Abstract
Primary ciliary dyskinesia (PCD; MIM 242650) is an autosomal recessive disorder of ciliary dysfunction with extensive genetic heterogeneity. PCD is characterized by bronchiectasis and upper respiratory tract infections, and half of the patients with PCD have situs inversus (Kartagener syndrome). We characterized the transcript and the genomic organization of the axonemal heavy chain dynein type 11 (DNAH11) gene, the human homologue of murine Dnah11 or lrd, which is mutated in the iv/iv mouse model with situs inversus. To assess the role of DNAH11, which maps on chromosome 7p21, we searched for mutations in the 82 exons of this gene in a patient with situs inversus totalis, and probable Kartagener syndrome associated with paternal uniparental disomy of chromosome 7 (patUPD7). We identified a homozygous nonsense mutation (R2852X) in the DNAH11 gene. This patient is remarkable because he is also homozygous for the F508del allele of the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Sequence analysis of the DNAH11 gene in an additional 6 selected PCD sibships that shared DNAH11 alleles revealed polymorphic variants and an R3004Q substitution in a conserved position that might be pathogenic. We conclude that mutations in the coding region of DNAH11 account for situs inversus totalis and probably a minority of cases of PCD.
Primary ciliary dyskinesia (PCD) (MIM 242650) is an autosomal recessive disorder with extensive genetic heterogeneity (1) that usually comes to medical attention because of recurrent upper respiratory tract infections (rhino-sinusitis, bronchitis, and bronchiectasis) or, in the adult male, because of infertility. In 50% of PCD patients there is dextrocardia, with or without situs inversus totalis; in this case the disorder is also referred to as Kartagener syndrome (MIM 244400). The prevalence of PCD has been estimated as 1 in 20,000–60,000.
The prevalence of situs inversus of any etiology appears to be in a range between 1 in 25,000 and 1 in 8,000. Twenty to twenty-five percent of these individuals with complete, mirror-image situs inversus have ciliary dyskinesia and respiratory symptoms (Kartagener syndrome) as associated findings (2).
In most of cases with PCD, electron microscopy reveals abnormalities of structural organization of the axoneme in cilia from respiratory epithelia and in spermatozoa. Yet other cases have structurally normal but dysmotile or immotile cilia. It has been suggested that subtle structural deficiencies of cilia may also be more common than previously estimated (3). The axoneme is composed of about 250 distinct proteins (4). Electron microscopy of the ciliary microtubules frequently reveals absence or abnormalities of the outer and/or inner dynein arms (5). These arms are multisubunit protein complexes with ATPase activity that promote sliding between adjacent microtubules, the basic action resulting in the beating of cilium and flagellum. The axonemal dynein arms are composed of heavy, intermediate, and light dynein chains (6). A defect in any one of these proteins could lead to an abnormal dynein arm and/or defective beating activity of the axoneme.
Our earlier linkage analyses in a large number of PCD families revealed extensive genetic heterogeneity (7). No single genomic region harboring a common PCD locus was identified, but several potential chromosomal regions that could harbor genes for PCD were localized (7).
To date, mutations in two genes have been associated with a minority of PCD/Kartagener syndrome cases. These are genes coding for the dynein axonemal intermediate chain 1 (DNAI1) (8–10) and the dynein axonemal heavy chain 5 (DNAH5) (11). Loss of function of the murine Dnah5 dynein gene also causes PCD in the mouse (12). Other genes coding for axonemal dyneins, such as the heavy chain DNAH9, the intermediate chain DNAI2, and the light chain LC8, were recently excluded as major causes of PCD (13, 14, ††). Moreover, the FOXJ1 gene, encoding a transcription factor involved in ciliary development, was also excluded as common cause of PCD (15).
In this study we characterized the human gene for axonemal heavy chain dynein DNAH11 and investigated its potential involvement in PCD. We detected a homozygous nonsense DNAH11 mutation in a patient with situs inversus and likely PCD and paternal uniparental isodisomy 7. Sequence analysis of the DNAH11 gene in an additional 6 selected PCD sibships that shared DNAH11 alleles revealed polymorphic variants and an R3004Q substitution in a conserved position that could be pathogenic. This study provides evidence that the DNAH11 gene is involved in the pathogenesis of situs inversus totalis and probable PCD, albeit in a small fraction of unrelated families.
Materials and Methods
Patients.
As part of an international collaboration, we collected well characterized families with PCD, 31 of which have two or more affected individuals; these families were described in ref 7. A microsatellite (CA)n repeat, D7S493, beginning at nucleotide +22 of intron 55 of the DNAH11 gene, was used as a marker to genotype the members of these 31 families. This marker was PCR-amplified with touchdown cycles from 50–60°C as the annealing temperature, using primers 5′-GGAAGTTCCCAGCCATAGTT-3′ and 5′-GAAAGCACTTACCTACTGAGGATTT-3′. Six families in which affected individuals shared both alleles at marker D7S493 were chosen for further mutation analysis.
DNA from a previously described male patient (C.C.) with significant respiratory disease, abnormal sweat chloride test, and homozygosity for the F508del mutation in the cystic fibrosis transmembrane conductance regulator (CFTR) gene (16) was also analyzed. The respiratory disease had an earlier age of onset and was considerably more severe than in patients with CFTR F508del homozygosity. At his current age of 7 years, the patient is on continuous oxygen and assisted ventilation and is considered to have end-stage lung disease. In addition to cystic fibrosis, this patient has dextrocardia with a structurally normal heart and visceral situs inversus with one structurally normal spleen. Biopsy of a clinically normal bronchial region (without inflammation) was performed to evaluate ciliary function and structure. No normal ciliary motion was observed, despite the lack of an inflammatory infiltrate. Electron microscopy demonstrated structurally normal cilia, with the typical 9 + 2 doublet microtubular configuration and normal inner and outer dynein arms. The likely diagnosis is the co-occurrence of two distinct but related phenotypes, those of cystic fibrosis and of PCD. Paternal uniparental disomy (UPD) for chromosome 7 was suspected and confirmed, as the F508del CFTR mutation was not present in the mother's DNA (16).
Genomic and cDNA Sequences of DNAH11.
We first identified 2 overlapping clone sequences contributed by the Genome Sequencing Center of the Washington University School of Medicine (GenBank accession nos. AC005078 and AC004002) coding for the central region of a putative outer arm axonemal beta heavy dynein chain that mapped to chromosome 7p21. Sequence comparison indicated that the putative protein was highly homologous to the mouse Dnah11/lrd predicted protein. Additional sequenced clones were added from the publicly available genome sequence (www.ncbi.nlm.nih.gov/genome/guide/human/): AC004595 corresponding to the 5′ end of the gene, AC73102, AC013481, and AC092097, and a larger contig with gaps was constructed. The public data were integrated with those of the Celera database (www.celera.com/) (clone GA_x2HTBL43YA41) to reconstruct the entire sequence of the region harboring the human DNAH11 gene.
The sequence data were used to determine the genomic structure and the exon–intron junctions, to recognize informative dinucleotide polymorphisms internal to the gene and to develop oligonucleotide primers for the cloning of the DNAH11 cDNA.
Detailed analysis of the genomic sequence was performed by using numerous programs, many of which interfaced with nix at Human Genome Mapping Project (www.hgmp.mrc.ac.uk/registered/webapp/NIX/).
Short partial cDNA sequences corresponding to the first P-loop of DNAH11, and a few expressed sequence tags were available in the early stages of the DNAH11 cDNA characterization. Additional cDNA sequences were determined by several methods, including 5′ and 3′ rapid amplification of cDNA ends (RACE), reverse transcription (RT)-PCR, and cDNA library screening.
Probes corresponding to different DNAH11 exons were used to screen approximately 600,000 clones from a human testis λgt10 cDNA library 5′ stretch (CLONTECH). Clones with sequences identical to the genomic sequence were further analyzed and used as probes to expand the cDNA cloning. PCR amplification of testis and fetal lung cDNA libraries with vector primers λgt10F or λgt10R and a DNAH11-specific primer was also used to obtain additional sequences of DNAH11. The 5′ and 3′ RACE reactions were performed on mRNA from human nasal epithelium after in vitro ciliogenesis (17), and were performed on fetal lung poly(A) RNA by using the Marathon cDNA amplification or the SMART-RACE cDNA kits (CLONTECH), according to the manufacturer's specifications.
The genomic sequence allowed the in silico determination of all DNAH11 exons by blast analysis using the mouse lrd/Dnah11 cDNA and the human cDNA sequences. The identity of putative exons was confirmed by RT-PCR from nasal epithelium, testis, and fetal lung cDNAs or cDNA libraries. Analysis of the putative protein sequence was performed with numerous computer programs available through the “Tools” option of ExPASy (www.expasy.org/tools/). Multiple sequence alignments with published full-length axonemal dyneins were performed by using the clustalw program (18) and shaded with genedoc (www.psc.edu/biomed/genedoc/).
Mutation Search.
The 82 exons of the DNAH11 gene were amplified by using DNA from one affected individual from each sibship showing concordance for both DNAH11 alleles, and from patient C.C. The amplicons were purified by Qiaquick columns (Qiagen) and directly sequenced by using standard protocols for the ABI377 automated sequencer. Sequence analyses and assembly were performed by using the gap4 of Staden package (19) and sequencher (Gene Codes) programs. When differences from the publicly available genomic sequence were found, the corresponding exons were amplified and sequenced in 10 unrelated controls from the Centre d'Etude du Polymorphisme Humain (CEPH) families (20), and in appropriate Hispanic controls.
Results and Discussion
In this study we investigated the potential involvement of a candidate gene, DNAH11, in situs inversus and PCD. The rationale for selecting the axonemal heavy chain dynein 11, DNAH11, gene is fourfold. First, various chains of axonemal dyneins are structural components of the dynein arms (6) within the ciliary microtubular structures of respiratory epithelia and sperm cells; these arms are often absent or defective in PCD patients (5). In addition, mutations of evolutionarily conserved dynein genes cause ciliary movement abnormalities in Chlamydomonas (21). Second, a missense mutation (E2271K) in the homologous mouse gene Dnah11 (lrd), has been described in the iv/iv (inversum viscerum) murine model that is characterized by situs inversus and immotile cilia in the embryonic node (22). The mutation lies in the motor domain, and might disrupt a microtubule-binding domain (23). Dnah11 is expressed in the node of the embryo at day 7.5, and is involved in left–right axis determination of the organs. Cilia in the node rotate rapidly, producing a flow of embryonic fluids; in the iv/iv mouse, it has been observed that the flow of fluid was missing (24). Targeted disruption of the mouse Dnah11 (lrd) gene results in randomization of laterality; the monocilia of mutant embryonic node cells were immotile (25). Third, a patient (C.C.) of Hispanic origin has been described with paternal uniparental isodisomy 7 and two different recessive disorders, namely cystic fibrosis (because of homozygosity of the CFTR gene F508del mutation), and most likely PCD with dextrocardia and situs inversus totalis (16). We hypothesized that PCD in this patient was caused by the presence of another recessive mutation in a gene on chromosome 7 brought to homozygosity through UPD7. Fourth, the DNAH11 gene maps to 7p15.3-21, a chromosomal region that in our genome-wide scan showed an NPL (nonparametric logarithm of odds) score of 1.44 for PCD families with dynein arm deficiency (7).
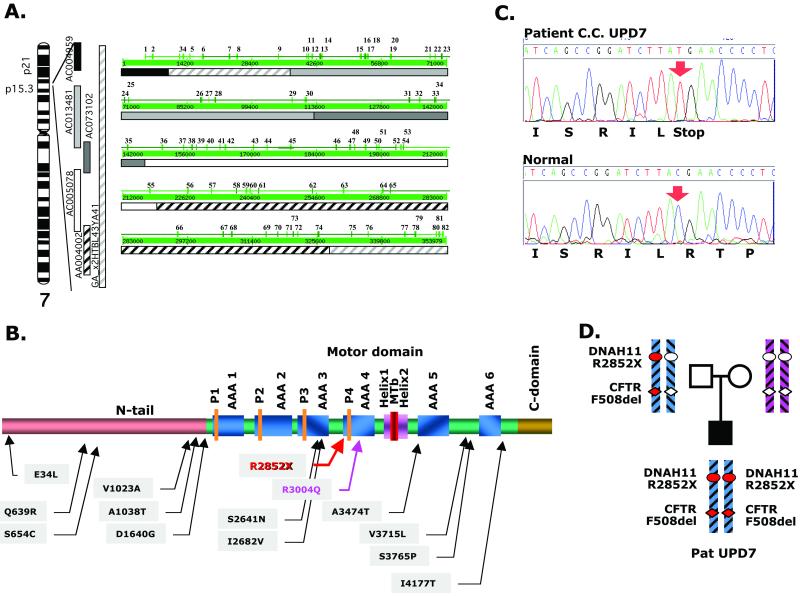
We used several different methods (cDNA library screening from human nasal epithelium and testis, RT-PCR, 5′ RACE, comparisons with the public and Celera genome sequences) to determine the full-length cDNA sequence of the DNAH11 gene, which consists of 13,670 nucleotides and contains an 13,569-nt ORF (GenBank accession no. AJ320497). Several alternatively spliced forms also exist. Comparison of the DNAH11 cDNA sequence to that of the overlapping bacterial artificial chromosomes (26) (GenBank accession nos. AC013481, AC005078, AC004595, AC092097, and AC004002) and Celera contig GA_x2HTBL43YA41 (27) allowed us to determine the genomic structure, intron–exon boundaries, and the precise mapping of DNAH11 within 7p21. The gene is composed of 82 exons, extending over 353 kb of genomic sequence (Fig. 1A). The longest DNAH11 cDNA that includes all identified exons, expressed in nasal epithelium or testis, encodes a putative protein of 4,523 amino acid residues. The putative protein (Fig. 1B and Fig. 2, which is published as supporting information on the PNAS web site, www.pnas.org) shows 84% identity and 91% similarity with the predicted mouse DNAH11 protein, as well as a high degree of homology (up to 77% identity) with previously described outer-arm axonemal β heavy chain dyneins from different species.
Fig 1.
(A) DNAH11 gene. Chromosomal localization, genomic clones, and exon distribution shown as the output of the nix program (http://menu.hgmp.mrc.ac.uk/menu-bin/Nix/Nix.pl). Exons are numbered. Nucleotide numbers are shown in the thick green line. Clones corresponding to the nucleotide sequences are also shown. (B) Schematic representation of the domain structure of DNAH11 (not exactly to scale). P1–P4, the four P-loops; AAA1–AAA6, the six AAA modules; Helix1-MT-Helix2, the B-link that includes the microtubule binding domain. The positions of the R2852X mutation and the other amino acid substitutions found are shown. (C) DNA sequence chromatogram of exon 52 of DNAH11 showing homozygosity for the nonsense R2852X mutation. (D) Nuclear pedigree of patient C.C. with paternal UPD7 and schematic representation of portions of chromosome 7 with mutations in the CFTR and DNAH11 genes. The mutant alleles are shown with red symbols, and the normal alleles are shown with clear symbols.
Mutation analysis (by direct sequencing of PCR products) of all 82 DNAH11 exons and the corresponding 160 intron-exon junctions of DNA from patient C.C. revealed a homozygous nonsense mutation (c.8554C→T; R2852X) in exon 52 (Fig. 1 C and D). The presence of the mutation was confirmed in a second sample. This mutation lies within the motor domain, 10 aa before the fourth P-loop. The mutant protein is predicted to contain a normal N-terminal tail domain, and three of the six AAA (ATPases Associated diverse cellular Activities) domains (28, 29). Because the N-terminal tail is predicted to be present, if stable, the mutant protein should be correctly incorporated into the dynein arm complex. The mutant protein would, however, not be expected to apply force to the adjacent microtubule doublet, given the absence of the microtubule binding domain. This hypothesis is compatible with the electron microscopic findings in the patient's cilia, which revealed normal axonemes and dynein arms (see figure 1 of ref. 16).
We also detected several sequence variants in patient C.C., all in homozygous form, some of which result in amino acid substitutions (V1023A, A1038T, S2641N, and S3765P). None of the other 54 unrelated PCD/Kartagener syndrome patients from our collection, nor the 140 DNAH11 Hispanic control alleles tested, had the R2852X nonsense codon. Analysis of patient C.C. provides convincing evidence that mutations in DNAH11 are associated with situs inversus totalis and probably one form of PCD. This is to our knowledge the first time that a patient with uniparental isodisomy led to the positional candidate cloning of a disease-related gene; moreover, the case of C.C. is unique because this patient is likely to have two distinct recessive disorders (cystic fibrosis and PCD), caused by different mutant genes on the same chromosome 7; an alternative, but less likely explanation is that the patient has cystic fibrosis and isolated situs inversus totalis. Aside from the laterality defect, the mucociliary clearance abnormality in patient C.C. could result from either or both disorders, cystic fibrosis and PCD.
After confirming the DNAH11 mutation in patient C.C., we subsequently selected 6 multiplex PCD families (GVA12, GVA15, GVA29, GVA30, UCL03, UCL09) in which affected individuals showed concordant inheritance of both alleles for the highly polymorphic D7S493 microsatellite marker within intron 55 of DNAH11. The DNA from one affected from each of these 6 families was screened for mutations in the 82 exons of DNAH11 and in the intron–exon junctions. We found 23 exonic changes (Table 1); 11 silent and 12 altering an amino acid. A further 19 intronic sequence variants within 50 nt of the exon boundaries were detected (Table 2). The 12 missense variants were further categorized as either benign variants or potentially pathogenic mutations. The likely polymorphisms were E34L, Q639R, S654C, V1023A, A1038T, D1640G, S2641N, I2682V, A3474T, V3715L, and I4177T.
Table 1.
Exonic variants in DNAH11: Pathogenic mutations vs. polymorphisms
| Exon | Codon change | AA change | Polymorphism vs. pathogenic mutation | Patients with variant |
|---|---|---|---|---|
| 1 | ACC → ACT | T18T | Polymorphism; no amino acid change, + controls | UCL09 |
| 1 | GAG → TTG | E34L | Polymorphism; + controls, | GVA12, GVA29, GVA30, UCL03 |
| 4 | AAT → AAC | N235N | Polymorphism; no amino acid change, + controls | UCL09 |
| 6 | CCA → CCG | P355P | Polymorphism; no amino acid change, + controls | GVA29 |
| 11 | CAG → CGG | Q639R | Polymorphism; mouse Dnah11 has R, + controls | GVA30, UCL09 |
| 11 | TCT → TGT | S654C | Polymorphism; + controls | GVA30, UCL09 |
| 14 | GCA → GCG | A818A | Polymorphism; no amino acid change, + controls | GVA29, GVA30, UCL09 |
| 16 | GTC → GCC | V1023A | Polymorphism; + controls | GVA12, GVA15, GVA30, UCL03 |
| 16 | GCT → ACT | A1038T | Polymorphism; + controls | C.C. hoz, GVA15, GVA12 |
| 25 | ATT → ATC | I1488I | Polymorphism; no amino acid change, + controls | GVA15, GVA29, GVA30 |
| 28 | GAT → GGT | D1640G | Polymorphism; + controls, no new splice site | GVA29 |
| 45 | TTC → TTT | F2437F | Polymorphism; no amino acid change, + controls | GVA12, GVA15 hoz, GVA29, GVA30, UCL03 hoz |
| 45 | TCG → TCA | S2452S | Polymorphism; no amino acid change, + controls | C.C. hoz, GVA29, GVA30 |
| 46 | ACG → ACA | T2549T | Polymorphism; no amino acid change, + controls | C.C. hoz, GVA12, GVA15 hoz, GVA29 hoz, GVA30, UCL03, UCL09 |
| 47 | CAC → CAT | H2599H | Polymorphism; no amino acid change, + controls | GVA29, UCL09 |
| 48 | AGT → AAT | S2641N | Polymorphism; + controls | C.C. hoz, UCL03, UCL09 |
| 49 | ATT → GTT | I2682V | Polymorphism; mouse Dnah11 has V, + controls | GVA29 |
| 52 | CGA → TGA | R2852X | Nonsense codon; pathogenic | C.C. hoz |
| 55 | CGG → CAG | R3004Q | Potential pathogenic mutation | GVA29 |
| 64 | GCC → ACC | A3474T | Polymorphism; mouse Dnah11 has T, + controls | GVA12 hoz, GVA29 hoz |
| 68 | TTG → GTG | L3715V | Polymorphism; mouse Dnah11 has V, + controls | C.C. hoz |
| 69 | TCT → CCT | S3765P | Polymorphism; + controls | C.C. hoz |
| 69 | CAT → CAC | H3773H | Polymorphism; no amino acid change, + controls | GVA12 |
| 77 | ATT → ACT | I4177T | Polymorphism; + controls | GVA12, GVA29, UCL03 |
| 80 | CTC → CTA | L4383L | Polymorphism; no amino acid change, + controls | GVA15 hoz, GVA29 hoz, GVA30, UCL03 hoz, UCL09 |
hoz, homozygous for the variant; all others were heterozygous.
+ controls: the variant was found in controls.
Splice site consensus checked at http://125.itba.mi.cnr.it/∼webgene/wwwspliceview.html and www.fruitfly.org/cgi-bin/seq_tools/splice.pl.
Table 2.
DNAH11 variants found in introns
| Intron | Base change |
|---|---|
| 4 | (−22) T → C |
| 16 | (−8) C → G |
| 24 | (+13) A → G |
| 24 | (−17) ins TTAAT |
| 28 | (−31) A → G |
| 37 | (−13) T → C |
| 38 | (−17) A → G |
| 45 | (+26) C → T |
| 45 | (−47) G → A |
| 47 | (−45) T → G |
| 48 | (+40) T → C |
| 53 | (+23) C → T |
| 53 | (−30) T → C |
| 55 | (+20) G → A |
| 55 | (+22) CA(n) |
| 58 | (−43) G → C |
| 62 | (−45) A → G |
| 71 | (−20) T(n) |
| 80 | (+23) A → C |
The missense mutation R3004Q was found in heterozygous form in patient GVA29 from the Canary Islands, who showed PCD (with dextrocardia and situs inversus totalis) with absence of the dynein arms (outer and inner). R3004, an amino acid of the fourth AAA domain, is a highly conserved residue in all β dynein chains (see Fig. 2). However, one α and one γ dynein heavy chain in Chlamydomonas have a Q at this position. None of the CEPH (20) DNAH11 alleles, nor 102 Hispanic control alleles analyzed had the Q3004 variant. It is therefore possible that the R3004Q DNAH11 mutation is pathogenic. However, no pathogenic mutation in the second allele of patient GVA29 could be identified.
The full molecular pathology of PCD has yet to be determined. Mutations have been found in the DNAI1 (intermediate axonemal dynein chain 1) (8–10) and DNAH5 (heavy axonemal γ dynein chain 5) (11) gene in a minority (less than 30%) of patients with PCD. A considerable number of dynein candidate genes need to be systematically analyzed for pathogenic PCD mutations. The human genome sequence annotation (http://www.ncbi.nlm.nih.gov/LocusLink/) contains 14 heavy chain genes. Moreover, the axonemal dynein arms are composed of 1–3 heavy chains (400–500 kDa), a variable number of intermediate chains (45–140 kDa), and one or more light chains (8–28 kDa) (31). To date, exclusion studies of the DNAH9 (14), DNAI2 (13, ††), and LC8 (††) genes have been reported. FOXJ1, a candidate gene (15), involved in the regulation of expression of dynein genes has also been excluded. Additional candidates are provided by mouse models such as the Dnah1/mdhc7 gene disruption mice (31). The elucidation of the molecular pathophysiology of PCD will provide a better understanding of the proteins important for ciliogenesis, ciliary movement, and left–right patterning of body structures.
In conclusion, we report the identification of pathogenic mutations in the human DNAH11 gene that cause one form of PCD, or, more conservatively, one form of situs inversus totalis. This result was accomplished by using a candidate gene approach, because the homologous murine DNAH11 gene is involved in a mouse syndrome with situs inversus (iv/iv). In addition, to our knowledge, a patient with UPD was instrumental for disease-gene “matchmaking” for the first time.
Supplementary Material
Acknowledgments
We thank the patients, their families, the United Kingdom PCD family support group, and the referring physicians: C. Barrazone, P. A Guerne, T. Rochat, and M. Zimmerman (Geneva); K. Brugger (Deissenhofen, Switzerland); C. Heili (Grub, Switzerland); E. Horak and M.H. Shöni (Davos, Switzerland); H. Hug-Batschelet and M. Ruthishauser (Basel); M. Kunzli and P. Eng (Aarau, Switzerland); W. Schäppi (Andelfingen, Switzerland); H.Walt and V. Dombi (Zurich); D.V. Schidlow (Philadelphia); R. Gershoni (Haifa, Israel); L. van Maldergem (Loverval, Belgium); and S. Amselem and B. Duriez (Creteil, France). We thank H. Omran (Freiburg, Germany) for communicating unpublished results, R. M. Gardiner (London) for encouragement and support, and G. Duriaux-Saïl for technical assistance. This work was supported by the Milena Carvajal-ProKartagener Foundation, Swiss Fonds National de la Recherche Scientifique Grant 31-63559.00, Office Fédéral de l'Education et de la Science Grant 95.0458-1, the Novartis Foundation (Switzerland), the Mental Retardation Research Center of Baylor College of Medicine, the Medical Research Council (United Kingdom), the Wellcome Trust, and Action Research (United Kingdom). L.B. was supported by a grant from the Blanceflor Ludovisi-Boncompagni née Bildt Foundation.
Abbreviations
AAA, ATPases associated diverse cellular A domains
RACE, rapid amplification of cDNA ends
UPD, uniparental disomy
CFTR, cystic fibrosis transmembrane conductance regulator
RT, reverse transcription
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ320497).
Bartoloni, L., Mitchison, H. M., Pazour, G. J., Maiti, A. K., Meeks, M., Chung, E., Dickert, B. L., Spiden, S., Gehrig, C., Rossier, C., et al. (2000) Eur. J. Hum. Genet. 8, Suppl. 1 (abstr.).
References
- 1.Afzelius B. A. & Mossberg, B. (1995) in The Metabolic and Molecular Bases of Inherited Disease, eds. Scriver, C. R., Beaudet, A. L., Sly, W. S. & Valle, D. (McGraw–Hill, New York), pp. 3943–3954.
- 2.Aylsworth A. S. (2001) Am. J. Med. Genet. 101, 345-355. [PubMed] [Google Scholar]
- 3.Teknos T. N., Metson, R., Chasse, T., Balercia, G. & Dickersin, G. R. (1997) Otolaryngol. Head Neck Surg. 116, 68-74. [DOI] [PubMed] [Google Scholar]
- 4.Dutcher S. K. (1995) Trends. Genet. 11, 398-404. [DOI] [PubMed] [Google Scholar]
- 5.Afzelius B. A. (1981) Am. J. Hum. Genet. 33, 852-864. [PMC free article] [PubMed] [Google Scholar]
- 6.Holzbaur E. L. & Vallee, R. B. (1994) Annu. Rev. Cell Biol. 10, 339-372. [DOI] [PubMed] [Google Scholar]
- 7.Blouin J. L., Meeks, M., Radhakrishna, U., Sainsbury, A., Gehring, C., Sail, G. D., Bartoloni, L., Dombi, V., O'Rawe, A., Walne, A., et al. (2000) Eur. J. Hum. Genet. 8, 109-118. [DOI] [PubMed] [Google Scholar]
- 8.Pennarun G., Escudier, E., Chapelin, C., Bridoux, A. M., Cacheux, V., Roger, G., Clement, A., Goossens, M., Amselem, S. & Duriez, B. (1999) Am. J. Hum. Genet. 65, 1508-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guichard C., Harricane, M. C., Lafitte, J. J., Godard, P., Zaegel, M., Tack, V., Lalau, G. & Bouvagnet, P. (2001) Am. J. Hum. Genet. 68, 1030-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zariwala M., Noone, P. G., Sannuti, A., Minnix, S., Zhou, Z., Leigh, M. W., Hazucha, M., Carson, J. L. & Knowles, M. R. (2001) Am. J. Respir. Cell Mol. Biol. 25, 577-583. [DOI] [PubMed] [Google Scholar]
- 11.Olbrich H., Haffner, K., Kispert, A., Volkel, A., Volz, A., Sasmaz, G., Reinhardt, R., Hennig, S., Lehrach, H., Konietzko, N., et al. (2002) Nat. Genet. 30, 143-144. [DOI] [PubMed] [Google Scholar]
- 12.Ibanez-Tallon I., Gorokhova, S. & Heintz, N. (2002) Hum. Mol. Genet. 11, 715-721. [DOI] [PubMed] [Google Scholar]
- 13.Pennarun G., Chapelin, C., Escudier, E., Bridoux, A. M., Dastot, F., Cacheux, V., Goossens, M., Amselem, S. & Duriez, B. (2000) Hum. Genet. 107, 642-649. [DOI] [PubMed] [Google Scholar]
- 14.Bartoloni L., Blouin, J. L., Maiti, A. K., Sainsbury, A., Rossier, C., Gehrig, C., She, J. X., Marron, M. P., Lander, E. S., Meeks, M., et al. (2001) Genomics 72, 21-33. [DOI] [PubMed] [Google Scholar]
- 15.Maiti A. K., Bartoloni, L., Mitchison, H. M., Meeks, M., Chung, E., Spiden, S., Gehrig, C., Rossier, C., Delozier-Blanchet, C. D., Blouin, J., et al. (2000) Cytogenet. Cell Genet. 90, 119-122. [DOI] [PubMed] [Google Scholar]
- 16.Pan Y., McCaskill, C. D., Thompson, K. H., Hicks, J., Casey, B., Shaffer, L. G. & Craigen, W. J. (1998) Am. J. Hum. Genet. 62, 1551-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jorissen M., Van der Schueren, B., Van den Berghe, H. & Cassiman, J. J. (1990) ORL J. Otorhinolaryngol. Relat. Spec. 52, 368-374. [DOI] [PubMed] [Google Scholar]
- 18.Thompson J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic. Acids. Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staden R. (1996) Mol. Biotechnol. 5, 233-241. [DOI] [PubMed] [Google Scholar]
- 20.Dausset J., Cann, H., Cohen, D., Lathrop, M., Lalouel, J. M. & White, R. (1990) Genomics 6, 575-577. [DOI] [PubMed] [Google Scholar]
- 21.Kamiya R. (1995) Cell Motil. Cytoskeleton 32, 98-102. [DOI] [PubMed] [Google Scholar]
- 22.Supp D. M., Witte, D. P., Potter, S. S. & Brueckner, M. (1997) Nature (London) 389, 963-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koonce M. P. (1997) J. Biol. Chem. 272, 19714-19718. [DOI] [PubMed] [Google Scholar]
- 24.Okada Y., Nonaka, S., Tanaka, Y., Saijoh, Y., Hamada, H. & Hirokawa, N. (1999) Mol. Cell 4, 459-468. [DOI] [PubMed] [Google Scholar]
- 25.Supp D. M., Brueckner, M., Kuehn, M. R., Witte, D. P., Lowe, L. A., McGrath, J., Corrales, J. & Potter, S. S. (1999) Development (Cambridge, U.K.) 126, 5495-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lander E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature (London) 409, 860-921. [DOI] [PubMed] [Google Scholar]
- 27.Venter J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304-1351. [DOI] [PubMed] [Google Scholar]
- 28.Asai D. J. & Koonce, M. P. (2001) Trends. Cell Biol. 11, 196-202. [DOI] [PubMed] [Google Scholar]
- 29.Mocz G. & Gibbons, I. R. (2001) Structure (London) 9, 93-103. [DOI] [PubMed] [Google Scholar]
- 30.Perrone C. A., Myster, S. H., Bower, R., O'Toole, E. T. & Porter, M. E. (2000) Mol. Biol. Cell 11, 2297-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neesen J., Kirschner, R., Ochs, M., Schmiedl, A., Habermann, B., Mueller, C., Holstein, A. F., Nuesslein, T., Adham, I. & Engel, W. (2001) Hum. Mol. Genet. 10, 1117-1128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.