13 | 30 | |||
| Cefotaxime | Positive | 0 | 0 | 0 | 1 |
| Negative | 15 | 30 | |||
The CLSI MIC breakpoints for ESBL screening for E. coli and Klebsiella spp. were 2 μg/ml or more for ceftazidime and cefotaxime.
A >3 twofold-concentration decrease in MIC for either antimicrobial agent tested in combination with clavulanic acid versus its MIC tested alone.
Use of cefepime susceptibility results as a marker for ESBL production appeared more useful (Table 6). All 13 isolates with cefepime MICs of 2 μg/ml or more were ESBL producers, and 15/18 (83%) of isolates with cefepime MICs of >0.25 μg/ml were ESBL producers. A confirmatory test with cefepime-clavulanic acid was only 73% sensitive using disk diffusion and 53% sensitive using ≥3 MIC reductions (Table 6).
TABLE 6.
Distribution of the results using cefepime as an indicator for ESBL production
| Test and result | Category (no. of isolates)
|
Sensitivity | Specificity | |
|---|---|---|---|---|
| ESBL | Non-ESBL | |||
| Disk diffusion screening testa | ||||
| Positive | 13 | 0 | 0.87 | 1 |
| Negative | 2 | 30 | ||
| Disk diffusion confirmatory testb | ||||
| Positive | 11 | 0 | 0.73 | 1 |
| Negative | 4 | 30 | ||
| Double disk diffusion test (20 mm) | ||||
| Positive | 13 | 0 | 0.87 | 1 |
| Negative | 2 | 30 | ||
| Double disk diffusion test (30 mm) | ||||
| Positive | 8 | 0 | 0.53 | 1 |
| Negative | 7 | 30 | ||
| MIC-based screening testc | ||||
| Positive | 14 | 0 | 0.93 | 1 |
| Negative | 1 | 30 | ||
| MIC-based confirmatory testd | ||||
| Positive | 8 | 0 | 0.53 | 1 |
| Negative | 7 | 30 | ||
Based on zone diameter of <27 mm as a screen for potential ESBL production.
A >5-mm increase in zone diameter of cefepime in combination with clavulanic acid versus cefepime tested alone.
Cefepime breakpoint based on our study was 1 μg/ml.
A >3 twofold-concentration decrease in MIC for either antimicrobial agent tested in combination with clavulanic acid versus its MIC tested alone.
DISCUSSION
There are now numerous reports of E. cloacae-producing ESBLs (1, 3, 7, 8, 10, 14, 16, 17), including several from the United States (5, 13, 21). The earliest reports of ESBL-producing E. cloacae strains from the United States were of TEM-12 and TEM-26 producers from Boston in 1988 (21) and SHV-3 producers from Boston in the 1990s (5). In a subsequent report, Levison and colleagues found SHV-7- and SHV-12-producing E. cloacae in three different hospitals in Philadelphia in 2000 and 2001 (13). In Pittsburgh, we have subsequently found SHV-2, -5, -7, -12, -14, and -30 in E. cloacae strains. In a 1-year period, 33% (15/45) of E. cloacae bloodstream isolates were found to be ESBL producers. Unfortunately, there are no surveillance data from the United States in order to determine whether this percentage is high or whether it is within the range found in similar hospitals. In a recently published study from Korea, 43% of E. cloacae blood culture isolates were found to be ESBL producers (16).
Wild-type E. cloacae strains obtained prior to the commercial release of cefepime had cefepime MICs of 0.06 to 0.5 μg/ml. It is uncertain whether any of the strains tested in these studies produced ESBLs. In our center, the MIC90 for cefepime for ESBL-producing strains was 64 μg/ml, while for non-ESBL producers it was 0.5 μg/ml. Studies examining outer membrane proteins, efflux pumps, and the expression levels and hydrolytic activities of SHV-type ESBLs against cefepime are currently under way. Thus, at the present time, we are unable to conclusively state that the production of ESBLs is responsible for the elevated cefepime MICs that we observed.
The clinical implications of this elevation in cefepime MIC are under evaluation, since some patients in our series failed cefepime therapy. Assessments of the pharmacokinetics and pharmacodynamics of cefepime presented at CLSI Antimicrobial Susceptibility Testing subcommittee meetings would suggest that cefepime may not be effective for treating serious infections when dosed at 1 g every 12 h for an organism with a MIC of 8 μg/ml or higher. Clinical data from patients with serious infections with ESBL-producing Klebsiella spp. or E. coli would support the concept that cefepime activity may be compromised against some ESBL-producing organisms with MICs in the current susceptible range (18, 27). Thus, it would appear prudent for cefepime breakpoints to be reconsidered.
Given that we have evaluated just 15 ESBL-producing isolates and 30 non-ESBL-producing isolates, we cannot categorically comment on ESBL detection methods for E. cloacae. Conventional methods using cefotaxime or ceftazidime are likely to be unreliable. However, a cefepime MIC of ≥2 μg/ml appears to be a consistently robust marker of ESBL production. While tests incorporating cefepime plus clavulanic acid may detect some ESBL producers with lower MICs, the tests were not more than 75% sensitive in our hands.
Why would SHV-type ESBLs evolve in an E. cloacae host? In our isolates, the SHV-type ESBLs are present in the company of a derepressed AmpC. Increased production of the AmpC beta-lactamase confers resistance to oxyiminocephalosporins and beta-lactam/beta-lactamase inhibitor combinations. The presence of an ESBL adds a selective advantage against cefepime. Alternatively, by production of large quantities of AmpC, protection against cephalosporins and beta-lactamase inhibitors may create a permissive microbiological environment within which ESBLs may evolve. It is known that ESBLs are not catalytically efficient against penicillins and are hypersusceptible to beta-lactamase inhibitors. This trade-off in catalytic activity would be compensated by a robust penicillinase, TEM-1, and a cephalosporinase that is inhibitor resistant. The net result would be expansion of resistance to cefepime.
In summary, in our center, ESBL production by E. cloacae isolates causing serious infections is common. Thirty-three percent of bloodstream isolates were ESBL producers. Occasionally, patients shared genotypically similar isolates, but most often ESBL-producing isolates appeared to arise de novo. Given clinical concerns regarding the efficacy of cefepime against some ESBL-producing strains, reassessment of cefepime breakpoints by CLSI would seem prudent.
Acknowledgments
We thank Robert Rae from Prolab for providing us with the cefepime-clavulanic acid disks.
The Veterans Affairs Merit Review Program and the National Institutes of Health (RO1 AI063517-01) supported R.A.B.
REFERENCES
- 1.Bell, J. M., J. D. Turnidge, R. N. Jones, and the SENTRY Asia-Pacific Participants. 2003. Prevalence of extended-spectrum β-lactamase-producing Enterobacter cloacae in the Asia-Pacific region: results from the SENTRY Antimicrobial Surveillance Program, 1998 to 2001. Antimicrob. Agents Chemother. 47:3989-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnet, R., J. L. M. Sampaio, R. Labia, C. De Champs, D. Sirot, C. Chanal, and J. Sirot. 2000. A novel CTX-M β-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil. Antimicrob. Agents Chemother. 44:1936-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouchillon, S. K., B. M. Johnson, D. J. Hoban, J. L. Johnson, M. J. Dowzicky, D. H. Wu, M. A. Visalli, and P. A. Bradford. 2004. Determining incidence of extended spectrum beta-lactamase producing Enterobacteriaceae, vancomycin-resistant Enterococcus faecium and methicillin-resistant Staphylococcus aureus in 38 centres from 17 countries: the PEARLS study 2001-2002. Int. J. Antimicrob. Agents 24:119-124. [DOI] [PubMed] [Google Scholar]
- 4.Chow, J. W., M. J. Fine, D. M. Shlaes, J. P. Quinn, D. C. Hooper, M. P. Johnson, R. Ramphal, M. M. Wagener, D. K. Miyashiro, and V. L. Yu. 1991. Enterobacter bacteremia: clinical features and emergence of antibiotic resistance during therapy. Ann. Intern. Med. 115:585-590. [DOI] [PubMed] [Google Scholar]
- 5.D'Agata, E., L. Venkataraman, P. DeGirolami, L. Weigel, M. Samore, and F. Tenover. 1998. The molecular and clinical epidemiology of enterobacteriaceae-producing extended-spectrum beta-lactamase in a tertiary care hospital. J. Infect. 36:279-285. [DOI] [PubMed] [Google Scholar]
- 6.Essack, S. Y., L. M. C. Hall, D. G. Pillay, M. L. McFadyen, and D. M. Livermore. 2001. Complexity and diversity of Klebsiella pneumoniae strains with extended-spectrum β-lactamases isolated in 1994 and 1996 at a teaching hospital in Durban, South Africa. Antimicrob. Agents Chemother. 45:88-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giakkoupi, P., L. S. Tzouvelekis, A. Tsakris, V. Loukova, D. Sofianou, and E. Tzelepi. 2000. IBC-1, a novel integron-associated class A β-lactamase with extended-spectrum properties produced by an Enterobacter cloacae clinical strain. Antimicrob. Agents Chemother. 44:2247-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho, P. L., R. H. Shek, K. H. Chow, R. S. Duan, G. C. Mak, E. L. Lai, W. C. Yam, K. W. Tsang, and W. M. Lai. 2005. Detection and characterization of extended-spectrum beta-lactamases among bloodstream isolates of Enterobacter spp. in Hong Kong, 2000-2002. J. Antimicrob. Chemother. 55:326-332. [DOI] [PubMed] [Google Scholar]
- 9.Jacoby, G. A., and L. S. Munoz-Price. 2005. The new beta-lactamases. N. Engl. J. Med. 352:380-391. [DOI] [PubMed] [Google Scholar]
- 10.Kartali, G., E. Tzelepi, S. Pournaras, C. Kontopoulou, F. Kontos, D. Sofianou, A. N. Maniatis, and A. Tsakris. 2002. Outbreak of infections caused by Enterobacter cloacae producing the integron-associated β-lactamase IBC-1 in a neonatal intensive care unit of a Greek hospital. Antimicrob. Agents Chemother. 46:1577-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaye, K. S., S. Cosgrove, A. Harris, G. M. Eliopoulos, and Y. Carmeli. 2001. Risk factors for emergence of resistance to broad-spectrum cephalosporins among Enterobacter spp. Antimicrob. Agents Chemother. 45:2628-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kessler, R. E., M. Bies, R. E. Buck, D. R. Chisholm, T. A. Pursiano, Y. H. Tsai, M. Misiek, K. E. Price, and F. Leitner. 1985. Comparison of a new cephalosporin, BMY 28142, with other broad-spectrum β-lactam antibiotics. Antimicrob. Agents Chemother. 27:207-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levison, M. E., Y. V. Mailapur, S. K. Pradhan, G. A. Jacoby, P. Adams, C. L. Emery, P. L. May, and P. G. Pitsakis. 2002. Regional occurrence of plasmid-mediated SHV-7, an extended-spectrum beta-lactamase, in Enterobacter cloacae in Philadelphia teaching hospitals. Clin. Infect. Dis. 35:1551-1554. [DOI] [PubMed] [Google Scholar]
- 14.Munday, C. J., G. M. Whitehead, N. J. Todd, M. Campbell, and P. M. Hawkey. 2004. Predominance and genetic diversity of community- and hospital-acquired CTX-M extended-spectrum beta-lactamases in York, UK. J. Antimicrob. Chemother. 54:628-633. [DOI] [PubMed] [Google Scholar]
- 15.National Nosocomial Infections Surveillance System. 2004. National Nosocomial Infections Surveillance (NNIS) System report, data summary from January 1992 through June 2004, issued October 2004. Am. J. Infect. Control 32:470-485. [DOI] [PubMed] [Google Scholar]
- 16.Pai, H., J. Y. Hong, J.-H. Byeon, Y.-K. Kim, and H.-J. Lee. 2004. High prevalence of extended-spectrum β-lactamase-producing strains among blood isolates of Enterobacter spp. collected in a tertiary hospital during an 8-year period and their antimicrobial susceptibility patterns. Antimicrob. Agents Chemother. 48:3159-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park, Y. J., S. Y. Park, E. J. Oh, J. J. Park, K. Y. Lee, G. J. Woo, and K. Lee. 2005. Occurrence of extended-spectrum beta-lactamases among chromosomal AmpC-producing Enterobacter cloacae, Citrobacter freundii, and Serratia marcescens in Korea and investigation of screening criteria. Diagn. Microbiol. Infect. Dis. 51:265-269. [DOI] [PubMed] [Google Scholar]
- 18.Paterson, D. L., W. C. Ko, A. Von Gottberg, J. M. Casellas, L. Mulazimoglu, K. P. Klugman, R. A. Bonomo, L. B. Rice, J. G. McCormack, and V. L. Yu. 2001. Outcome of cephalosporin treatment for serious infections due to apparently susceptible organisms producing extended-spectrum beta-lactamases: implications for the clinical microbiology laboratory. J. Clin. Microbiol. 39:2206-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paterson, D. L., L. B. Rice, and R. A. Bonomo. 2001. Rapid method of extraction and analysis of extended-spectrum beta-lactamases from clinical strains of Klebsiella pneumoniae. Clin. Microbiol. Infect. 7:709-711. [PubMed] [Google Scholar]
- 20.Rhomberg, P. R., R. N. Jones, H. S. Sader, et al. 2004. Results from the Meropenem Yearly Susceptibility Test Information Collection (MYSTIC) Programme: report of the 2001 data from 15 United States medical centres. Int. J. Antimicrob. Agents 23:52-59. [DOI] [PubMed] [Google Scholar]
- 21.Rice, L. B., S. H. Willey, G. A. Papanicolaou, A. A. Medeiros, G. M. Eliopoulos, R. C. Moellering, Jr., and G. A. Jacoby. 1990. Outbreak of ceftazidime resistance caused by extended-spectrum beta-lactamases at a Massachusetts chronic-care facility. Antimicrob. Agents Chemother. 34:2193-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanders, C. C., W. E. Sanders, Jr., and R. V. Goering. 1982. In vitro antagonism of beta-lactam antibiotics by cefoxitin. Antimicrob. Agents Chemother. 21:968-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanders, W. E., Jr., J. H. Tenney, and R. E. Kessler. 1996. Efficacy of cefepime in the treatment of infections due to multiply resistant Enterobacter species. Clin. Infect. Dis. 23:454-461. [DOI] [PubMed] [Google Scholar]
- 24.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuji, A., A. Maniatis, M. A. Bertram, and L. S. Young. 1985. In vitro activity of BMY-28142 in comparison with those of other beta-lactam antimicrobial agents. Antimicrob. Agents Chemother. 27:515-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vuye, A., and J. Pijck. 1985. In vitro antibacterial activity of BMY-28142, a new extended-spectrum cephalosporin. Antimicrob. Agents Chemother. 27:574-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zanetti, G., F. Bally, G. Greub, J. Garbino, T. Kinge, D. Lew, J.-A. Romand, J. Bille, D. Aymon, L. Stratchounski, L. Krawczyk, E. Rubinstein, M.-D. Schaller, R. Chiolero, M.-P. Glauser, A. Cometta, and the Cefepime Study Group. 2003. Cefepime versus imipenem-cilastatin for treatment of nosocomial pneumonia in intensive care unit patients: a multicenter, evaluator-blind, prospective, randomized study. Antimicrob. Agents Chemother. 47:3442-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]