Abstract
Use of a common set of human immunodeficiency virus type 1 (HIV-1) RNA standards eliminated differences among absolute HIV-1 RNA copy number estimates made with three commercially available assays. The relative changes in the viral RNA levels determined by the commercial assays were similar and were unaffected by the use of a common set of standards.
Quantitation of human immunodeficiency virus type 1 (HIV-1) RNA is being used to manage HIV-1-infected patients, to approve antiretroviral agents for licensure, and as entry criteria, endpoints, and change points for AIDS clinical trials (1, 2, 4, 9, 13, 19). Many of these uses rely on relative changes between measurements and increasingly on absolute viral RNA values (2). The differences between the values obtained by laboratories and kits with spiked samples were significantly reduced through the use of a common set of standards (20). However, spiked plasma samples do not reflect the actual variation in viral composition and in the plasma matrix. Previous assessments of kit differences in the measurement of patient RNA copy number have been limited by small sample sizes and the use of kit assay standards alone (12, 14, 17). The objective of this study was to assess the feasibility of reducing kit-related differences in HIV-1 RNA copy number estimates by utilizing a common set of external standards.
These analyses included two sets of data. The first analysis consisted of estimates from a three-way comparison of the Chiron Enhanced Sensitivity branched DNA (ES bDNA) assay (Chiron Corporation, Inc., Emeryville, Calif.), the reverse transcription (RT)-PCR amplification Monitor assay (Roche Molecular Systems, Branchburg, N.J.), and the Organon Teknika Corporation (OTC) NASBA-QT (Advanced BioScience Laboratories, Incorporated, Kensington, Md.). In the three-way comparison, 90 specimens from 22 pregnant women coenrolled in the Women and Infants Transmission Study (15) and Pediatric AIDS Clinical Trials Group protocol 076 were selected (5). The second analysis consisted of estimates from a two-way comparison of the ES bDNA and RT-PCR assays. This two-way comparison of ES bDNA and RT-PCR assay results was based on 912 specimens from 479 women enrolled in Pediatric AIDS Clinical Trials Group protocol 076. In both analyses, absolute levels and changes in RNA level from baseline to labor and delivery were compared among kits. Determinations of HIV RNA in all plasma specimens in both sets of comparisons were performed by the manufacturers themselves to minimize laboratory variation and to focus on the contribution of the kits to variation.
Specimens were assayed in accordance with each manufacturer’s instructions (7, 10, 18) by using both kit and National Institute of Allergy and Infectious Diseases (NIAID)-sponsored virology quality assurance (VQA) standards. The VQA standards have been previously described (8, 20). They consist of supernatants from HIV-1-infected patient cultures spiked into seronegative plasma, which was then characterized by multiple parameters for determination of absolute HIV-1 RNA copy numbers. The RT-PCR assay specimens were pretreated with heparinase (16). Specimens analyzed by Chiron were tested with the ES bDNA assay (7). Depending on the available volume, either 1.0 or 0.5 ml of plasma was assayed by this procedure. For the NASBA-QT assay, 10-fold-diluted calibrators were used to increase sensitivity (17); the sensitivity was similar to that of the newer NucliSens version of the assay. All samples from the same patient were assessed by batch assay to eliminate interassay variability and focus only on differences among kits.
For this analysis, the limit of assay sensitivity and the lowest observed RNA copy number were, respectively, 200 and 466 copies/ml for the RT-PCR assay of 0.2-ml samples, 1,000 and 1,100 copies/ml for the NASBA-QT assay of 0.1-ml samples, and 500 and 561 copies/ml for the ES-bDNA assay of 1.0-ml samples. Absolute copy numbers for pairwise comparisons were based on total specimens above the limit of detection for both assays, while relative-change pairwise comparisons were based on patients with estimates above the limit of detection for both specimens with both assays.
Results were compared among kits both before and after adjustment to the VQA standards. Adjustment for all kits was accomplished by using regressions of the estimated RNA concentration on the nominal log10 concentration for the VQA standards. The mean, median, and number of values above or below the assay cutoffs were calculated. Paired t tests were used to test the null hypothesis that the average difference in estimated RNA concentration between kits was zero.
The viral RNA copy numbers estimated by the kits were similar when either kit-based or VQA-adjusted standards were used (Table 1). However, differences in estimated copy numbers between kits were statistically significant at P < 0.001 (Table 2). The VQA standards for the group analysis eliminated differences among all of the assays. The results of the larger comparison between the RT-PCR and ES bDNA assays were similar to the results of the smaller three-way comparison.
TABLE 1.
Estimates of log10 RNA concentration for both the kit-adjusted and VQA-adjusted standards
| Standard and assay | No.a | Minimum | Q1b | Median | Q3c | Maximum |
|---|---|---|---|---|---|---|
| Kit-based standard | ||||||
| Chiron ES bDNA | 52 | 2.75 | 3.24 | 3.49 | 3.99 | 4.85 |
| OTC NASBA-QT | 49 | 3.04 | 3.76 | 4.00 | 4.46 | 5.32 |
| RT-PCR | 64 | 2.67 | 3.45 | 3.83 | 4.17 | 5.57 |
| VQA-adjusted standard | ||||||
| Chiron ES bDNA | 52 | 2.95 | 3.51 | 3.86 | 4.26 | 5.16 |
| OTC NASBA-QT | 49 | 2.64 | 3.47 | 3.88 | 4.14 | 5.45 |
| RT-PCR | 64 | 2.50 | 3.31 | 3.70 | 4.19 | 5.69 |
Number of specimens.
Q1, 25th percentile.
Q3, 75th percentile.
TABLE 2.
Differences in log10 RNA concentration among estimates based on VQA or kit standards for determining nominal copy numbera
| Kit comparison | Kit standard-based difference
|
VQA standard-based difference
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Median | SD | P value | No.b | Mean | Median | SD | P value | No.b | |
| Monitor vs ES bDNA | 0.38 | 0.37 | 0.28 | <0.001 | 42 | 0.04 | 0.03 | 0.32 | 0.42 | 42 |
| Monitor vs NASBA-QT | −0.18 | −0.15 | 0.29 | <0.001 | 45 | 0.02 | 0.03 | 0.39 | 0.76 | 45 |
| NASBA-QT vs ES bDNA | 0.57 | 0.56 | 0.39 | <0.001 | 37 | 0.01 | 0.10 | 0.48 | 0.91 | 37 |
| Monitor vs ES bDNA (all) | 0.32 | 0.36 | 0.36 | <0.001 | 311 | 0.04 | 0.04 | 0.37 | 0.11 | 293 |
| Monitor vs ES bDNA (placebo) | 0.31 | 0.34 | 0.38 | <0.001 | 159 | 0.03 | 0.03 | 0.43 | 0.37 | 152 |
| Monitor vs ES bDNA (zidovudine) | 0.32 | 0.37 | 0.33 | <0.001 | 152 | 0.04 | 0.04 | 0.30 | 0.13 | 141 |
The upper section represents the subset of samples used for a three-way comparison, while the lower section is the entire data set obtained with a two-way comparison. The P value is that obtained by testing the null hypothesis that the average difference was zero.
Number of specimens.
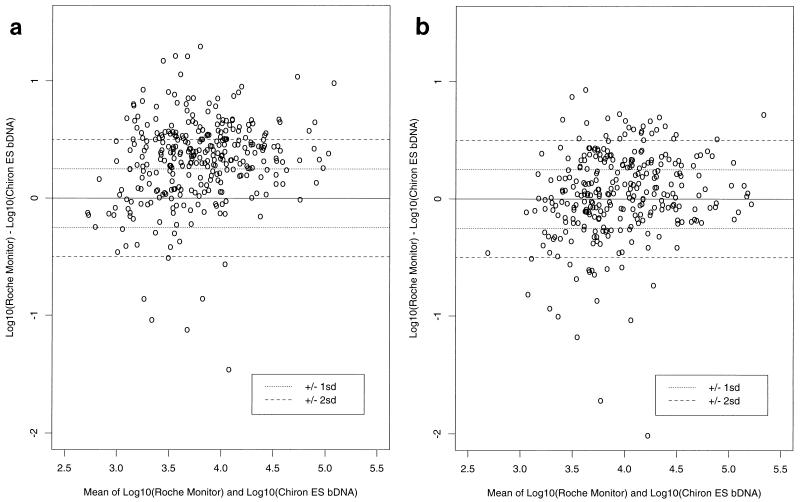
For all kits, the standard deviation (SD) was determined to be 0.18 log10 HIV-1 RNA copies per ml based on the performance of the VQA standard with a copy number of 1,500. Thus, the 95% confidence limits for the difference between two estimates was equivalent to ±0.5 log10. Although there was individual-subject variability in the HIV-1 RNA copy numbers estimated by the kits (Fig. 1a and b), the use of VQA standards reduced the number of patients within ±2 SD from 29.6 to 14.6% and increased the number of patients ±1 SD from 30.5 to 53.9% (Table 3).
FIG. 1.
Difference between log10 estimated RNA concentrations from the RT-PCR and Chiron ES bDNA assays plotted against the log-transformed mean of the two estimates. A positive difference indicates that the RT-PCR copy number was higher than the Chiron ES bDNA copy number for that patient. a, kit-based copy number estimates; b, VQA-adjusted copy number estimates.
TABLE 3.
Frequency and percentage of points falling in each of the SD categories for the differences between log10 estimated RNA concentrations from the RT-PCR and Chiron ES bDNA assays
| SD category | Kit-based data
|
VQA-adjusted data
|
||
|---|---|---|---|---|
| Frequency | % | Frequency | % | |
| >+2 | 85 | 27.3 | 25 | 8.5 |
| >+1, ≤+2 | 116 | 37.3 | 60 | 20.5 |
| ≥−1, ≤+1 | 95 | 30.5 | 158 | 53.9 |
| ≥−2, <−1 | 8 | 2.6 | 32 | 10.9 |
| <−2 | 7 | 2.3 | 18 | 6.1 |
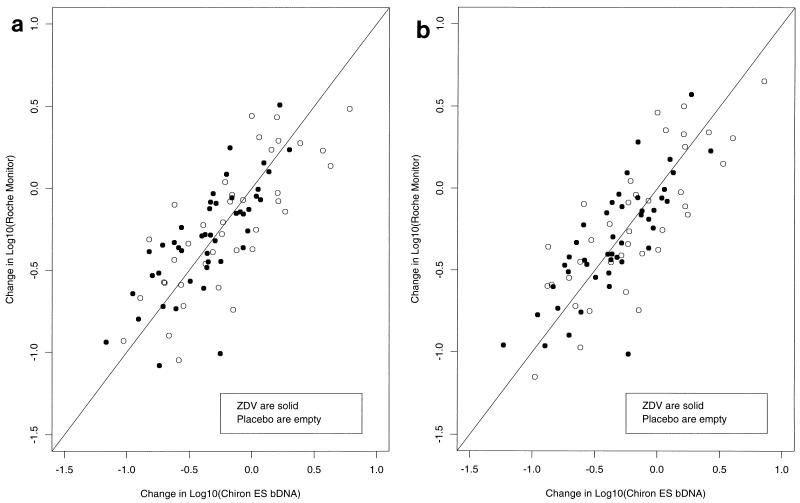
No statistically significant differences between kits in the mean relative change in HIV-1 RNA level were detected when either kit-based or VQA-adjusted estimates of RNA copy number were used (Table 4). Relative changes in viral RNA level for the individual subjects were strongly correlated between the RT-PCR and ES bDNA assays (Fig. 2a and b). Changes in opposite directions were identified in 12 (14%) of 83 patients. However, the difference in relative change between kits varied widely among the patients. The SD of the difference between relative changes for the RT-PCR and ES bDNA kits was 0.26, and the 10th and 90th percentiles of the differences were −0.30 and +0.30 log10. Thus, 20% of the time, there was at least a twofold difference between the relative change in the results obtained with one kit and the relative change in those obtained with the other.
TABLE 4.
Differences for within-individual log10 changes in RNA based on VQA or kit standards for determining nominal copy numbera
| Kit comparison | Kit standard-based difference
|
VQA standard-based difference
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Minimum | Mean | Maximum | P value | No.b | Minimum | Mean | Maximum | P value | No.b | |
| NASBA-QT vs Monitor | −0.33 | 0.01 | 0.49 | 0.90 | 11 | −0.48 | −0.02 | 0.58 | 0.86 | 11 |
| NASBA-QT vs ES bDNA | −0.87 | −0.03 | 0.33 | 0.77 | 11 | −0.44 | −0.001 | 0.40 | 0.99 | 11 |
| Monitor vs ES bDNA | −0.66 | −0.02 | 0.42 | 0.83 | 11 | −0.54 | −0.02 | 0.44 | 0.87 | 11 |
| Monitor vs ES bDNA (all) | −0.52 | −0.01 | 0.76 | 0.78 | 83 | −0.52 | −0.01 | 0.78 | 0.84 | 82 |
| Monitor vs ES bDNA (placebo) | −0.52 | 0.04 | 0.59 | 0.41 | 38 | −0.52 | 0.03 | 0.60 | 0.58 | 37 |
| Monitor vs ES bDNA (zidovudine) | −0.43 | −0.05 | 0.76 | 0.19 | 45 | −0.43 | −0.03 | 0.78 | 0.35 | 45 |
The upper section represents the subset of samples used for a three-way comparison, while the lower section is the entire data set obtained with the two-way comparison. The P value is that obtained by testing the null hypothesis that the average difference was zero.
Number of patients.
FIG. 2.
Change in log10 RNA concentration as measured by RT-PCR assay versus change in log10 RNA concentration as measured by the Chiron ES bDNA assay. a, kit-based copy number estimates; b, VQA-adjusted copy number estimates. ZDV, zidovudine.
In this study, each assay was performed by the respective manufacturer; thus, interlaboratory difference within kits was not assessed. The kit differences observed suggest that accurate determination of an absolute HIV-1 RNA copy number in patient plasma will be significantly affected by the kit used for the assessment. It was possible, however, to make the absolute copy numbers among the kits equivalent if they were adjusted to a common set of viral RNA standards. A common set of standards was unnecessary for assessment of the relative change in patient plasma RNA levels, since these values did not differ significantly among the three assays. It is possible that greater or lesser kit-related differences in relative change may occur with more potent antiviral therapy, since the magnitude of the changes in virus load in this study were modest (+0.76 to −0.66 log10).
In summary, the use of absolute viral levels is problematic for individual patient management because the assessment of the absolute viral RNA level is dependent upon the test method. However, the ability to adjust for kit differences in absolute copy number by using external standards has important implications for cross-protocol study analysis and possibly for individual patient management when kits are changed. Relative change in viral RNA level can be used for therapeutic management without adjustment. Given the lack of a universal standard, longitudinal assessment of HIV-1 RNA level should be done with the same manufacturer’s assay kit.
Acknowledgments
We acknowledge Joe Romano, Robert Kuzma, and Helen Payne for technical assistance and Shirley Traite and Michael Posner for computer programming.
This work was supported under NIAID contracts NO-AI-35172 (D.B. and S.L.), NO1-AI-35161 (S.L.), and NO1-AI-95030 (D.E.S.) and NIAID grants AI-27664, AI-30731 (R.W.C.), AI-25883-09 (G.M.), AI-34840 (C.H.), and AI-34858 (G.H.).
REFERENCES
- 1.BHIVA Guidelines Co-ordinating Committee. British HIV association guidelines for antiretroviral treatment of HIV seropositive individuals. Lancet. 1997;349:1086–1092. [PubMed] [Google Scholar]
- 2.Carpenter C C J, Fischl M A, Hammer S M, Hirsch M S, Jacobsen D M, Katzenstein D A, Montaner J S G, Richman D D, Saag M S, Schooley R T, Thompson M A, Vella S, Yeni P G, Volberding P A for the International AIDS Society—USA. Antiretroviral therapy for HIV-1 infection in 1996. JAMA. 1996;276:146–154. [PubMed] [Google Scholar]
- 3.Connor E M, Sperling R S, Gelber R, Kiselev P, Scott G, O’Sullivan M J, VanDyke R, Bey M, Shearer W, Jacobson R L, Jimenez E, O’Neill E, Bazin B, Delfraissy J, Culnane M, Coombs R, Elkins M, Moye J, Stratton P, Balsley J for The Pediatric AIDS Clinical Trials Group Protocol 076 Study Group. Reduction of maternal-infant transmission of human immunodeficiency virus type 1 with zidovudine treatment. N Engl J Med. 1994;331:1173–1180. doi: 10.1056/NEJM199411033311801. [DOI] [PubMed] [Google Scholar]
- 4.Coombs R W. HIV-1 burden as a marker of disease progression and clinical response to therapy in AIDS. Clin Lab Med. 1994;14:301–311. [PubMed] [Google Scholar]
- 5.Garcia F, Vidal V, Gatell J M, Miro J M, Sorino A, Pumarola T. Viral load in asymptomatic patients with CD4+ lymphocyte counts above 500 × 106/l. AIDS. 1997;11:53–57. doi: 10.1097/00002030-199701000-00008. [DOI] [PubMed] [Google Scholar]
- 6.Holodniy M, Mole L, Winters M, Merigan T C. Diurnal and short-term stability of HIV virus load as measured by gene amplification. J Acquired Immune Defic Syndr. 1994;7:363–368. [PubMed] [Google Scholar]
- 7.Kern D, Collins M, Fultz T, Detmer J, Hamren S, Peterkin J J, Sheridan P, Urdea M, White R, Yeghaizarian T, Todd J. An enhanced-sensitivity branched-DNA assay for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996;34:3196–3202. doi: 10.1128/jcm.34.12.3196-3202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin H J, Myers L E, Yen-Lieberman B, Hollinger F B, Henrard D, Hooper C J, Kokka R, Kwok S, Rasheed S, Vahey M, Winters M A, McQuay L J, Nara P L, Reichelderfer P, Coombs R W, Jackson J B. Multicenter evaluation of methods for the quantitation of plasma HIV-1 RNA. J Infect Dis. 1994;170:553–562. doi: 10.1093/infdis/170.3.553. [DOI] [PubMed] [Google Scholar]
- 9.Merigan, T. 1995. Individualization of therapy using viral markers. J. Acquired Immune Defic. Syndr. Hum. Retrovirol. 10(Suppl. 1):S41–S46. [PubMed]
- 10.Mulder J, McKinney N, Christopherson C, Sninsky J, Greenfield L, Kwok S. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J Clin Microbiol. 1994;32:292–300. doi: 10.1128/jcm.32.2.292-300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paxton W B, Coombs R W, McElrath M J, Keefer M C, Hughes J, Sinangil F, Chernoff D, Demeter L, Williams B, Corey L for The National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. Longitudinal analysis of quantitative virologic measures in human immunodeficiency virus-infected subjects with >400 CD4 lymphocytes: implications for applying measurements to individual patients. J Infect Dis. 1997;175:247–254. doi: 10.1093/infdis/175.2.247. [DOI] [PubMed] [Google Scholar]
- 12.Revets H, Marissens D, De Wit S, Lacor P, Clumeck N, Lauwers S, Zissis G. Comparative evaluation of NASBA HIV-1 RNA QT, AMPLICOR-HIV Monitor, and QUANTIPLEX HIV RNA assay, three methods for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996;34:1058–1064. doi: 10.1128/jcm.34.5.1058-1064.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saag, M. S., M. Holodniy, D. R. Kuritzkes, W. A. O’Brien, R. Coombs, M. E. Poscher, D. M. Jacobson, G. M. Shaw, D. D. Richman, and P. A. Volberding. HIV viral load markers in clinical practice. Nat. Med. 2:625–629. [DOI] [PubMed]
- 14.Schuurman R, Descamps D, Weverling G J, Kaye S, Tijnagel J, Williams I, van Leeuwen R, Tedder R, Boucher C A B, Brun-Vezinet F, Loveday C. Multicenter comparison of three commercial methods for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996;34:3016–3022. doi: 10.1128/jcm.34.12.3016-3022.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheon A R, Fox H E, Rich K C, Stratton P, Diaz C, Tuomala R, Mendez H, Carrington J, Alexander G. The Women and Infants Transmission Study (WITS) of maternal-infant HIV transmission: study design, methods, and baseline data. J Women’s Health. 1996;5:69–78. [Google Scholar]
- 16.Sperling R S, Shapiro D E, Coombs R W, Todd J A, Herman S A, McSherry G D, O’Sullivan M J, Van Dyke R B, Jimenez E, Rouzioux C, Flynn P M, Sullivan J L for The Pediatric AIDS Clinical Trials Group Protocol 076 Study Group. Maternal viral load, zidovudine treatment, and the risk of transmission of human immunodeficiency virus type 1 from mother to infant. N Engl J Med. 1996;335:1621–1629. doi: 10.1056/NEJM199611283352201. [DOI] [PubMed] [Google Scholar]
- 17.Vandamme A-M, Schmit J-C, van Dooren S, van Laethem K, Gobbers E, Kok W, Goubau P, Witvrouw M, Peetermans W, de Clercq E, Desmyter J. Quantification of HIV-1 RNA in plasma: comparable results with NASBA HIV-1 RNA QT and the Amplicor HIV Monitor test. J Acquired Immune Defic Syndr. 1996;13:127–139. doi: 10.1097/00042560-199610010-00003. [DOI] [PubMed] [Google Scholar]
- 18.van Gemen B, Kievits T, Schukkink R, van Strijp D, Malek L T, Sooknanan R, Huisman H G, Lens P. Quantification of HIV-1 RNA in plasma using NASBA during HIV-1 primary infection. J Virol Methods. 1993;43:177–188. doi: 10.1016/0166-0934(93)90075-3. [DOI] [PubMed] [Google Scholar]
- 19.Volberding P. HIV quantification: clinical applications. Lancet. 1996;347:71–73. doi: 10.1016/s0140-6736(96)90205-6. [DOI] [PubMed] [Google Scholar]
- 20.Yen-Lieberman B, Brambilla D, Jackson B, Bremer J, Coombs R, Cronin M, Herman S, Katzenstein D, Leung S, Lin H J, Palumbo P, Rasheed S, Todd J, Vahey M, Reichelderfer P. Evaluation of a quality assurance program for quantification of human immunodeficiency virus type 1 RNA in plasma by the AIDS Clinical Trials Group virology laboratories. J Clin Microbiol. 1996;34:2695–2701. doi: 10.1128/jcm.34.11.2695-2701.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]