Abstract
We report the development and evaluation of a human immunodeficiency virus type 1 testing algorithm consisting of three rapid antibody detection tests. Stored serum samples from Uganda were utilized with a final algorithm sensitivity of 100% and a specificity of 98.9% (95% confidence interval, 98.6% to 99.3%).
Providing humanitarian or medical assistance in resource-limited environments poses challenges for health care workers, including occupational exposure to blood-borne pathogens. Evaluating the risk of human immunodeficiency virus type 1 (HIV-1) transmission includes the timely determination of source patient seroreactivity. Traditional HIV-1 serology by enzyme immunoassay (EIA) and Western blot analysis is very time-consuming and may be unavailable in resource-limited settings. Reflexive initiation of postexposure prophylaxis (PEP) consumes potentially limited stocks of antiretroviral drugs with the potential for toxicity. Rapid HIV-1 testing can assist PEP decision-making (13) in these time-critical and supply-constrained situations (3, 5, 6).
While the risk of occupational transmission is related to dose, route of exposure, and viral load of the source material, insufficient clinical evidence exists to definitively address the timing of PEP following exposure (2). Animal model data suggest that the decision to use antiretrovirals must be made efficiently. Dendritic cells are infected within 24 h in macaques mucosally exposed to simian immunodeficiency virus, with migration to regional lymph nodes in the subsequent 24 to 48 h (9). PEP failed in this model when initiated at a time beyond 48 h (11). HIV-1 screening in voluntary counseling and testing settings requires diagnostic algorithms based on HIV-1 prevalence, assay operating characteristics, and cost constraints. Postexposure testing must also be timely to complement PEP decisions.
Four HIV-1 rapid EIAs were evaluated for use in a multitest algorithm. We chose specimens from among 14,000 HIV-1 EIA-nonreactive and 1,500 HIV-1 EIA-reactive cryopreserved serum samples collected from an HIV-1 seroprevalence study conducted in the Rakai District of Uganda (14) with an HIV-1 prevalence of 16.9% (8). These samples were shipped frozen to the Walter Reed Army Institute of Research (WRAIR) in Rockville, MD, where the serum was stored at −70°C. The Rakai project, study number M-1356, was approved by the human-use review boards of the WRAIR, the U.S. Army Medical Research and Material Command, the Uganda Virus Research Institute, and the AIDS Research Subcommittee of the Uganda Council for Science and Technology.
All reference and rapid serologic testing was performed at the WRAIR. The OraQuick HIV-1 rapid antibody test (OraSure Technologies, Bethlehem, PA), Determine HIV-1/2 (Abbott Laboratories, Inc., Abbott Park, IL), Hema-Strip HIV-1/2 (Saliva Diagnostic Systems, Inc., Medford, NY), and InstantScreen rapid HIV-1/2 assay (German-American Institute For Applied Biomedical Research, Potsdam, Germany) were performed according to the directions on the package inserts. Test results were read by study investigators who were unaware of sample seroreactivity. Serum specimens were then subjected to EIA screening, confirmatory Western blotting (reference serology), and quantitative viral RNA testing (if indicated) (1, 7) essentially as described previously (4). Operating characteristics, predictive values, statistical simulation of the various permutations of rapid tests, and the performance of the final testing algorithm were calculated using STATA version 7.0 (STATA Corporation, College Station, TX).
Specimens for HIV-1 testing were subjected to testing in two phases. The first phase included 1,000 samples randomly selected from the pool of 15,500 samples. The operating characteristics of the rapid tests were compared to reference testing, and descriptive epidemiological characteristics of the tests were calculated (Table 1). The Hema-Strip HIV-1/2 was insensitive (92.5% sensitivity; 95% confidence interval [CI], 90.8 to 94.1%) compared to the 100% sensitivities observed for the other three rapid tests. The OraQuick test demonstrated the best specificity (97.6% specificity; 95% CI, 96.6 to 98.5%), with both the Determine HIV-1/2 and InstantScreen assays demonstrating specificities of <92%.
TABLE 1.
Rapid HIV-1 test operating characteristicsa
| Test | Sensitivity
|
Specificity
|
PPV
|
NPV
|
||||
|---|---|---|---|---|---|---|---|---|
| % | 95% CI | % | 95% CI | % | 95% CI | % | 95% CI | |
| OraQuick | 100 | 100 | 97.6 | 96.6-98.5 | 80.9 | 78.4-83.3 | 100 | 100 |
| Hema-Strip | 92.5 | 90.8-94.1 | 94.5 | 93.1-95.9 | 63.2 | 60.3-66.2 | 99.2 | 98.6-99.8 |
| InstantScreen | 100 | 100 | 91.8 | 90.1-93.5 | 55.7 | 52.6-58.8 | 100 | 100 |
| Determine | 100 | 100 | 91.7 | 90.0-93.4 | 55.4 | 52.3-58.4 | 100 | 100 |
Testing was accomplished through the use of 1,000 randomly selected Ugandan serum samples drawn from a panel of 15,500 samples (1,500 seropositive and 14,000 seronegative). Resultant HIV-1 seroprevalence in this phase was 9.3%.
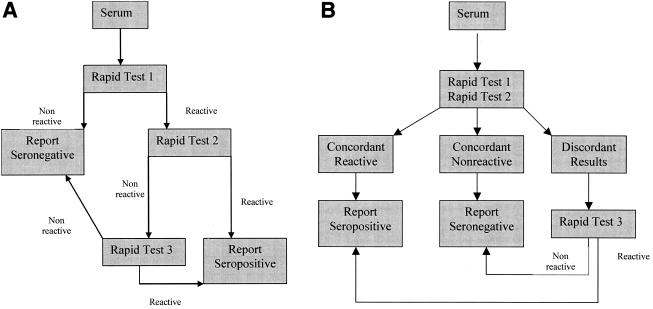
Hypothetical evaluations of three-test serial and parallel designs were performed. The serial design (Fig. 1A) employs a single screening test. Should that test be nonreactive, the sample was considered HIV-1 negative. Should the first test be reactive, a second test was conducted. An HIV-1-positive final result was assigned in the case of concordance between the results of the first two tests. A third test was performed in cases of discordance, the result of which determined the HIV-1 status. The parallel design (Fig. 1B) utilized two tests performed simultaneously, and concordant reactive or nonreactive results would indicate HIV-1-positive or -negative results, respectively. The third test was utilized for discordant test pairs, the result of which would determine the HIV-1 status. All of the designs, including the combination of the OraQuick, InstantScreen, and Determine HIV-1/2 assays, had equivalent performances.
FIG. 1.
(A) Serial design. The sample is subjected to a single rapid test in which a nonreactive result is reported as seronegative. A reactive sample is subjected to a second rapid test, in which a concordant reactive result is reported as seropositive. Discordance is resolved by the reactivity of a third test, and serologic status is reported by best-of-three agreement. (B). Parallel design. The sample is subjected simultaneously to two rapid tests. Concordant seroreactivity or nonreactivity is reported as seropositive or seronegative, respectively. Discordance is resolved by the reactivity of a third test, and serologic status is reported by best-of-three agreement. This design was selected for use in the algorithm evaluation phase of this study.
Final algorithm selection was based on time to results and concern for false negatives seen with chronic HIV-1 infections (13) that are manifest in resource-limited areas. Therefore, the parallel design was chosen to diversify the first-line antigen detection by utilizing two different proprietary systems. The best-performing algorithms displayed the following performance characteristics: sensitivity, 100%; specificity, 99.6% (95% CI, 99.5 to 100%); positive predictive value, 97.9% (95% CI, 97.0 to 98.8%); and negative predictive value, 100% (prevalence, 9.3%). The decision to utilize the OraQuick-Determine-InstantScreen algorithm was based on the ease of use and reduced time to results (22 min versus 35 min) to complete each algorithm (Table 2). Use of the higher-specificity test earlier in the hypothetical algorithm made no difference in the results.
TABLE 2.
Performance of hypothetical algorithms calculated by statistical modeling (STATA version 7.0) of individual test operating characteristics derived from 1
| Design | Algorithma | % Sensitivity (95% CI) | % Specificity (95% CI) | Time to resultsb(min) |
|---|---|---|---|---|
| Parallel | OraQuick-Determine-InstantScreen | 100 (100) | 99.6 (99.0-99.9) | 22 |
| Determine-InstantScreen-OraQuick | 100 (100) | 99.6 (99.0-99.9) | 35 | |
| Serial | Determine-InstantScreen-OraQuick | 100 (100) | 99.6 (99.2-100) | 37 |
| InstantScreen-OraQuick-Determine | 100 (100) | 99.6 (99.0-99.9) | 37 | |
| OraQuick-Determine-InstantScreen | 100 (100) | 99.6 (99.0-99.9) | 37 |
Test order is given from left to right.
Time to results by algorithm was calculated by actual performance of each algorithm.
Thirty-five hundred samples were randomly selected from the remaining pool of 14,500 specimens and subjected to the OraQuick-Determine-InstantScreen parallel algorithm, and the results were compared to those of reference serology. Of these, 474 specimens were concordantly HIV-1 positive, 2,993 specimens were concordantly HIV-1 negative, and 33 specimens were false positive by the rapid-test algorithm (Table 3). There were no false-negative results. This describes a sensitivity of 100% (95% CI, 100%) and a specificity of 98.9% (95% CI, 98.6 to 99.3%). The PPV and NPV, based on a prevalence of 13.5%, were 93.5% (95% CI, 92.7 to 94.3%) and 100% (95% CI, 100%), respectively. All rapid-test-algorithm false-positive sample results were below detection by viral RNA testing (data not shown).
TABLE 3.
Results of the parallel testing algorithm OraQuick-Determine-Instant-Screena
| Rapid-test algorithm results | Double EIA-Western blot algorithm results
|
||
|---|---|---|---|
| No. of samples positive | No. of samples negative | Total no. | |
| Positive | 474 | 33 | 507 |
| Negative | 0 | 2,993 | 2,993 |
| Total | 474 | 3,026 | 3,500 |
Test order is given left to right. A total of 3,500 serum samples were randomly selected from the remaining panel of 14,500 samples (1,000 had been selected for the first phase of the study), with a resultant HIV-1 seroprevalence of 13.5%. Results of the parallel rapid-test algorithm are compared with those of serial EIA and confirmatory Western blotting with the resulting performance characteristics: sensitivity, 100% (95% CI, 100%); specificity, 98.9% (95% CI, 98.6 to 99.3); PPV, 93.5% (95% CI, 92.7 to 94.3); NPV, 100% (95% CI, 100%).
Parallel testing is not indicated for routine HIV-1 screening, especially in areas of high endemicity (15), as it is not cost-effective. However, postexposure HIV-1 testing addresses different goals. An ideal postexposure testing algorithm would include diverse antigen sources and different test principles (10, 12) to eliminate the possibility of false-negative results (13) and to provide accurate results within the shortest time (2, 15). The OraQuick-Determine-InstantScreen algorithm described here would cost approximately $12 versus $8 for testing with OraQuick alone, but it demonstrated increased specificity and would reduce the less easily defined costs of unnecessary administration of antiretrovirals with their attendant toxicities. This approach, in combination with risk factor analysis, might be especially relevant in resource-limited environments where available antiretroviral drugs are scarce.
Acknowledgments
We thank Jose L. Sanchez for review of the manuscript.
The views expressed here are the private opinions of the authors and are not to be considered as official or reflecting the views of the U.S. Army or the U.S. Department of Defense. Use of trade names is for identification only and does not imply endorsement by the U.S. Government.
This work was supported in part by Cooperative Agreement No. DAMD17-93-V-3004 between the U.S. Army Medical Research and Materiel Command and the Henry Jackson Foundation for the Advancement of Military Medicine.
REFERENCES
- 1.Birx, D. L., L. D. Loomis-Price, N. Aronson, J. Brundage, C. Davis, L. Deyton, R. Garner, F. Gordin, D. Henry, W. Holloway, T. Kerkering, R. Luskin-Hawk, J. McNeil, N. Michael, P. Foster Pierce, D. Poretz, S. Ratto-Kim, P. Renzullo, N. Ruiz, K. Sitz, G. Smith, C. Tacket, M. Thompson, E. Tramont, B. Yangco, R. Yarrish, R. R. Redfield, et al. 2000. Efficacy testing of recombinant human immunodeficiency virus (HIV) gp160 as a therapeutic vaccine in early-stage HIV-1-infected volunteers. J. Infect. Dis. 181:881-889. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2001. Updated U.S. Public Health Service guidelines for the management of occupational exposures to HBV, HCV, and HIV and recommendations for postexposure prophylaxis. MMWR Recomm. Rep. 50:1-52. [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 1998. Update: HIV counseling and testing using rapid tests-United States, 1995. Morb. Mortal. Wkly. Rep. 47:211-215. [PubMed] [Google Scholar]
- 4.Foglia, G., G. D. Royster IV, K. M. Wasunna, R. Kibaya, J. A. Malia, E. K. Calero, W. Sateren, P. O. Renzullo, M. L. Robb, D. L. Birx, and N. L. Michael. 2004. Use of rapid and conventional testing technologies for human immunodeficiency virus type 1 serologic screening in a rural Kenyan reference laboratory. J. Clin. Microbiol. 42:3850-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giles R. E., K. R. Perry, and J. V. Parry. 1999. Simple/rapid test devices for anti-HIV screening: do they come up to the mark? J. Med. Virol. 59:104-109. [PubMed] [Google Scholar]
- 6.Henderson, D. K. 1999. Postexposure chemoprophylaxis for occupational exposures to the human immunodeficiency virus. JAMA 281:931-936. [DOI] [PubMed] [Google Scholar]
- 7.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 8.Nalugoda, F., R. H. Gray, D. Serwadda, N. K. Sewankambo, F. Wabwire-Mangen, N. Kiwanuka, T. Lutalo, G. Kigozi, C. Li, F. Makumbi, M. Kiddugavu, L. Paxton, S. Zawedde, and M. Wawer. 2004. Burden of infection among heads and non-head of rural households in Rakai, Uganda. AIDS Care. 16:107-115. [DOI] [PubMed] [Google Scholar]
- 9.Spira, A. I., P. A. Marx, B. K. Patterson, J. Mahoney, R. A. Koup, S. M. Wolinsky, and D. D. Ho. 1996. Cellular targets of infection and route of viral dissemination after an intravaginal inoculation of simian immunodeficiency virus into rhesus macaques. J. Exp. Med. 183:215-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stickle, D. F., S. J. Pirruccello, S. Swindells, and S. H. Hinrichs. 2002. Discrepant results of 2 screening tests for anti-HIV antibody. Clin. Infect. Dis. 35:773-774; author reply 774-775. [DOI] [PubMed] [Google Scholar]
- 11.Tsai, C.-C., P. Emau, K. E. Follis, T. W. Beck, R. E. Benveniste, N. Bischofberger, J. D. Lifson, and W. R. Morton. 1998. Effectiveness of postinoculation (R)-9-(2-phosphonylmethoxypropyl)adenine treatment for prevention of persistent simian immunodeficiency virus SIVmne infection depends critically on timing of initiation and duration of treatment. J. Virol. 72:4265-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urassa, W., K. Godoy, J. Killewo, G. Kwesigabo, A. Mbakileki, F. Mhalu, and G. Biberfeld. 1999. The accuracy of an alternative confirmatory strategy for detection of antibodies to HIV-1: experience from a regional laboratory in Kagera, Tanzania. J. Clin. Virol. 14:25-29. [DOI] [PubMed] [Google Scholar]
- 13.van den Berk, G. E. L., P. H. J. Frissen, R. M. Regez, and P. J. G. M. Rietra. 2003. Evaluation of the rapid immunoassay Determine HIV 1/2 for detection of antibodies to human immunodeficiency virus types 1 and 2. J. Clin. Microbiol. 41:3868-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wawer, M. J., N. K. Sewankambo, S. Berkley, D. Serwadda, S. D. Musgrave, R. H. Gray, M. Musagara, R. Y. Stallings, and J. K. Konde-Lule. 1994. Incidence of HIV-1 infection in a rural region of Uganda. BMJ 308:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.World Health Organization. 2004. Rapid HIV tests: guidelines for use in HIV testing and counselling services in resource-constrained settings. [Online.] http://www.who.int/hiv/pub/vct/en/rapidhivtestsen.pdf.