Abstract
Escherichia coli O174 and O177 are newly described O serogroups which were reported as human pathogens. Identification of these strains by serotyping has been restricted, as the required sera are not commercially available. In this study, a collection of 13 E. coli O174 strains and 12 E. coli O177 strains was studied on the O:H serotypes and virulence markers. The O-antigen gene clusters of E. coli O174 and O177 were sequenced, and associated genes were assigned functions on the basis of homology. Two genes, each specific for E. coli O174 and O177, were identified. PCR assays based on the O-antigen-specific genes were developed and tested on 25 clinical and environmental isolates of those two serogroups as well as 26 isolates of other O serogroups. As little as 1 pg per μl of chromosomal DNA and as few as 0.1 CFU per g of pork and water samples were detected for either strain. The PCR assays established in this study were shown to be highly sensitive and reliable and could be the method of choice for detection of these two human pathogens from clinical, food, and other environmental samples.
Escherichia coli is a species that includes both commensal and pathogenic clones of strains, which are normally identified by serological typing of their O (lipopolysaccharide), H (flagellar), and, in some cases, K (capsular) surface antigens. At present, 176 O and 53 H antigens, which can occur in different combinations in wild-type isolates of strains, have been described for E. coli (17, 20, 23). E. coli clones causing disease in humans, such as enteropathogenic E. coli (EPEC) and enterohemorrhagic E. coli (EHEC), are frequently associated with certain O:H types, for example, EHEC O157:H7 and EPEC O55:H6 strains. A growing number of E. coli serotypes are associated with the EPEC or EHEC group, and among them are the newly described O serogroups O174 (formerly designated as OX3 or OX174) and O177 (formerly called OX177) (3, 10, 23, 24). Typing sera for detection of the eight novel E. coli O groups from O174 to O181 are not commercially available, and their serological detection is restricted to a small number of E. coli reference laboratories. Studies from reference centers indicate that domestic animals serve as a reservoir for these strains, which were reported as human pathogens in different countries and on different continents (3, 8, 19, 21, 23, 25, 26).
More recently, DNA-based typing methods were employed for the detection of E. coli O-antigen-encoding genes, and correspondent PCR protocols for the identification of pathogenic E. coli strains such as O157, O111, O114, and O172 were developed (12, 14, 28, 30). The O antigen, as a part of the lipopolysaccharide moiety of gram-negative bacteria, consists of many repeats of an oligosaccharide unit (O unit) (9). The genes for O-antigen synthesis are normally clustered between the two housekeeping genes galF and gnd on the E. coli chromosome, and a conserved 39-bp JUMPStart sequence, which is required for the regulation of downstream genes by RfaH (15), is located in the intergenic region between galF and the O-antigen gene cluster (18). The genes in the O-antigen gene cluster are normally classified into three main classes: genes for synthesis of nucleotide sugar precursors, genes encoding glycosyltransferases, and genes for O-unit processing, including the flippase (Wzx) and polymerase (Wzy) genes (9). Genes in the last two classes are O-antigen specific since they are specific to sugar donors, sugar acceptors, and linkages between the sugars (30). The differences between O-antigen forms are almost entirely due to genetic variations in the O-antigen gene clusters (9).
PCR methods for the detection of O-antigen-specific genes such as wzx and wzy represent a reliable, rapid, and sensitive alternative to serotyping, particularly for E. coli strains that belong to serogroups that are not covered by the panel of commercially available antisera (5, 14, 22, 27). Moreover, PCR detection of O-antigen-specific genes allows detection of strains which are serologically rough for their O antigens, and last but not least, serological cross-reactions between different O antigens that may cause difficulties in O typing are ruled out.
In this study, a collection of 13 E. coli O174 strains and 12 E. coli O177 strains was studied on the O:H serotypes and virulence markers. The O-antigen gene clusters of the E. coli reference strains for the newly defined serogroups O174 and O177 were sequenced, and the associated genes were identified on the basis of homology. By screening against all 186 E. coli O serotypes (including Shigella) strains, genes specific for E. coli serogroups O174 and O177 were identified. The PCR assays that were developed in this study for detection of O-antigen-specific genes were shown to be highly specific and sensitive and were shown to be useful for the detection of E. coli O174 and O177 strains from clinical, food, and environmental samples.
MATERIALS AND METHODS
Bacterial strains.
The reference strains for E. coli O174 (2531-54) and O177 (E40874-85) used for nucleotide sequence analysis of their O-antigen clusters were described elsewhere previously (23). A collection of 13 E. coli O174 and 13 O177 strains which were characterized for their serotypes, phenotypical traits, and virulence markers was used for evaluation of the O-antigen gene-specific PCR (see below). The strains originated from feces of humans and animals and from food and were collected at different time periods and geographical locations investigated (Table 1). Other E. coli and Shigella type strains used in this study were previously described (12).
TABLE 1.
Relevant properties of E. coli O174 and O177 strains
| Strain no. | Strain (reference) | Serotypea | Origin (yr of isolation) | VCAb | Hlyc | Positive for virulence gened | Diseasee |
|---|---|---|---|---|---|---|---|
| G1609 | 2531-54 (23) | O174:H27 | Human; E. coli O174 reference strain | − | None | None | D |
| G1610 | CB9135 (25) | O174:[H8] | Sheep, Norway (2001) | + | None | stx1c, stx2c | AS |
| G1611 | CB9534 | O174:[H8] | Raw Cheese, France (2002) | + | Ehly | ehxA, stx1c | None |
| G2032 | CB8293 (3) | O174:H16 | Human, Germany (1999) | + | None | stx2c | ND |
| G2033 | CB9043 (25) | O174:H8 | Sheep, Norway (2001) | + | Ehly | ehxA, stx1c, stx2c | AS |
| G2034 | CB8377 | O174:H2 | Ground meat, Germany (2000) | + | Ehly | ehxA, stx1, stx2 | None |
| G2035 | CB7950 (3) | O174:H2 | Human, Germany (1999) | + | Ehly | ehxA, stx1, stx2 | BD |
| G2036 | CB8958 | O174:H21 | Human, Germany (2001) | + | None | stx2c | D |
| G2037 | CB7855 (3) | O174:H21 | Human, Germany (1998) | + | Ehly | ehxA, stx2d | BD |
| G2038 | CB7033 (3) | O174:H2 | Human, Germany (1997) | + | Ehly | ehxA, stx1, stx2 | BD |
| G2039 | CB9042 (25) | O174:H8 | Sheep, Norway (2001) | + | Ehly | ehxA, stx1c, stx2c | AS |
| G2040 | CB8185 | O174:H28 | Minced meat, Germany (1999) | + | Ehly | ehxA, stx2 | None |
| G2041 | CB7398 | O174:H43 | Human, France (1995) | − | None | None | HUSf |
| G1606 | E40874-85 (23) | O177:H25 | Cattle; E. coli O177 reference strain | + | None | stx2, eae-β | ND |
| G1607 | CB9861 | O177:H11 | Cattle, Brazil (2003) | − | Ehly | eae-β1, ehxA | AS |
| G1608 | CB9268 | O177:H11 | Cattle, Germany (1990) | + | Ehly | ehxA, stx2, eae-β | AS |
| G2042 | CB7137 | O177:H25 | Cattle, Switzerland (1997) | + | Ehly | ehxA, stx2, stx2c, eae-β | AS |
| G2043 | CB7135 | O177:[H25] | Cattle, Switzerland (1997) | + | Ehly | ehxA, stx2c, eae-β | AS |
| G2044 | CB9714 | O177:H25 | Human, Germany (2003) | − | Ehly | eae-β, ehxA | D |
| G2045 | CB1138 | O177:H11 | Calf, Germany (1990) | − | Ehly | eae-β, ehxA | D |
| G2046 | CB1139 | O177:H11 | Calf, Germany (1990) | − | Ehly | ehxA, eae-β | D |
| G2047 | CB8989 | O177:[nt] | Human, Germany (2001) | + | None | stx1, eae-α | D |
| G2048 | CB8995 | O177:[nt] | Human, Germany (2001) | + | None | stx1, eae-α | D |
| G2049 | CB7134 | O177:[H25] | Cattle, Switzerland (1997) | + | Ehly | ehxA, stx2, stx2c, eae-β | AS |
| G2068 | CB9657 | O177:H26 | Human, Thailand (2003) | − | None | eae-β | D |
In fliC genotypes of nonmotile strains, [nt] indicates that a serotype could not be assigned to known fliC RFLP type.
VCA, Vero cell toxicity assay.
Hemolytic phenotype. Ehly, enterohemolytic (6).
Virulence markers investigated by PCR. ehxA, EHEC hemolysin A-encoding gene; stx, Shiga toxin-encoding gene; eae, intimin-encoding gene; EAF plasmid, plasmid-specific 397-bp region.
AS, no signs of disease; D, nonbloody diarrhea; BD, bloody diarrhea; HUS, hemolytic uremic syndrome; ND, no data.
Strain was originally reported to carry an stx2 gene (F. Grimont, Institut Pasteur, Paris, France, personal communication).
Serotyping and molecular typing of flagellar (fliC) genes.
Serotyping of E. coli O and H antigens was performed as described previously (5). Specific antisera for detection of O antigens O174 and O177 were produced at the Robert Koch Institute with reference strains 2531-54 (O174:H27) and E40874-85 (O177:H25), respectively (Table 1). Agglutination reactions were performed in twofold dilution steps of antiserum with boiled bacteria as described previously (20) Agglutinating titers with homologous strains were 1:3,200, and no significant cross-reactions (<1:200) were observed with other E. coli O antigens. The O174 and O177 test strains did not differ for more than 1 titer step in their agglutination reactions compared to their respective reference strains. Nonmotile E. coli strains were investigated for their flagellar (fliC) genes by PCR and analysis of HhaI restriction fragment length polymorphisms (RFLPs) of PCR products as described previously (4).
Phenotypical and genotypical characterization of virulence markers.
The E. coli O174 and O177 strains were investigated for the production of cytotoxins with the Verocell assay and for hemolysins on enterohemolysin agar (Oxoid, Wesel, Germany) (6). Detection and subtyping of genes encoding the EPEC adherence factor (EAF) plasmid, intimins (eae), Shiga toxins (stx), and EHEC-hemolysin (ehxA) were performed by PCR and RFLP analysis using primers and restriction enzymes described previously (3, 13). An updated nomenclature of Shiga toxin types was used as described recently (3).
Construction of DNase I shotgun bank, sequencing, and analysis of O-antigen gene cluster.
Chromosomal DNA was prepared as previously described (2). The primer pairs wl-1098 (5′-ATT GGT AGC TGT AAG CCA AGG GCG GTA GCG T-3′) and wl-2211 (5′-CAC TGC CAT ACC GAC GAC GCC GAT CTG TTG CTT GG-3′), based on the JUMPStart sequence and gnd, respectively (30), were used to amplify the O-antigen gene clusters of E. coli O174 and O177 type strains 2531-54 and E40874-85 (Table 1). The PCR was performed for 30 cycles as follows: a denaturation step at 94°C for 10 s, an annealing step at 60°C for 30 s, and an extension step at 68°C for 15 min. Shotgun banks for each strain were constructed as described previously (30). Sequencing was carried out using an ABI 3730 automated DNA sequencer, and sequence data were analyzed using computer programs as described previously (11).
Specificity and sensitivity test of O-serogroup-specific PCR assay.
Chromosomal DNA was prepared from each of 186 type strains to represent a broad range of O antigens of E. coli and Shigella and was used to make DNA pools as described previously (12). A total of 13 pools were made, each containing DNA from 12 to 19 strains (12). Either E. coli strain 2531-54 (O174) or strain E40874-85 (O177) and the strains of pool 12 were used to make up pool 13. Primer pairs (see Table 4) specific for wzx and wzy genes of E. coli O174 and O177 were used to screen the DNA pools. The PCR was performed as follows: a denaturation step at 95°C for 30 s, an annealing step at 45°C for 30 s, and an extension step at 72°C for 1 min for 30 cycles. Template DNA from 50 clinical E. coli isolates including 12 E. coli O174, 11 E. coli O177 (Table 1), and 26 E. coli strains belonging to other O serogroups (data not shown) were prepared as described previously (14) and screened with the primers listed in Table 4 in a double-blind test.
TABLE 4.
Primers used for PCR detection of strains of E. coli O serogroups O174 and O177
| Serogroup | Gene | Base positions | Forward primer/reverse primer | Length of PCR product (bp) |
|---|---|---|---|---|
| O174 | wzx | 805-2076 | wl-4397 (positions 891-908) (5′-TCTAGGACCTGTAGAACA-3′)/wl-4398 (positions 1529-1546) (3′-GTAGTTGATCTGAGCGAT-5′) | 656 |
| wl-4421 (positions 1479-1498) (5′-AGTAATGATCGGAACTATGC-3′)/wl-4422 (positions 1861-1878) (3′-TTTTGGAATAACCGTCGT-5′) | 400 | |||
| wzy | 2057-3196 | wl-4399 (positions 2421-2439) (5′-TATGGGTCCTATTACTTTC-3′)/wl-4400 (positions 3160-3179) (3′-GTATCGGAGATCATTATTAC-5′) | 759 | |
| wl-4423 (positions 2512-2529) (5′-TCTAGGGACTGTTGTTAC-3′)/wl-4424 (positions 2944-2961) (3′-TCTTTAACCCGACTTATC-5′) | 450 | |||
| O177 | wzx | 3030-4295 | wl-4393 (positions 3528-3545) (5′-GTTGCGTTGCCTGCTGTA-3′)/wl-4394 (positions 4190-4207) (3′-GGTAAAGCCCTATCATCC-5′) | 680 |
| wl-4417 (positions 3170-3187) (5′-TATGGCATTAGTGGGTTA-3′)/wl-4418 (positions 3532-3549) (3′-GCAACGGACGACATTCAT-5′) | 380 | |||
| wzy | 4855-6141 | wl-4395 (positions 4946-4963) (5′-TTTTATTAGGGTCAGGAG-3′)/wl-4396 (positions 5419-5436) (3′-CACAACGACGGATTATCA-5′) | 491 | |
| wl-4419 (positions 5386-5403) (5′-ATTATTGCCGATACACCG-3′)/wl-4420 (positions 5798-5815) (3′-CAGGACAAGACCCATGAC-5′) | 430 |
The sensitivity of the PCR assays was tested with 10-fold serial dilutions of chromosomal DNA of E. coli strain 2531-54 (O174) or E40874-85 (O177) with the primer pairs designed (see Table 4). Primer pairs wl-4397/wl-4398 and wl-4399/wl-4400 from E. coli O174 and wl-4393/wl-4394 and wl-4395/wl-4396 from E. coli O177 were also used to screen for E. coli strains of these two O serogroups in pork and water samples using a method described previously (14).
Nucleotide sequence accession numbers.
The DNA sequences of the E. coli O174 and O177 O-antigen gene clusters have been deposited in GenBank under the accession numbers DQ008592 and DQ008593, respectively.
RESULTS AND DISCUSSION
Serotypes and virulence markers of E. coli O174 and O177 strains.
The 13 E. coli O174 strains originated from diseased humans, healthy sheep, and food. They were distributed over seven H types (H2, H8, H16, H21, H27, H28, and H43) (Table 1). All strains were negative for an intimin-encoding gene (eae), which is in accordance with previous investigations made with the E. coli O174 group (3, 7, 8, 19, 25). Twelve of the O174 strains from our study produced Shiga toxins, and toxin types 1 and 2 were associated with O174:H2 and O174:H28 strains, types 1c and 2c were associated with O174:H8 and O174:H16 strains, and type 2d was associated with O174:H21 strains. Eight of the O174 strains showed an enterohemolytic phenotype and carried an ehxA gene, which is a typical trait of many EHEC strains (3). Toxin type 2 E. coli O174:H2 and type 2d O174:H21 strains were associated with bloody diarrhea in humans. Studies of E. coli O174 in animals have indicated that cattle are a reservoir for O174:H2 and O174:H21 strains and that sheep are a reservoir for O174:H8 strains (7, 21, 25).
The 11 E. coli O177 strains originated from diarrheagenic humans and calves as well as from healthy cattle. They were distributed over three H types (H11, H25, and H26), except for two nonmotile strains, which could not be typed for their fliC genes (Table 1). In contrast to E. coli O174, all E. coli O177 strains investigated were positive for an eae gene. Those distributed over H types H11, H25, and H26 carried an eae-β gene, and the two nonmotile strains carried an eae-α gene. Seven of the E. coli O177 strains produced Shiga toxins (types 1 and/or 2). Nine were positive for enterohemolysin and the ehxA gene. According to their virulence profiles, E. coli O177:H11 and O177:H25 were associated with two pathotypes such as atypical EPEC (eae positive) and EHEC (eae and stx positive) (24). None of the E. coli O177 and O174 strains were positive for sequences of the EAF plasmid, which is harbored by typical (class I) EPEC strains (24).
Nucleotide sequence analysis of the E. coli O174 O-antigen gene cluster.
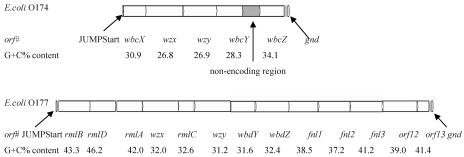
A sequence of 5,660 bases between the JUMPStart sequence and gnd was obtained from E. coli O174, and five open reading frames (ORFs) were found (Fig. 1). By comparing these to related genes in nucleotide sequence databases, all ORFs were assigned functions and shown to be related to O-antigen biosynthesis (Table 2).
FIG. 1.
O-antigen gene clusters of E. coli O174 and E. coli O177. All genes are transcribed in a direction from the JUMPStart sequence to gnd.
TABLE 2.
Putative genes in the E. coli O174 O-antigen gene cluster
| Gene name | Location in sequence | G+C content (%) | Similar protein(s), species (GenBank accession no.) | % Identical aa/% similar aa (no. of aa overlap)a | Putative function of protein |
|---|---|---|---|---|---|
| wbcX | 51-794 | 30.9 | LgtD, Rickettsia prowazekii (CAA14903) | 6/43 (209) | Glycosyltransferase |
| wzx | 797-2068 | 26.8 | Wzx, Yersinia enterocolitica (type 0:8) (AAC60766) | 27/49 (381) | O-unit flippase |
| wzy | 2049-3188 | 26.9 | Wzy, Bacillus cereus ATCC 10987 (AAS44289) | 24/46 (243) | O-unit polymerase |
| wbcY | 3191-4075 | 28.3 | Putative glycosyltransferase, Bacillus halodurans C-125 (BAB07432) | 32/55 (232) | Glycosyltransferase |
| wbcZ | 4661-5482 | 34.1 | Putative glycosyltransferase, Salmonella enterica (AAG09520) | 52/70 (265) | Glycosyltransferase |
aa, amino acid.
(i) O-unit-processing genes.
Both Wzx and Wzy are typical inner membrane proteins with more than nine transmembrane segments, and Wzy also contains a large periplasmic loop of more than 30 amino acids. Orf2 has 11 predicted transmembrane segments and shares 49% similarity to Wzx of Yersinia enterocolitica (type 0:8). It belongs to the protein family PF01943 (E value, 1.4 × e−5), and members of this family are involved in the export of the O antigen and teichoic acid. Therefore, orf2 was proposed to be an O-unit flippase gene (wzx) and was named accordingly. Orf3 has 12 predicted transmembrane segments and a large periplasmic loop of 61 amino acid residues. It shares 46% similarity to the putative Wzy of Bacillus cereus. Therefore, orf3 was proposed to be an O-antigen polymerase gene (wzy) and was named accordingly.
(ii) Glycosyltransferase genes.
Orf1, Orf4, and Orf5 belong to the glycosyltransferase family 2 (pfam00535 E values, 5 × e−5, 1.8 × e−13, and 6.0 × e−25, respectively) and share 43 to 70% similarity with other putative glycosyltransferases (Table 2). Therefore, orf1, orf4, and orf5 were proposed to be glycosyltransferase genes and were named wbcX, wbcY, and wbcZ, respectively.
The O-antigen structure of E. coli O174 has not been characterized. The absence of sugar synthesis genes in the O-antigen gene cluster indicates that the O unit of E. coli O174 consists of common sugars such as Glc, Gal, GlcNAc, and GalA. Genes for the synthesis of common sugars are outside the O-antigen gene cluster (9). Glycosyltransferase genes are highly specific to sugar donors, sugar receptors, and the linkages between them. The wecA gene, which is located outside the O-antigen gene cluster, is responsible for the transfer of GlcNAc presented as the first sugar in most E. coli O units (1). Three glycosyltransferase genes were found in the E. coli O174 O-antigen gene cluster. Therefore, E. coli O174 O antigen was predicted to be composed of tetrasaccharide repeating units.
A large nonencoding region between orf4 and orf5 (positions 4084 to 4668) was found. It contains numbers of stop codons and shares no similarity to other genes in the databases. It appears that this intergenic region underwent numerous changes under selection pressure.
Nucleotide sequence analysis of the E. coli O177 O-antigen gene cluster.
A sequence of 13,198 bases between the JUMPStart sequence and gnd was obtained from E. coli O177, and 13 ORFs were found (Fig. 1). By comparing these to related genes in nucleotide sequence databases, all ORFs were assigned functions and were shown to be related to O-antigen biosynthesis (Table 3).
TABLE 3.
Putative genes in the E. coli O177 O-antigen gene cluster
| Gene name | Location in sequence | G+C content (%) | Similar protein(s), strain(s) (GenBank accession no.) | % Identical aa/% similar aa (no. of aa overlap)a | Putative function of protein |
|---|---|---|---|---|---|
| rmlB | 108-1193 | 43.3 | RmlB, Escherichia coli K-12 (AAC75102) | 97/98 (361) | dTDP-d-glucose-4,6-dehydratase |
| rmlD | 1193-2092 | 46.2 | RmlD, Escherichia coli K-12 (AAC75101) | 97/98 (299) | dTDP-6-deoxy-l-mannose-dehydrogenase |
| rmlA | 2150-3040 | 42.0 | RmlA, Escherichia coli K-12 (AAC75100) | 91/96 (290) | Glucose-1-phosphate thymidylyltransferase |
| wzx | 3030-4295 | 32.0 | Wzx, Shigella flexneri 2a strain 301 (AAN43639) | 51/74 (407) | O-unit flippase |
| rmlC | 4308-4874 | 32.6 | RmlC, Escherichia coli K-12 (AAN60457) | 65/79 (174) | dTDP-6-deoxy-d-glucose-3,5-epimerase |
| wzy | 4855-6141 | 31.2 | Wzy, Shigella flexneri 2a strain 301 (AAN43636) | 25/52 (175) | O-unit polymerase |
| wbdY | 6138-7010 | 31.6 | Rhamnosyltransferase I, Shigella dysenteriae type 1 (AAA16937) | 30/48 (294) | Glycosyltransferase |
| wbdZ | 6982-8169 | 32.4 | Glycosyltransferase, Shigella boydii type 13 (AAR24277) | 26/45 (398) | Glycosyltransferase |
| fnlA | 8174-9208 | 38.5 | Fnl1, Escherichia coli O26 (AAN60461) | 92/98 (344) | l-Fucosamine synthetase |
| fnlB | 9192-10316 | 37.2 | Fnl2, Escherichia coli O26 (AAN60462) | 96/98 (374) | l-Fucosamine synthetase |
| fnlC | 10331-11443 | 41.2 | Fnl3, Escherichia coli O26 (AAN60463) | 98/98 (370) | l-Fucosamine synthetase |
| wbuB | 11443-12651 | 39.0 | Putative l-fucosamine transferase, Escherichia coli O26 (AAN60464) | 96/97 (402) | l-Fucosamine transferase |
| wbuC | 12642-13049 | 41.4 | WbuC, Escherichia coli O26 (AAN60465) | 98/100 (135) | Unknown |
aa, amino acid.
(i) Sugar synthesis genes.
Orf1, Orf2, Orf3, and Orf5 share 97, 97, 91, and 65% identity to RmlB, RmlD, RmlA, and RmlC, respectively, of the E. coli K-12 O-antigen gene cluster. They also share high-level identity to other known Rml proteins, RmlB, RmlD, RmlA, and RmlC, of E. coli and Shigella strains (data not shown). The Rml proteins are enzymes for the synthesis of dTDP-l-rhamnose and have been well characterized in many bacterial strains. orf1, orf2, orf3, and orf5 were identified as rmlB, rmlD, rmlA, and rmlC, respectively. The presence of the rml genes indicates that the E. coli O177 O antigen contains a rhamnose moiety. In most E. coli and Shigella strains, the rmlABCD genes are arranged as a group at the 5′ end of the O-antigen gene cluster. However, in E. coli O177, the rmlC gene is separated from other rml genes by wzx. The same gene order was found in the Shigella boydii type 9 O-antigen gene cluster (29). As in the case of S. boydii type 9, the atypical position of rmlC also indicates that the E. coli O177 O-antigen gene cluster was assembled more recently by lateral transfer.
Orf9, Orf10, and Orf11 share 80, 55, and 70% identity to FnlA (WbjB), FnlB (WbjC), and FnlC (WbjD) of the Pseudomonas aeruginosa O11 O-antigen gene cluster, respectively, which were experimentally identified as UDP-l-FucNAc biosynthesis pathway enzymes (16). They also share 92, 96, and 98% identity to FnlA (Fnl1), FnlB (Fnl2), and FnlC (Fnl3) of the E. coli O26 O-antigen gene cluster, respectively. Therefore, orf9, orf10, and orf11 were identified as fnlA, fnlB, and fnlC and were named accordingly. The presence of the fnl genes indicates that the E. coli O177 O antigen contains a fucosamine moiety.
(ii) O-unit-processing genes.
Orf4 has 10 predicted transmembrane segments and shares 74% similarity to Wzx of S. flexneri 2a strain 301. It belongs to the Pfam family PF01943 (E value, 3.1 × e−52), and members of this family are involved in the export of the O antigen and teichoic acid. Therefore, orf4 was proposed to be an O-unit flippase gene (wzx) and was named accordingly. Orf6 has 10 predicted transmembrane segments and a large periplasmic loop of 56 amino acid residues. It also shares 52% similarity to Wzy of S. flexneri 2a. Therefore, orf6 was proposed to be an O-antigen polymerase gene (wzy) and was named accordingly.
(iii) Glycosyltransferase genes.
Orf7 and Orf8 belong to glycosyltransferase family 2 (PF00535; E value, 1.7 × e−2) and glycosyltransferase family 1 (PF00534; E value, 6.6 × e−7). They also share 48 and 45% similarity to glycosyltransferases of Shigella dysenteriae type 1 and Shigella boydii type 13, respectively. orf7 and orf8 were proposed to be glycosyltransferase genes and were named wbdY and wbdZ, respectively. Orf12 belongs to glycosyltransferase family 1 (PF00534; E value, 4.5 × e−3) and shares 96% identity to a putative l-fucosamine transferase, WbuB, of E. coli O26 (9); based on this high-level identity, orf12 was proposed to be an l-Fucosamine transferase gene and was named wbuB.
(iv) Nonfunctional gene.
Orf13 shares 98% identity to a proposed remnant gene product, WbuC, of E. coli O26 (9). orf13 was proposed to be nonfunctional and was named wbuC.
The O-antigen structure of E. coli O177 has not been characterized. Based on the number of glycosyltransferase genes found in the O-antigen gene cluster, E. coli O177 O antigen was predicted to be composed of tetrasaccharide O-unit containing rhamnose and FucNAc.
Specific identification of E. coli O174 and O177 serogroup-specific genes by PCR.
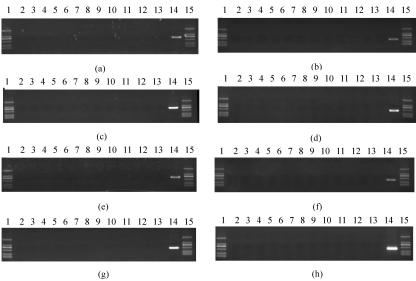
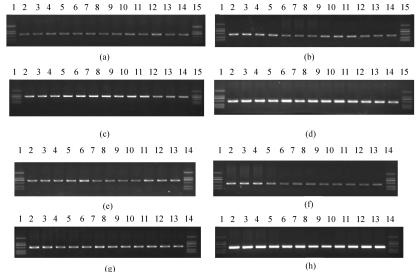
Two pairs of primers specific for wzx and wzy genes were designed for E. coli O174 (Table 4) and used to screen DNA pools containing representatives of the 186 known O-antigen forms of E. coli and Shigella strains. Except for the pools containing E. coli O174, which gave PCR products of the expected size, no PCR products were obtained with all other pools tested (Fig. 2). The same high primer specificity was found with primers designed for E. coli O177 (Table 4). The primer pairs listed in Table 4 were further used in double-blind tests on E. coli clinical isolates including 12 E. coli O174 strains, 11 E. coli O177 strains (Table 1), and 26 E. coli strains of other O serogroups (Fig. 3). All of the E. coli O174 and O177 strains gave the expected PCR products corresponding to primer pairs used, and no PCR products were obtained from strains belonging to other O serogroups. The PCR products amplified from all O174 and O177 strains by the primers wl-4421/wl-4422 and wl-4417/wl-4418 were sequenced, and it was found that all the sequences of different strains of O174 or O177 share identity of more than 99.5% (at most, 1 base different). The results show that the primers for E. coli O174 and O177 are highly specific and suitable for the development of PCR assays for the identification and detection of E. coli O174 or O177 strains.
FIG. 2.
Amplification products obtained by PCR of 13 pools of DNA including 186 E. coli and Shigella O-antigen type strains and E. coli O174 strain G1609 or E. coli O174 strain G1606 using primer pairs wl-4397/wl-4398 (a), wl-4421/wl-4422 (b), wl-4399/wl-4400 (c), wl-4423/wl-4424 (d), wl-4393/wl-4394 (e), wl-4417/wl-4418 (f), wl-4395/wl-4396 (g), and wl-4419/wl-4420 (h). Lanes 1 and 15, DNA marker, bands with lengths of 100 bp, 250 bp, 500 bp, 750 bp, 1 kb, and 2 kb; lanes 2 to 14, pool 1 to pool 13, respectively.
FIG. 3.
Amplification products obtained by PCR of 13 clinical isolated E. coli O174 strains using primer pairs wl-4397/wl-4398 (a), wl-4421/wl-4422 (b), wl-4399/wl-4400 (c), and wl-4423/wl-4424 (d). Lanes 1 and 15, DNA marker, bands with lengths of 100 bp, 250 bp, 500 bp, 750 bp, 1 kb, and 2 kb; lane 2, G1609; lane 3, G1610; lane 4, G1611; lane 5, G2032; lane 6, G2033; lane 7, G2034; lane 8, G2035; lane 9, G2036; lane 10, G2037; lane 11, G2038; lane 12, G2039; lane 13, G2040; lane 14, G2041. Also shown are amplification products obtained by PCR of 12 clinically isolated E. coli O177 strains using primer pairs wl-4393/wl-4394 (e), wl-4417/wl-4418 (f), 4395/wl-4396 (g), and wl-4419/wl-4420 (h). Lanes 1 and 14, DNA marker, bands with lengths of 100 bp, 250 bp, 500 bp, 750 bp, 1 kb, and 2 kb; lane 2, G1606; lane 3, G1607; lane 4, G1608; lane 5, G2042; lane 6, G2043; lane 7, G2044; lane 8, G2045; lane 9, G2046; lane 10, G2047; lane 11, G2048; lane 12, G2049; lane 13, G2068.
Sensitivity of the O-serogroup-specific PCR assays.
The primer pairs described in Table 4 were used to amplify serially diluted chromosomal DNA prepared from the E. coli reference strains for O174 (2531-54) and O177 (E40874-85). Positive PCR results were obtained from as little as 1 pg per μl of DNA for either of the type strains.
wzx-specific primer pairs wl-4397/wl-4398/wl-4399/wl-4400 (O174) and wl-4393/wl-4394/wl-4395/wl-4396 (O177) were used for the detection of serogroup O174 and O177 reference strains from spiked samples of pork and water. By this, as few as 103 CFU per g was detected in samples that were examined directly, and 0.1 CFU per g was detected in samples that were further incubated at 37°C for 12 h.
The O-antigen gene-specific PCR assays developed in this study were found to be highly suitable for the detection of E. coli strains belonging to the novel serogroups O174 and O177. E. coli groups O174 and O177 were shown to comprise Shiga toxin-producing E. coli, EHEC, and atypical EPEC strains which were isolated from diseased humans, animals, and food in different parts of the world. At present, the O-antigen-specific PCR is the only method generally applicable for screening of E. coli O174 and O177 strains, since other diagnostic tools for their detection are not yet commercially available. Detection of O174 and O177 strains by PCR could be the method of choice for epidemiological investigations of outbreaks and sporadic infections and for the investigation of animals and the food chain. Last but not least, this method could be useful to investigate the association of certain O174 strains with specific animal hosts for the development of possible prevention strategies against the spread of these pathogens.
Acknowledgments
This work was supported by the Chinese National Science Fund for Distinguished Young Scholars (grant number 30125001), NSFC General Program (grant numbers 30270029, 30370339, and 30370023), and the 863 Program (grant number 2002AA2Z2051) and by funding from the Science and Technology Committee of Tianjin City (grant number 013181711) to L.F. and L.W. L.B. and G.K. were supported by funds from the European Commission project “Attaching and Effacing Escherichia coli Infections” (reference number QLK2-CT-2000-00600). L.L. was supported from Brazil by the “Fundação Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES)” (grant number BEX 1686/03-8) and by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (grant number 03/01823-9).
We are grateful to H. Steinrück, BfR, Berlin, Germany, for helpful discussions.
REFERENCES
- 1.Alexander, D. C., and M. A. Valvano. 1994. Role of the rfe gene in the biosynthesis of the Escherichia coli O7-specific lipopolysaccharide and other O-specific polysaccharides containing N-acetylglucosamine. J. Bacteriol. 176:7079-7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bastin, D. A., and P. R. Reeves. 1995. Sequence and analysis of the O antigen gene (rfb) cluster of Escherichia coli O111. Gene 164:17-23. [DOI] [PubMed] [Google Scholar]
- 3.Beutin, L., G. Krause, S. Zimmermann, S. Kaulfuss, and K. Gleier. 2004. Characterization of Shiga toxin-producing Escherichia coli strains isolated from human patients in Germany over a 3-year period. J. Clin. Microbiol. 42:1099-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beutin, L., E. Strauch, S. Zimmermann, S. Kaulfuss, C. Schaudinn, A. Mannel, and H. R. Gelderblom. 2005. Genetical and functional investigation of fliC genes encoding flagellar serotype H4 in wildtype strains of Escherichia coli and in a laboratory E. coli K-12 strain expressing flagellar antigen type H48. BMC Microbiol. 5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beutin, L., J. Tao, L. Feng, G. Krause, S. Zimmermann, K. Gleier, Q. Xia, and L. Wang. 2005. Sequence analysis of the Escherichia coli O15 antigen gene cluster and development of a PCR assay for rapid detection of intestinal and extraintestinal pathogenic E. coli O15 strains. J. Clin. Microbiol. 43:703-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beutin, L., S. Zimmermann, and K. Gleier. 1996. Rapid detection and isolation of Shiga-like toxin (verocytotoxin)-producing Escherichia coli by direct testing of individual enterohemolytic colonies from washed sheep blood agar plates in the VTEC-RPLA assay. J. Clin. Microbiol. 34:2812-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanco, M., J. E. Blanco, A. Mora, G. Dahbi, M. P. Alonso, E. A. González, M. I. Bernárdez, and J. Blanco. 2004. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from cattle in Spain and identification of a new intimin variant gene (eae-ξ). J. Clin. Microbiol. 42:645-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanco, M., N. L. Padola, A. Kruger, M. E. Sanz, J. E. Blanco, E. A. Gonzalez, G. Dahbi, A. Mora, M. I. Bernardez, A. I. Etcheverria, G. H. Arroyo, P. M. Lucchesi, A. E. Parma, and J. Blanco. 2004. Virulence genes and intimin types of Shiga-toxin-producing Escherichia coli isolated from cattle and beef products in Argentina. Int. Microbiol. 7:269-276. [PubMed] [Google Scholar]
- 9.D'Souza, J. M., L. Wang, and P. R. Reeves. 2002. Sequence of the Escherichia coli O26 antigen gene cluster and identification of O26 specific genes. Gene 297:123-127. [DOI] [PubMed] [Google Scholar]
- 10.Ewing, W. H. 1986. Edwards and Ewing's identification of the Enterobacteriaceae, 4th ed. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 11.Feng, L., S. N. Senchenkova, J. Yang, A. S. Shashkov, J. Tao, H. Guo, G. Zhao, Y. A. Knirel, P. Reeves, and L. Wang. 2004. Structural and genetic characterization of the Shigella boydii type 13 O antigen. J. Bacteriol. 182:383-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng, L., W. Wang, J. Tao, H. Guo, G. Krause, L. Beutin, and L. Wang. 2004. Identification of Escherichia coli O114 O-antigen gene cluster and development of an O114 serogroup-specific PCR assay. J. Clin. Microbiol. 42:3799-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franke, J., S. Franke, H. Schmidt, A. Schwarzkopf, L. H. Wieler, G. Baljer, L. Beutin, and H. Karch. 1994. Nucleotide sequence analysis of enteropathogenic Escherichia coli (EPEC) adherence factor probe and development of PCR for rapid detection of EPEC harboring virulence plasmids. J. Clin. Microbiol. 32:2460-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo, H., L. Feng, J. Tao, C. Zhang, and L. Wang. 2004. Identification of Escherichia coli O172 O-antigen gene cluster and development of a serogroup-specific PCR assay. J. Appl. Microbiol. 97:181-190. [DOI] [PubMed] [Google Scholar]
- 15.Hobbs, M., and P. R. Reeves. 1994. The JUMPstart sequence: a 39 bp element common to several polysaccharide gene clusters. Mol. Microbiol. 12:855-856. [DOI] [PubMed] [Google Scholar]
- 16.Kneidinger, B., K. O'Riordan, J. Li, J. Brisson, J. Lee, and J. Lam. 2003. Three highly conserved proteins catalyze the conversion of UDP-N-acetyl-D-glucosamine to precursors for the biosynthesis of O antigen in Pseudomonas aeruginosa O11 and capsule in Staphylococcus aureus type 5. Implications for the UDP-N-acetyl-L-fucosamine biosynthetic pathway. J. Biol. Chem. 278:3615-3627. [DOI] [PubMed] [Google Scholar]
- 17.Lior, H. 1994. Classification of Escherichia coli, p. 31-72. In C. L. Gyles (ed.), Escherichia coli in domestic animals and humans. CAB International, Wallingford, United Kingdom.
- 18.Marolda, C. L., and M. A. Valvano. 1998. Promoter region of the Escherichia coli O7-specific lipopolysaccharide gene cluster: structural and functional characterization of an upstream untranslated mRNA sequence. J. Bacteriol. 180:3070-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meichtri, L., E. Miliwebsky, A. Gioffre, I. Chinen, A. Baschkier, G. Chillemi, B. E. Guth, M. O. Masana, A. Cataldi, H. R. Rodriguez, and M. Rivas. 2004. Shiga toxin-producing Escherichia coli in healthy young beef steers from Argentina: prevalence and virulence properties. Int. J. Food Microbiol. 96:189-198. [DOI] [PubMed] [Google Scholar]
- 20.Orskov, F., and I. Orskov. 1984. Serotyping of Escherichia coli, vol. 14. Academic Press, London, United Kingdom.
- 21.Padola, N. L., M. E. Sanz, J. E. Blanco, M. Blanco, J. Blanco, A. I. Etcheverria, G. H. Arroyo, M. A. Usera, and A. E. Parma. 2004. Serotypes and virulence genes of bovine Shigatoxigenic Escherichia coli (STEC) isolated from a feedlot in Argentina. Vet. Microbiol. 100:3-9. [DOI] [PubMed] [Google Scholar]
- 22.Perelle, S., F. Dilasser, J. Grout, and P. Fach. 2002. Identification of the O-antigen biosynthesis genes of Escherichia coli O91 and development of a O91 PCR serotyping test. J. Appl. Microbiol. 93:758-764. [DOI] [PubMed] [Google Scholar]
- 23.Scheutz, F., T. Cheasty, D. Woodward, and H. R. Smith. 2004. Designation of O174 and O175 to temporary O groups OX3 and OX7, and six new E. coli O groups that include verocytotoxin-producing E. coli (VTEC): O176, O177, O178, O179, O180 and O181. APMIS 112:569-584. [DOI] [PubMed] [Google Scholar]
- 24.Trabulsi, L. R., R. Keller, and T. A. T. Gomes. 2002. Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 8:508-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urdahl, A. M., L. Beutin, E. Skjerve, S. Zimmermann, and Y. Wasteson. 2003. Animal host associated differences in Shiga toxin-producing Escherichia coli isolated from sheep and cattle on the same farm. J. Appl. Microbiol. 95:92-101. [DOI] [PubMed] [Google Scholar]
- 26.Vernozy-Rozand, C., M. P. Montet, Y. Bertin, F. Trably, J. P. Girardeau, C. Martin, V. Livrelli, and L. Beutin. 2004. Serotyping, stx2 subtyping, and characterization of the locus of enterocyte effacement island of Shiga toxin-producing Escherichia coli and E. coli O157:H7 strains isolated from the environment in France. Appl. Environ. Microbiol. 70:2556-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, L., C. E. Briggs, D. Rothemund, P. Fratamico, J. B. Luchansky, and P. R. Reeves. 2001. Sequence of the E. coli O104 antigen gene cluster and identification of O104 specific genes. Gene 270:231-236. [DOI] [PubMed] [Google Scholar]
- 28.Wang, L., H. Curd, W. Qu, and P. R. Reeves. 1998. Sequencing of Escherichia coli O111 O-antigen gene cluster and identification of O111-specific genes. J. Clin. Microbiol. 36:3182-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, L., W. Qu, and P. R. Reeves. 2001. Sequence analysis of four Shigella boydii O-antigen loci: implication for Escherichia coli and Shigella relationships. Infect. Immun. 69:6923-6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, L., and P. R. Reeves. 1998. Organization of Escherichia coli O157 O-antigen gene cluster and identification of its specific genes. Infect. Immun. 66:3545-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]