Abstract
The design principles of spider dragline silk, nature's high-performance fiber, are still largely unknown, in particular for the noncrystalline glycine-rich domains, which form the bulk of the material. Here we apply two-dimensional solid-state NMR to determine the distribution of the backbone torsion angles (φ,ψ) as well as the orientation of the polypeptide backbone toward the fiber at both the glycine and alanine residues. Instead of an “amorphous matrix,” suggested earlier for the glycine-rich domains, these new data indicate that all domains in dragline silk have a preferred secondary structure and are strongly oriented, with the chains predominantly parallel to the fiber. As proposed previously, the alanine residues are predominantly found in a β sheet conformation. The glycine residues are partly incorporated into the β sheets and otherwise form helical structures with an approximate 3-fold symmetry.
Spider dragline silk is a high-performance fiber with mechanical properties rivaling the best man-made materials (1–3). Progress in genetic engineering (4–7) and the emerging understanding of the spinning process (7–10) have created the prospect of producing artificial spider silk or silk-like high-performance materials (6, 11). The molecular structure that leads to the functional properties of the material and the mechanism by which nature assembles this structure, however, are far from being understood.
Silk is a composite material with a hierarchical structure. Its primary structure, the amino acid sequence, has been optimized over millions of years of biological evolution (12). Spider dragline silk is thought to be composed mainly of two proteins, Spidroin I and II (13, 14). These proteins can be described, in a simplified picture, as block copolymers with poly-Ala blocks (consisting of approximately eight monomers) and of glycine-rich domains, with (Gly-Gly-X) as most abundant motive in Spidroin I. Here X denotes a variable amino acid. The loosely conserved motif Gly-Gly-X is repeated approximately 10 times (15). Glycine-rich repeats in Spidroin II include (Gly-Pro-Gly-Gly-X) and (Gly-Pro-Gly-Gln-Gln) (14).
There is increasing evidence that the structures higher up in the hierarchy are not solely determined by the primary structure, but that the spinning process itself also has an important influence (16, 17). Already the secondary structure, the conformation of the amino acids, was shown to be strongly influenced by the spinning in a related type of silk. The primary structure of the cocoon silk from the Eri silkworm Samia cynthia ricini follows the design principles of spider dragline silk with the typical poly-Ala and Gly-Gly-X blocks. The aqueous protein solution produced by the silk glands of S. cynthia ricini was shown to adopt an α-helical conformation for the poly-Ala segments when dried into a film. The native fibrous silk features alanine in β sheet conformation, however (18). Such polymorphic behavior is often found in proteins (19). Structures in silk, beyond the secondary structure, include tertiary structure and the possible formation of supramolecular helices, microcrystals and a microfibrillar organization of the silk fiber (11, 20, 21).
In this paper, we address the secondary structure of dragline silk as well as the orientation of the polypeptide backbone with respect to the fiber direction at both the glycine and alanine residues. The poly-Ala domains have previously been characterized by NMR and x-ray and were found to be predominantly in β sheet conformation (22–24) and to organize into microcrystals with sizes of at least 2 × 5 × 7 nm (25, 26). Considerably larger domains were seen by transmission electron microscopy (27); the size of the ordered domains hints that these must incorporate some glycine. The major constituent of silk, the glycine-rich domains, has not been observed in diffraction experiments and has, in the absence of atomistic structural information, been described as an “amorphous” or “rubber-like” matrix (2, 28). NMR data have suggested that 31-helical structures may be present (24), and that the Gly-Leu-Gly-X-Gln-Gly motif, also present in these domains, forms β turn (29). β Spirals, a succession of β turns, were postulated for the Gly-Pro-Gly-Gly-X and Gly-Pro-Gly-Gln-Gln segments (14, 30).
Materials and Methods
All samples were 13C labeled at the carbonyl functionality of selected amino acids. Silk from the spider Nephila edulis was collected as described in ref. 31. During the labeling procedure, the spiders were kept on a low diet of Tenebrio meal worms (once per week). One week before and during collection of the labeled silk, the spiders were fed daily doses of an aqueous amino acid solution, containing either the 100% labeled amino acids of interest or the 100% labeled amino acids together with a mixture of unlabeled amino acids. The mixture roughly followed the composition of dragline silk and consisted of glycine, alanine, glutamine, serine, tyrosine, proline, and leucine in the molar ratios 45:22:11:7:4:3:2. During the first week, the spiders were silked daily to empty the silk glands. Oriented silk samples were obtained by reeling directly from the spider onto a holder such that the part of the sample inside the NMR coil was uniaxially oriented.
The isotopic enrichment was determined by using mass spectroscopy. After hydrolysis with 6 M HCl at 108°C for 16 hr, the silk was derivatized with Nα-(2,4-dinitro-5-fluorophenyl)-l-alaninamide. The mixture of derivates was separated on a Hewlett–Packard HP1100 LC-MSD machine (Agilent, Palo Alto, CA). For each derivatized amino acid, the typical isotopic pattern was determined with a commercially available standard by extracting and integrating the relevant ions at the predetermined retention times. The isotopic pattern was recorded for the [M+], [M+H+], [M+1+H+], [M+2+HP], [M+3+H+] ions and was compared to the pattern of an unlabeled silk sample that was used as standard and control. The enrichment was determined by using software that fitted the pattern for each amino acid in the silk to the corresponding pattern of the amino acid standard. The procedure was repeated three times for each silk sample and the enrichment averaged. The serine enrichment could not be determined accurately by using this method. More experimental details will be given elsewhere. The approximate relative abundances of the amino acids in silk from N. edulis were determined separately by using a standard analysis. The results are given in Table 1.
Table 1.
13C labeling percentages of silk samples from N. edulis
| AA | DOQSY, 1-13C Ala, % | DOQSY, 1-13C Gly, % | DECODER, 1-13C Ala, % | DECODER, 1-13C Gly, % | Relative abundance of amino acids, % |
|---|---|---|---|---|---|
| Ala | 27.2 (17) | 10.0 (7) | 11.8 (6) | 10.1 (24) | 29 |
| Gly | 3.9 (10) | 59.4 (2) | 1.9 (6) | 48.1 (23) | 40 |
| Pro | 1.9 (1) | 1.6 (8) | 1.6 (6) | 1.4 (4) | 3 |
| Tyr | −1.1 (23) | −1.0 (25) | −1.0 (19) | 1.5 (5) | 4 |
| Glu | 4.0 (9) | 3.5 (2) | 1.9 (3) | 2.9 (4) | 10 |
The absolute labeling degree of the five most abundant amino acids in silk is given in percentage, together with the relative abundance of the respective amino acids in molar percentage. The title of each column refers to the experiment and the intended labeling. SDs are given in brackets.
M. Hronsky, personal communication.
Experimental conditions for double-quantum single-quantum correlation experiment for static samples (DOQSY) spectra were as described in ref. 18, except for the spectrum of glycine-labeled silk, which was taken at a magnetic field of 7.0 T. Direction exchange with correlation for orientation–distribution evaluation and reconstruction (DECODER) spectra were obtained at 9.4 T by using a home-built setup. Radio frequency field strengths were approximately 50 kHz on both channels. To improve the stability of the analysis, the probability density functions (PDFs) from DECODER spectra are the result of a fit that included not only the two-dimensional (2D) DECODER spectra presented below but also spectra of the same samples, but with a reorientation from 90 to 45°. Samples of about 25 mg were used, corresponding to approximately 1019 13C spins. The spectra of Figs. 1 B and C, and 3 A and B had signal-to-noise ratios of 33, 67, 45, and 63, respectively, for measuring times of 2 days per spectrum.
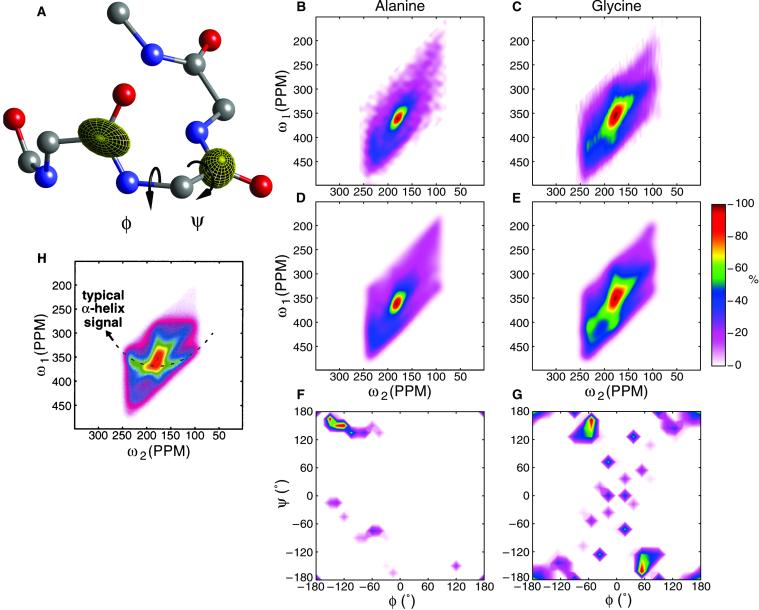
Fig 1.
(A) The DOQSY spectrum is determined by the relative orientation of the 13C O chemical-shift tensors, which depend on backbone torsion angles φ and ψ. (B) Experimental 2D DOQSY spectra of 1-13C alanine and (C) 1-13C glycine-labeled N. edulis silk samples taken with 1.5 and 1.0 ms excitation times, respectively. D and E show the corresponding best fits. The fit shown for the alanine-labeled sample includes the 13Cα chemical-shift bias (18). F and G show the probability P(φ,ψ) for the alanine- and glycine-labeled samples. H shows a simulated DOQSY spectrum of a random-coil conformation.
O chemical-shift tensors, which depend on backbone torsion angles φ and ψ. (B) Experimental 2D DOQSY spectra of 1-13C alanine and (C) 1-13C glycine-labeled N. edulis silk samples taken with 1.5 and 1.0 ms excitation times, respectively. D and E show the corresponding best fits. The fit shown for the alanine-labeled sample includes the 13Cα chemical-shift bias (18). F and G show the probability P(φ,ψ) for the alanine- and glycine-labeled samples. H shows a simulated DOQSY spectrum of a random-coil conformation.
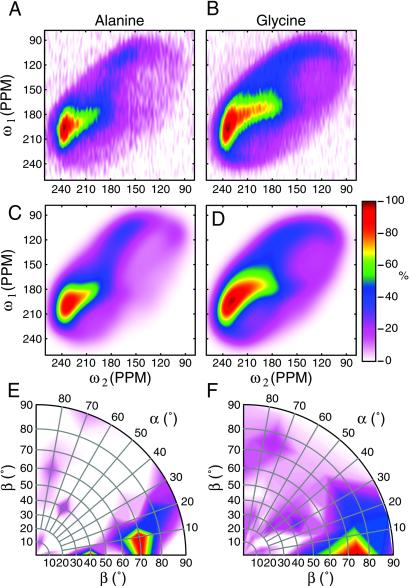
Fig 3.
A and B show experimental 2D DECODER spectra for 1-13C alanine and 1-13C glycine-labeled silk samples and C and D the best fits. The orientation of the sample was flipped from 60 to 30° in the mixing time of the experiment. E and F are polar projections of the probability density P(αF, βF) for both samples.
To address the ill-posed nature of the problem, spectra were fitted with a Tikhonov regularization algorithm, which used a small norm of the probability density P(φ,ψ) as additional constraint (18):
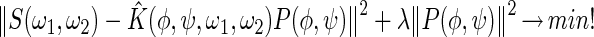 |
Here, S(ω1, ω2) denotes the experimental spectrum and K̂(φ, ψ, ω1, ω2), the kernel describing the experiment. Nonnegativity was imposed by using a non-negative least squares algorithm and the optimum regularization parameter λ determined by using the self consistent method (32). The results of fitting the DOQSY spectra to a single set of torsion angles are published as supporting information on the PNAS web site, www.pnas.org. The chemical-shift tensors in the fits were assumed such that the most-shielded component is perpendicular to the peptide plane and the intermediately shielded component is 5° off from the C O bond in the direction of the Cα atom (18). From a number of model studies, this orientation is estimated to be accurate within 5°.
O bond in the direction of the Cα atom (18). From a number of model studies, this orientation is estimated to be accurate within 5°.
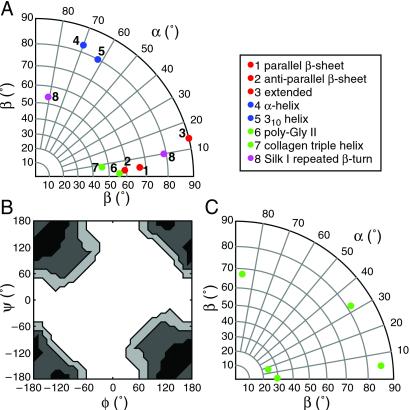
The regular molecular structures of Fig. 4 were created by concatenating 20 repeating units of 1 or more residues. The axis direction of the resulting structure was put along the fiber direction. For the examples, conformations with the following (φ,ψ) angle pairs were taken: 1 = (−119°, 113°), 2 = (−139°, 135°), 3 = (−180°, 180°), 4 = (−57°, −47°), 5 = (−49°, −26°), 6 = (−79°, 150°), 7 = (−51°, 153°), 8 = (−60°, 130°), and (70°, 30°). The angles for the right-handed β spiral were taken from ref. 33 as a model for the proposed β spiral in ref. 34.
Fig 4.
Regular model structures with the chain direction (helix axis) along the fiber yield sharply peaked distributions for (αF, βF). (A) The expected center of the distributions for various common conformations. More details are given in Materials and Methods. B shows the regions of torsion angles that are compatible with a regular structure along the fiber direction and (αF, βF) = (10°, 70°). Contour levels are set 10, 20, and 30° maximum absolute difference. Darker color indicates smaller values for the deviation. (C) A right-handed β spiral structure is expected to show a broad range of angles (αF, βF).
Results and Discussion
Measurement of Local Structure in Silk.
In a recent study, we have shown that the distribution of backbone torsion angles (φ,ψ) in a heterogeneous solid protein can be determined with solid-state NMR (18). DOQSY (35) was adapted to proteins enriched at the carbonyl moiety. Using this technique, it is possible to measure the relative orientation of the carbonyl chemical-shift tensors of amino acids neighboring in the primary structure (Fig. 1A). From the resulting spectra, the (φ,ψ) angles can be obtained simultaneously by assuming standard bond lengths and angles for the peptide unit (36) and the orientation of the chemical-shift tensor in the molecule. Because the solid-state NMR spectrum does not resolve the different residues of the primary structure, it is not possible to get a full structure. Through application of selective labeling, however, we were able to measure the (φ,ψ) values in an amino-acid-specific manner. In general, the (φ,ψ) angles will not have a fixed value but are given by a PDF P(φ,ψ). Using a regulatory data analysis (18, 37), “model-free” in the functional shape of P(φ,ψ), we can identify the secondary structure elements in silk of the spider N. edulis at the alanine and glycine residues, which make up almost 70% of the primary sequence. Note that we selectively determine the angles for Ala-Ala and Gly-Gly pairs in this way.
The isotopic enrichment of the four silk samples has been investigated by using mass spectroscopy to detect crosslabeling effects (see Table 1). We estimate from these data and from the predicted sequences for Spidroin I and II (13, 14) that the spectra of Figs. 1 B and C and 3 A and B will show perturbing signals, compared to spectra of ideally labeled samples, in the order of <3, 4, 30, and 13%, respectively.
Fig. 1 B and C show the experimental DOQSY spectra of samples labeled at the alanine and glycine residues, respectively. Fitting these spectra yielded the PDF shown in Fig. 1 F and G. Alanine shows a clear preference for the β sheet region with most abundant conformation (φ,ψ) = (−135°, 150°), which, within the grid resolution of 15° used corresponds well with known β sheet structures. Seventy-three percent of all torsion angles are in the β sheet region. The uniqueness of the PDFs is an inherent concern, because the analysis pertains to the solution of so-called ill-posed problems. Monte Carlo simulations have shown that strong peaks are stable in position, but the minor intensities in the PDF must be interpreted with care, as they can be artifacts. Such small peaks are often highly correlated with other conformations and induced by a low signal-to-noise ratio and systematic errors in the principal values of the chemical-shift tensor. The latter is known to be (φ,ψ)-dependent, although not large compared to the line width, and neglecting this dependence introduces systematic errors (18). The isotropic chemical shift of the Cα carbon of alanine was used as a further constraint to distinguish between (φ,ψ) and (−φ,−ψ), because these are indistinguishable in the DOQSY experiment (18). The Cα chemical shift was determined from a 2D 15N-13C HETCOR experiment of a U-13C, 15N-alanine labeled silk sample (data not shown).
The glycine PDF shows a more complex structure. As glycine is not chiral, the chemical shift cannot be used to distinguish between torsion angle pairs (φ,ψ) and (−φ,−ψ). The PDF therefore shows inversion symmetry around the center. The distribution is peaked with two maxima at (∓60°, ±135°) and (±180°, ±180°). The position of the peak at (±180°, ±180°) is less stable than the signal around (∓60°, ±135°) and can be shifted by one grid point, corresponding to 15° in both φ and ψ on varying of fitting conditions, which is a consequence of the complexity of the spectrum. We interpret the two conformations as a helix with three amino acids per turn (31-helix)∥ and an extended β sheet conformation, respectively. Note that we measured only the Gly-Gly torsion angles, and that further justification of this interpretation is based also on the results obtained with oriented silk, described below. The spectrum does not support a random-coil model, though. In such a disordered structure, the PDF is expected to be solely determined by the conformational energy E(φ,ψ) via a Boltzmann distribution (38). A PDF has been calculated on the basis of the conformational energy of glycine as given in chapter VII of ref. 38, and the corresponding spectrum is given in Fig. 1H. The signature of an α-helical conformation, which must give a sizeable contribution to a random-coil structure, clearly lacks in the experimental spectrum and PDF.
These DOQSY spectra represent the relative orientation of carboxylic tensors of residues, neighboring in the primary sequence, in the limit of short double-quantum excitation times only. For longer times, the effects of multispin and non-next-neighbor contributions become important, and the excitation times should be taken as short as possible. Because of the decreasing signal-to-noise ratios, a compromise must be found, though, and we have used 1.5 and 1.0 ms for the alanine and glycine-labeled silks, respectively. On the basis of a regular poly-Ala β sheet structure (39), we then expect approximately 30% of the intensity for alanine at (−180°, 180°) from parallel tensors (both inter- and intrastrand), which is indeed observed. From the labeling degree and the sequences of Spidroin I and II, the ratio of contributions to the spectrum from (Ala)2 and (Ala)3 was determined to be 5:1, which is sufficient to neglect (Ala)3 in the analysis. No (Gly)3 units are predicted for Spidroin I and II, but interchain distances are known to be in the range of 3.5 Å for a poly-Gly β sheet structure. Such short distances are not expected in silk, because of the bulkier residues surrounding the glycine residues, but the effects of close non-next-neighboring residues are difficult to predict. It can be safely assumed, however, that they are farther apart than neighbors. The high labeling degree of the glycine-enriched sample allowed the use of shorter excitation times (1.0 and even 0.5 ms; data not shown). The results of the analysis for glycine did not vary significantly for DOQSY spectra taken with 1.5, 1.0, or 0.5 ms, demonstrating the stability of the approach.
Orientation Toward the Silk Fiber.
Complementary to the local structure, we determined the orientation of the peptide chain toward the fiber, as described by a pair of Euler angles (αF, βF) (see Fig. 2A). The upper three traces of Fig. 2 B and C show one-dimensional 13C spectra for macroscopically oriented spider silk samples labeled at the alanine and glycine residues. The deviations of the spectra compared to a nonoriented sample show that both domains have significant order of the polypeptide chain direction with respect to the fiber direction. The Ala and Gly 13C O tensors have similar principal values, which correspond to a rigid sample without large-amplitude internal dynamics. The chemical-shift values were determined to be [242, 182, 90] and [242, 178, 96] ppm for the alanine and glycine-labeled silk, respectively, with an estimated accuracy of ±2 ppm. The corresponding values for the crystalline amino acids are [242, 185, 106] ppm (40) and [248, 179, 104] ppm (41). Clearly the 13C
O tensors have similar principal values, which correspond to a rigid sample without large-amplitude internal dynamics. The chemical-shift values were determined to be [242, 182, 90] and [242, 178, 96] ppm for the alanine and glycine-labeled silk, respectively, with an estimated accuracy of ±2 ppm. The corresponding values for the crystalline amino acids are [242, 185, 106] ppm (40) and [248, 179, 104] ppm (41). Clearly the 13C O tensors in silk are not significantly averaged by motions fast at the NMR time scale, which, in this case, is 10s of kilohertz. The glycine-rich domains, therefore, are not elastomeric or rubber-like,** as postulated in some models (2, 28).
O tensors in silk are not significantly averaged by motions fast at the NMR time scale, which, in this case, is 10s of kilohertz. The glycine-rich domains, therefore, are not elastomeric or rubber-like,** as postulated in some models (2, 28).
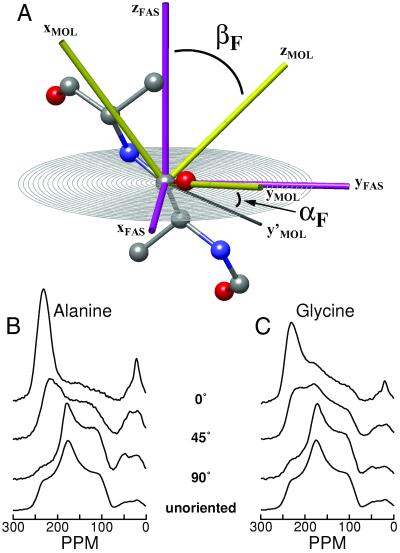
Fig 2.
(A) Definition of the axis systems used to describe the DECODER spectrum, where MOL is the molecular axis system, i.e., the peptide plane, with the z axis perpendicular to the peptide plane. FAS denotes the fiber axis system, with the z axis along the fiber direction. The orientation of a peptide plane toward the fiber axis is given by two Euler angles (αF, βF). Note that in the extended conformation of the peptide shown, the xMOL is approximately along the chain direction. In less-extended β sheet conformations, an angle of up to 23° may be observed. (B and C) Experimental 1D 13C spectra of 1-13C alanine and 1-13C glycine-labeled spider dragline silk, respectively, of oriented samples from N. edulis, for three sample orientations. The bottom trace is from an unoriented sample of 1-13C glycine-labeled silk and serves as an example of a nonoriented sample. Note that DECODER is insensitive to the size of (possibly) ordered microdomains but gives an integral over the entire sample volume.
From the series of 1D spectra shown in Fig. 2, we tried to extract the orientational distribution function P(αF, βF) by using a regulatory approach equivalent to that used for the DOQSY spectra. The regulatory analysis did not yield a stable result, though, and we resorted to measuring 2D spectra, which correlate the 1D spectra at two different sample orientations [DECODER spectroscopy (42, 43)]. Experimental DECODER spectra, fits, and PDFs for an experiment 60° → 30° are shown in Fig. 3. An isotropic distribution would yield a uniformly colored PDF. At both the alanine and glycine residues, the polypeptide chain is found to have a substantial degree of order with respect to the fiber direction. For the alanine, 70% of the intensity is found in a narrow region around the maximum at (αF, βF) = (10°, 70°). The glycine-rich domains show a somewhat broader peak centered around the same area and containing 61% of the total intensity. The peak intensity was calculated by summing all signals within the 15% contour (of the maximum of the PDF).
Structural Model.
The distribution functions obtained from DOQSY and DECODER provide complementary information but are uncorrelated.†† Nonetheless, the data may be compared to simulated data of simple model structures. The appearance of a single strong maximum for P(αF, βF) indicates that Ala and Gly appear predominantly in a regular arrangement with the chain axis parallel to the fiber direction. Typical (αF, βF) angles, for regular structures formed from some well known secondary-structure elements, are shown in Fig. 4A. For irregular structures, or regular structures with a chain axis deviating from the fiber axis direction by more than 20°, typically a broad distribution of the (αF, βF) angles is observed.
The strongly peaked distribution around (αF, βF) = (10°, 70°), combined with the (φ,ψ) distribution, are in perfect agreement with a regular β sheet structure for the poly-Ala domains (see also Fig. 4B). Such a structure was also found already by Simmons et al. (23), who determined the angle of the alanine Cα—Cβ bond vector toward the oriented fiber, in terms of Gaussian distributions. They found a 40:60 mixture of a highly oriented part [full width half height (FWHH) = 5°] and a less-oriented part (FWHH = 75°) and obtained a value of the orientation function 〈cos2φ′〉 = 0.760.‡‡ From x-ray data of major ampulate silk from Araneus marmoreus and Nephila cruentata, the orientation function 〈cos2φ′〉 was calculated to be 0.776 and 0.788, respectively (44). If we reduce our distribution to the angular correlation function, we obtain 0.742.§§
For glycine, the DOQSY experiment indicates that there are two conformations. We interpret the peak at (±180°, ±180°) as the combined signals for the inter- and intrachain contacts of an extended β sheet conformation, suggesting that some Gly-Gly pairs are part of the larger ordered β sheet regions, previously observed by electron microscopy (27). The conformation (∓60°, ±135°) could be one of the several angle pairs of a β spiral (30), but this structure is expected to show a broad range of angles (αF, βF) in the DECODER distribution (Fig. 4C). The DECODER experiment is expected to be sensitive to the presence of several (αF, βF) values, because it probes all glycines, which means approximately 50% of all residues in the glycine-rich domains. We cannot exclude β spirals existing in small numbers, but our data do not support a dominant presence in dragline silk. Instead of such a short and bulky β spiral (33), we propose a regular helical structure along the fiber direction, with an approximate 3-fold symmetry. A similar structure was found in other materials, e.g., poly-Gly II (45), poly-(L-Ala-Gly-Gly-Gly) II (46), poly-(L-Ala-Gly-Gly) II (46) and poly-(lactic acid), an ester analog of polypeptides (38, 47). The stability of these structures is given by the interchain hydrogen-bonding (45). This model is supported by both the DOQSY and DECODER distributions.¶¶ The 31-helical structure is elongated, consistent with the elongational flow of the silk dope solution during fiber spinning (11), and imagined to allow for efficient packing by virtue of interstrand hydrogen bonds. Note that previous results have indicated that the local structure is not significantly altered on supercontraction (48).
Recently, evidence for the existence of fibrillar substructures in silk thread has been presented (20, 21). Such substructures are consistent with our data, as long as the fibrils are parallel and oriented along the fiber (20), which was indeed observed. Much broader distributions in the DECODER experiment would be observed when not fulfilling this constraint.
A schematic summary of the structural information obtained in this study and data from literature is shown in Fig. 5. Peptide chains of Spidroin I and II are highly oriented along the fiber direction, possibly induced by the elongational flow inside the duct during spinning. The majority of the alanine residues are incorporated in regular β sheet structures, which predominantly form microcrystalline domains, which include glycine residues, tentatively assigned to be the GGX regions. The remaining glycine-rich parts were found to predominantly have observed a well-defined 31-helical secondary structure. Note that the observed glycine residues not only are ordered, they are also rigid on the NMR time scale, as are the alanines.
Fig 5.
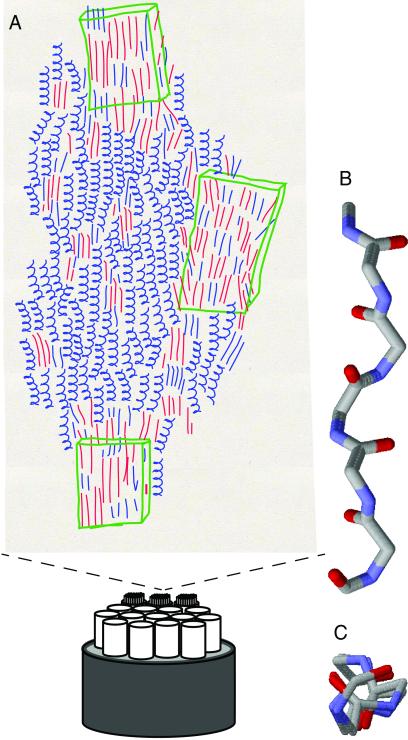
(A) Proposed model for spider dragline silk: a skin-core organization, with a multitude of fibrillar substructures and covered by a hard skin, forms the fiber (20, 21). The molecular structure consists of β sheet regions, containing alanine (red lines) and glycine (blue lines), interleaved with predominantly 31-helical parts (blue curls), which do not contain alanine. All chains tend to be parallel. The crystallites are irregular and strongly depend on processing [e.g., drawing speed (49)]. Depending on the method, larger or smaller ordered domains can be identified as crystalline (green boxes) (B and C) Side and top projections of a repetitive model peptide with dihedral angles (−60, 135) to show the approximate 3-fold symmetry.
Supplementary Material
Acknowledgments
Else Bomholt Rasmussen and Helene Jørgensen are acknowledged for preparing the silk samples and Gerrit Janssen, Jan van Os, Hans Janssen, Andreas Hunkeler, and Markus Andrist for technical support. M. Hronsky is acknowledged for supplying the amino acid composition data of N. edulis. This research was supported by the Swiss National Science Foundation and the European Science Foundation through the Network: Silk, Properties, and Production.
Abbreviations
DOQSY, double-quantum single-quantum correlation experiment for static samples
PDF, probability density function
nD, n-dimensional
DECODER, direction exchange with correlation for orientation–distribution evaluation and reconstruction
This paper was submitted directly (Track II) to the PNAS office.
Creating an idealized repetitive structure with the conformation (∓60°, ±135°) will generate a helical array with an approximately 3-fold rotational symmetry (see Fig. 5).
Elastomeric and rubbery polymers are characterized by considerable internal motion and have significantly reduced amplitudes of all anisotropic spin-system parameters.
Only a direct correlation in a 3D NMR experiment would yield the abundance and orientation of each conformation (φ,ψ) toward the fiber direction. Practically, this is currently not yet feasible due to the demands on the hardware.
φ′ is the angle of the chain direction toward the fiber direction.
To calculate the orientation function, 〈cos2φ〉 P(αF, βF) was integrated over αF and βF. An average angle of 23.5° between zMAS and the true normal of the β sheet chain direction was assumed. This angle was determined from simulated β sheet structures with torsion angles around those observed in the DOQSY PDF.
The DECODER spectra can be explained reasonably well by simulation of a regular fragment containing three carbonyl tensors and a fixed approximate 31-helical conformation, by using a Gaussian distributions centered at αF = 10° [full width half height (FWHH) 15°], βF = 75° (FWHH 20°).
References
- 1.Denny M. W. (1980) Mech. Prop. Biol. Mater. 34, 247-272. [PubMed] [Google Scholar]
- 2.Gosline J. M., DeMont, M. E. & Denny, M. W. (1986) Endeavor 10, 37-43. [Google Scholar]
- 3.Kaplan D. L., Adams, W. W., Viney, C., Farmer, B. L. & ACS Symposium Series (1994) in Silk Polymers—Material Science and Biotechnology, eds. Kaplan, D., Adams, W. W., Farmer, B. & Viney, C. (Am. Chem. Soc., Washington, DC), Vol. 544, pp. 2–16. [Google Scholar]
- 4.Fahnestock S. R. & Bedzyk, L. A. (1997) Appl. Microbiol. Biotechnol. 47, 33-39. [DOI] [PubMed] [Google Scholar]
- 5.Fahnestock S. R. & Irwin, S. R. (1997) Appl. Microbiol. Biotechnol. 47, 23-32. [DOI] [PubMed] [Google Scholar]
- 6.O'Brien J. P., Fahnestock, S. R., Termonia, Y. & Gardner, K. H. (1998) Adv. Mater. 10, 1185-1195. [Google Scholar]
- 7.Lazaris A., Arcidiacono, S., Huang, Y., Zhou, J.-F., Duguay, F., Chretien, N., Welsh, E. A., Soares, J. W. & Karatzas, C. N. (2002) Science 295, 472-476. [DOI] [PubMed] [Google Scholar]
- 8.Trabbic K. A. & Yager, P. (1998) Macromolecules 31, 462-471. [Google Scholar]
- 9.Seidel A., Liivak, O. & Jelinski, L. W. (1998) Macromolecules 31, 7633-7636. [Google Scholar]
- 10.Seidel A., Liivak, O., Calve, S., Adaska, J., Ji, G., Yang, Z., Grubb, D., Zax, D. B. & Jelinsky, L. W. (2000) Macromolecules 33, 775-780. [Google Scholar]
- 11.Vollrath F. & Knight, D. P. (2001) Nature (London) 410, 541-548. [DOI] [PubMed] [Google Scholar]
- 12.Gatesy J., Hayashi, C., Motriuk, D., Woods, J. & Lewis, R. V. (2001) Science 291, 2603-2605. [DOI] [PubMed] [Google Scholar]
- 13.Xu M. & Lewis, R. V. (1990) Proc. Natl. Acad. Sci. USA 87, 7120-7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinman M. B. & Lewis, R. V. (1992) J. Biol. Chem. 267, 19320-19324. [PubMed] [Google Scholar]
- 15.Lewis R. V. (1992) Acc. Chem. Res. 25, 392-398. [Google Scholar]
- 16.Calvert P. (1998) Nature (London) 393, 309-311. [Google Scholar]
- 17.Vollrath F., Knight, D. P. & Hu, X. W. (1999) R. Soc. 266, 817-820. [Google Scholar]
- 18.van Beek J. D., Beaulieu, L., Schäfer, H., Demura, M., Asakura, T. & Meier, B. H. (2000) Nature (London) 405, 1077-1079. [DOI] [PubMed] [Google Scholar]
- 19.Wilson D., Valuzzi, R. & Kaplan, D. (2000) Biophys. J. 78, 2690-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Putthanarat S., Stribeck, N., Fossey, S. A., Eby, R. K. & Adams, W. W. (2000) Polymer 41, 7735-7747. [Google Scholar]
- 21.Augsten K., Mühlig, P. & Herrmann, C. (2000) Scanning 22, 12-15. [DOI] [PubMed] [Google Scholar]
- 22.Simmons A., Ray, E. & Jelinski, L. W. (1994) Macromolecules 27, 5235-5237. [Google Scholar]
- 23.Simmons A. H., Michal, C. A. & Jelinski, L. W. (1996) Science 271, 84-87. [DOI] [PubMed] [Google Scholar]
- 24.Kümmerlen J., van Beek, J. D., Vollrath, F. & Meier, B. H. (1996) Macromolecules 29, 2920-2928. [Google Scholar]
- 25.Grubb D. T. & Jelinski, L. W. (1997) Macromolecules 30, 2860-2867. [Google Scholar]
- 26.Riekel C., Bränden, C., Craig, C., Ferrero, C., Heidelbach, F. & Müller, M. (1999) Int. J. Biol. Macromol. 24, 179-186. [DOI] [PubMed] [Google Scholar]
- 27.Thiel B. L., Guess, K. B. & Viney, C. (1997) Biopolymers 41, 703-719. [DOI] [PubMed] [Google Scholar]
- 28.Gosline J. M., Denny, M. W. & DeMont, M. E. (1984) Nature (London) 309, 551-552. [Google Scholar]
- 29.Michal C. A. & Jelinski, L. W. (1998) J. Biol. NMR 12, 231-241. [DOI] [PubMed] [Google Scholar]
- 30.Hayashi C. Y. & Lewis, R. V. (1998) J. Mol. Biol. 275, 773-784. [DOI] [PubMed] [Google Scholar]
- 31.Work R. W. & Emerson, P. D. (1982) J. Arachnol. 10, 1-10. [Google Scholar]
- 32.Honerkamp J. & Weese, J. (1990) Cont. Mech. Thermodyn. 2, 17-30. [Google Scholar]
- 33.Venkatachalam C. M. & Urry, D. W. (1981) Macromolecules 14, 1225-1229. [Google Scholar]
- 34.Hayashi C. Y., Shipley, N. H. & Lewis, R. V. (1999) Int. J. Biol. Macromol. 24, 271-275. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt-Rohr K. (1996) Macromolecules 29, 3975-3981. [Google Scholar]
- 36.Pauling L., Corey, R. B. & Branson, H. R. (1951) Proc. Natl. Acad. Sci. USA 37, 205-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schäfer H., Sternin, E., Stannarius, R., Arndt, M. & Kremer, F. (1996) Phys. Rev. Lett. 76, 2177-2180. [DOI] [PubMed] [Google Scholar]
- 38.Flory P. J., (1969) Statistical Mechanics of Chain Molecules (Wiley, New York).
- 39.Arnott S., Dover, S. D. & Elliott, A. (1967) J. Mol. Biol. 30, 201-208. [DOI] [PubMed] [Google Scholar]
- 40.Naito A., Ganapathy, S., Akasaka, K. & McDowell, C. A. (1981) J. Chem. Phys. 74, 3190-3197. [Google Scholar]
- 41.Haberkorn R. A., Stark, R. E., van Willigen, H. & Griffin, R. G. (1981) J. Am. Chem. Soc. 103, 2534-2539. [Google Scholar]
- 42.Schmidt-Rohr K., Hehn, M., Schaefer, D. & Spiess, H. W. (1992) J. Chem. Phys. 97, 2247-2262. [Google Scholar]
- 43.Chmelka B. F., Schmidt-Rohr, K. & Spiess, H. W. (1993) Macromolecules 26, 2282-2296. [Google Scholar]
- 44.Work R. W. & Morosoff, N. (1982) Text. Res. J. 52, 349-356. [Google Scholar]
- 45.Crick F. H. C. & Rich, A. (1955) Nature (London) 176, 780-781. [DOI] [PubMed] [Google Scholar]
- 46.Lotz B. & Keith, H. D. (1971) J. Mol. Biol. 61, 201-215. [DOI] [PubMed] [Google Scholar]
- 47.Kang S., Hsu, S. L., Stidham, H. D., Smith, P. B., Leugers, M. A. & Yang, X. (2001) Macromolecules 34, 4542-4548. [Google Scholar]
- 48.van Beek J. D., Kümmerlen, J., Vollrath, F. & Meier, B. H. (1999) Int. J. Biol. Macromol. 24, 173-178. [DOI] [PubMed] [Google Scholar]
- 49.Riekel C. & Vollrath, F. (2001) Int. J. Biol. Macromol. 29, 203-210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.