Abstract
A total of 66 isolates of Salmonella javiana isolated from food, food handlers, and patrons that were epidemiologically linked to an outbreak of gastroenteritis were analyzed by pulsed-field gel electrophoresis. Analysis with restriction endonucleases XbaI and SpeI supported the epidemiologic association and suggested a pathway of transmission among food, food handlers, and patrons.
During the months of June and July 1996, a sudden increase in the number of cases involving Salmonella group D was observed in a suburb west of Boston, Mass. Investigation by state public health epidemiologists linked these cases of gastroenteritis to a single restaurant. Stool samples and/or subcultures from ill patrons and food handlers, as well as a portion of a chicken sandwich that was partially consumed by one of the ill patrons, were submitted to the Massachusetts State Laboratory Institute for culture, identification or confirmation, serotyping, and strain typing. Salmonella javiana was isolated from 66 of the 95 clinical specimens submitted, representing 55 patrons and 11 restaurant employees. In addition, S. javiana was isolated from the leftover chicken sandwich.
In the Commonwealth of Massachusetts, S. javiana is a rarely encountered organism. The average reported incidence of S. javiana for the 5 years preceding this outbreak was 15.2/year. In the 3-week period during the outbreak, the total number of S. javiana isolates received increased dramatically to 75.
Laboratory and epidemiological methods used previously to establish chains of transmission in food-borne infections have been limited to serotyping (subtyping) and classical epidemiological case studies, all of which are not sufficiently sensitive to distinguishing individual strains. The application of pulsed-field gel electrophoresis (PFGE) provides precise information that can be used to accept or reject epidemiologic associations to a high degree of confidence (4–6). PFGE is believed to possess a discriminating capacity greater than those of ribotyping and other probe-based restriction fragment length polymorphism methods (7). We report here the first use of PFGE to confirm the chain of transmission of S. javiana from restaurant food handlers or food to patrons.
Bacterial strains.
Sixty-seven isolates of S. javiana were isolated from specimens from 55 ill restaurant patrons, 11 food handlers, and one chicken sandwich that had been partially consumed by one of the ill patrons. The chicken sandwich isolate was used as a control strain that was included on each gel. Included were 13 non-outbreak-related S. javiana strains which were temporally and/or geographically different from that involved in the outbreak. A slight modification of standard serotyping methods was used to confirm the positive isolates as S. javiana (2). All isolates were stored on nutrient agar slants at room temperature.
DNA preparation.
Intact genomic DNA was prepared by a modification of the method described by Barrett et al. (1). Briefly, single-colony isolates were selected from purity blood agar plates and grown at 37°C in Luria-Bertani nutrient broth to mid-log phase. One milliliter of suspension was harvested, centrifuged, washed with 1 ml of SE wash buffer, and then resuspended in 0.5 ml of SE wash buffer. An equal volume of 1.2% plug agarose (1.2% Bio-Rad chromosomal grade agarose, 10 mM Tris-HCl [pH 7.4], 0.1 mM EDTA [pH 8.0]) was added to the suspension, and the mixture was injected into molds to form plugs. The agarose plugs were incubated in lysis solution containing 1.0 mg of proteinase K per ml in a 50°C water bath overnight. The proteinase K was then removed from the plugs by four washes in 5 ml of Tris-EDTA buffer. DNA plugs can then be stored in Tris-EDTA buffer at 4°C.
Restriction endonuclease digestion and PFGE.
A portion (1 by 3.5 by 5 mm) of each plug was cut and washed in restriction buffer. The slices were digested overnight with selected restriction enzymes. The enzyme concentrations, buffers, and incubation periods used were those recommended by the manufacturer. Selection of restriction enzymes was based on the recognition sites of the enzymes XbaI (5′-TCTAGA-3′) and SpeI (5′-ACTAGT-3′) (New England Biolabs, Inc., Beverly, Mass.) and the 50 to 54% G+C content of previously reported Salmonella species (3). Restriction fragments of DNA were separated by PFGE with the CHEF-Mapper apparatus (Bio-Rad) through a 1.0% gel (1.0% Bio-Rad pulsed-field-certified agarose, 0.5× Tris-borate-EDTA buffer). Electrophoresis was performed over a period of 22 h (initial switch time, 2.16 s; final switch time, 54.17 s) at 6 V/cm and 14°C in 0.5× Tris-borate-EDTA buffer. A lambda ladder standard (Bio-Rad) was used as a molecular size marker. The gels were stained with ethidium bromide and then illuminated on a UV transilluminator (302 nm; Bio-Rad). Results of the XbaI restriction were confirmed by using the second enzyme SpeI. Isolates were considered to be genetically indistinguishable only if their restriction pattern profiles had the same numbers of bands and the corresponding bands were of the same apparent sizes when both restriction enzymes were used.
DNA analysis.
DNA fragment patterns were electronically scanned directly into a computer via a charge-coupled device video camera directly connected to a personal computer. Once the images had been acquired, they were compared both visually and with the assistance of analysis software (Molecular Analyst Fingerprinting Plus by Bio-Rad) to determine relatedness.
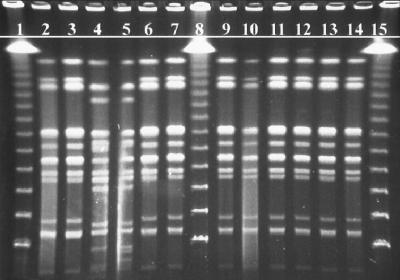
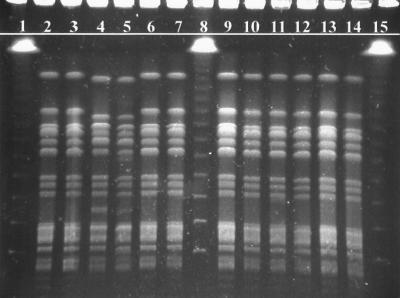
Results. Restriction profiles of the isolates’ genomic DNA by XbaI restriction digestion yielded 10 to 15 fragments ranging in size from ∼40 to ∼550 kbp (Fig. 1). Isolates of S. javiana which were epidemiologically linked to the outbreak (Fig. 1, lanes 3, 6, 7, 9, 10, 11, 12, 13, and 14) and the outbreak control strain (Fig. 1, lane 2) all demonstrated patterns with identical numbers and sizes of fragments. Samples not epidemiologically associated with the outbreak due to separation by time and/or geography (Fig. 1, lanes 4 and 5) demonstrated significant multiband differences. Lambda ladder standards (Fig. 1, lanes 1, 8, and 15) provided approximate molecular size references. Restriction profiles of the isolates’ genomic DNA by SpeI restriction digestion yielded 13 to 15 fragments also ranging in size from ∼40 to ∼550 kbp (Fig. 2). The isolates were arranged in the same order as in Fig. 1. The results of the XbaI restriction digestion were confirmed by the second restriction digestion with SpeI.
FIG. 1.
PFGE fingerprints of S. javiana isolates from a suspected outbreak after digestion with XbaI. Lanes: 1, 8, and 15, lambda molecular size marker ladder (Bio-Rad); 4 and 5, unrelated S. javiana isolates; 2, isolate from the leftover chicken sandwich (control strain); 3 and 11 to 14, representative sampling of results for all patrons; 6, 7, 9 and 10, isolates from food handlers.
FIG. 2.
PFGE fingerprints of S. javiana isolates from a suspected outbreak after digestion with SpeI. Lanes: 1, 8, and 15, lambda molecular size marker ladder (Bio-Rad); 4 and 5, unrelated S. javiana isolates; 2, isolate from the leftover chicken sandwich (control strain); 3 and 11 to 14, representative sampling of results for all patrons; 6, 7, 9, and 10, isolates from food handlers.
Discussion.
This report describes an outbreak investigation whereby identification of the implicated organism by PFGE analysis of DNA content allowed us to trace a genetically indistinguishable strain of S. javiana from food handlers to leftover food and finally to patrons who were epidemiologically linked to the outbreak.
As part of a concurrent investigation, the Massachusetts Department of Public Health (MDPH) Epidemiologic Division obtained case report forms from the MDPH laboratory. In addition, public health nurses assisted in acquiring completed case surveys from all residents with culture-confirmed cases of S. javiana. During this time, the MDPH Food and Drug Division inspected the restaurant and interviewed its employees. When the management of the restaurant was made aware of the large number of S. javiana cases associated with their establishment, they voluntarily closed the facility and proceeded to have the restaurant sanitized and discarded all remaining food. Stool specimens were required from all employees before they could return to work. The local board of health organized a personal hygiene class for all employees. After a final inspection of the facility, resulting in a satisfactory rating, the restaurant subsequently reopened with no further incidence of the disease.
All positive cases associated with the outbreak demonstrated identical pattern profiles through restriction digestion analysis utilizing two distinct endonucleases (XbaI and SpeI). Isolates not epidemiologically associated with the outbreak demonstrated pattern profiles that were conclusively different from the outbreak patterns.
Transmission of enteric pathogens in a restaurant environment from food or a food handler to a patron represents a breakdown in accepted food handling practices. Typically, investigations of such food-borne or similar situational outbreaks are carried out by epidemiological studies and laboratory identification of the implicated organism to the subtype level through serotyping. This study demonstrates that PFGE analysis represents a powerful and very valuable epidemiologic tool for the characterization of S. javiana and for investigation of food-borne outbreaks.
REFERENCES
- 1.Barrett T J, Lior H, Green J H, Khakharia R, Wells J G, Bell B P, Greene K D, Lewis J, Griffin P M. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J Clin Microbiol. 1994;12:3013–3017. doi: 10.1128/jcm.32.12.3013-3017.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ewing W H. Identification of Enterobacteriaceae. 4th ed. New York, N.Y: Elsevier Science Publishing Co., Inc.; 1986. [Google Scholar]
- 3.Jones D, Sneath P H A. Genetic transfer and bacterial taxonomy. Bacteriol Rev. 1970;34:40–81. doi: 10.1128/br.34.1.40-81.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwartz D C, Cantor C R. Separation of yeast chromosome-sized DNAs by pulsed-field gradient gel electrophoresis. Cell. 1984;37:67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- 5.Smith C L, Cantor C R. Purification, specific fragmentation, and separation of large DNA molecules. Methods Enzymol. 1987;155:449–467. doi: 10.1016/0076-6879(87)55030-3. [DOI] [PubMed] [Google Scholar]
- 6.Smith C L, Klco S R, Cantor C R. Pulsed-field gel electrophoresis and the technology of large DNA molecules. In: Davies K, editor. Genome analysis: a practical approach. Oxford, England: IRL Press; 1988. pp. 41–72. [Google Scholar]
- 7.Swaminathan B, Matar G M. Molecular typing methods. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: American Society for Microbiology; 1993. pp. 26–50. [Google Scholar]